Abstract
Background
It is well established that adipose tissue plays a key role in energy storage and release but is also a secretory organ and a source of stem cells. Among different lineages, stem cells are able to differentiate into adipocytes and osteoblasts. As secreted proteins could regulate the balance between both lineages, we aimed at characterizing the secretome of human multipotent adipose-derived stem cell (hMADS) at an early step of commitment to adipocytes and osteoblasts.
Results
A proteomic approach, using mono-dimensional electrophoresis and tandem mass spectrometry, allowed us to identify a total of 73 proteins at day 0 and day 3 of adipocyte and osteoblast differentiation. Analysis of identified proteins showed that 52 % corresponded to classical secreted proteins characterized by a signal peptide, that 37 % previously described in the extracellular compartment were devoid of signal peptide and that 11 % neither exhibited a signal peptide nor had been previously described extracellularly. These proteins were classified into 8 clusters according to their function. Quantitative analysis has been performed for 8 candidates: PAI-1, PEDF, BIGH3, PTX3, SPARC, ENO1, GRP78 and MMP2. Among them, PAI-1 was detected at day 0 and day 3 of osteoblast differentiation but never in adipocyte secretome. Furthermore we showed that PAI-1 mRNA was down-regulated in the bone of ovariectomized mice.
Conclusion
Given its regulation during the early events of hMADS cell differentiation and its status in ovariectomized mice, PAI-1 could play a role in the adipocyte/osteoblast balance and thus in bone diseases such as osteoporosis.
Background
Adipose tissue is no longer considered as a mere energy reservoir but it plays also an endocrine role, releasing a panoply of secreted molecules, i.e. adipokines such as leptin, adiponectin, plasminogen activator inhibitor 1 (PAI-1), vaspin and tumor necrosis factor α (TNFα) [1,2]. Furthermore, adipose tissue is a source of stem cells, representing a promising tool for pharmacological studies and clinical applications [3]. A balanced development of adipose tissue is of crucial importance to ensure some of the most important physiological functions, including reproduction, haemostasis, angiogenesis, blood pressure and immune function [1,4]. Alterations of fat cell number and size are present in lipodystrophy and obesity that are associated to type 2 diabetes [5]. Another condition altering fat cell formation is osteoporosis, where an imbalance between adipocytes and osteoblasts in bone marrow is observed. Aging, menopause, glucocorticoid treatment or alcohol abuse can lead to an increase in bone marrow adiposity [6,7]. To date, several issues are still pending, for instance whether infiltration of fat in bone marrow causes low bone mass or is due to bone loss [6,7]. Since adipocytes and osteoblasts share the same mesenchymal precursor, the study of the adipocyte/osteoblast balance represents a worthy challenge to treat adipose tissue and bone disorders. It is well described that secreted leptin and adiponectin can affect bone formation both directly on osteoblastogenesis and indirectly by acting on osteoclastogenesis [6,8-10]. Several molecules secreted by osteoblasts such as Wnt and bone morphogenetic protein favor osteogenesis at the expense of adipogenesis [7,11], thus pointing out a crosstalk between both lineages.
During the last two decades, a large number of molecular regulators of adipogenesis and osteogenesis have been described. Among them, peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding proteins (C/EBPs) are well recognized factors that play major roles in adipogenesis [12], whereas runt-related transcription factor 2 (runx2), distal-less homeobox 5 (dlx5), muscle segment homeobox 2 (msx2) and osterix represent master regulators of osteogenesis [6,13].
In the present work, we aimed at identifying molecules secreted at early step of differentiation of human mesenchymal stem cells towards adipocytes and osteoblasts. To address this point, we used a cellular model recently established in our laboratory, termed hMADS cells (human multipotent adipose tissue-derived stem cells). hMADS cells, isolated from the adipose tissue of young donors, present extensive capacities of self-renewal, clonogenicity and multipotency, as they fully differentiate into adipocytes, osteoblasts, myoblasts and chondrocytes while exhibiting a normal karyotype [14-17].
Recently, proteomic approaches have been applied to study rodent and human adipose tissue secretome using cellular models that focused mainly on late events of adipogenesis [18-22]. Moreover, with respect to early events of osteogenesis, a characterization of secreted molecules from osteoblasts has not been so far reported.
Herein we have identified 73 proteins by a proteomic approach and confirmed these findings by Western-blot for 8 candidates. These proteins and their involved pathways, in particular the plasminogen system, could play an important role in regulating the adipocyte/osteoblast balance.
Results
Characterization of the secretome of hMADS cells committed towards adipocytes and osteoblasts
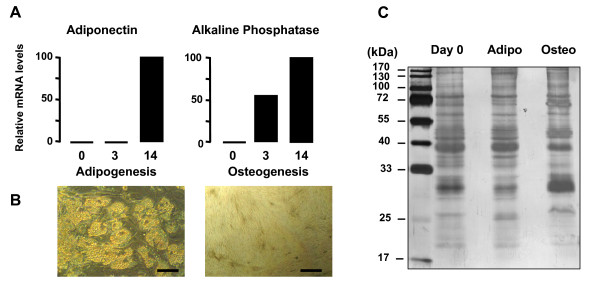
hMADS cells differentiate into adipocytes and osteoblasts, as shown in Figure 1A. Gene expression of representative markers of adipocyte (adiponectin) and osteoblast differentiation (alkaline phosphatase) are reported and in agreement with previously published data [14-17]. The terminal differentiation of hMADS cells into adipocytes and osteoblasts is also illustrated by the typical cellular morphology as shown in Figure 1B.
Figure 1.
Analysis of terminal differentiation of hMADS cells into adipocytes and osteoblasts and Coomassie Blue staining of hMADS cell secretome. A. qRT-PCR analysis of mRNA levels of specific adipogenic (adiponectin) and osteogenic (alkaline phosphatase) markers at day 3 and day 14 of differentiation as compared to day 0. The data are representative of three independent experiments. B. Microphotographs of hMADS cells differentiated into adipocytes and osteoblasts at day 14. Bar scale = 50 μm. C. Representative gel of secreted proteins from hMADS cells at day 0 and day 3 of differentiating adipocytes (adipo) and osteoblasts (osteo) after 6 h of incubation. The gel is representative of 3 independent experiments.
In order to analyze the secretome at early step of adipogenesis and osteogenesis of hMADS cells, secretion media at day 3 of differentiation into adipocytes and osteoblasts were compared with those of cells at day 0. Cells were washed 3 times with PBS and incubated for 6 h with serum-free culture medium, containing 0.1 μg/ml of transferrin without differentiation inducers. At the end of this incubation period, media were harvested, filtered and concentrated. Very low cell death or cell lysis occurred during the incubation period, as checked by microscopic analysis and lactate dehydrogenase activity measurements [see Additional file 1]. Four μg of each protein sample were loaded onto one-dimensional SDS-PAGE and stained with Bio-Safe Coomassie Stain. A representative gel is reported in Figure 1C. The excised bands from the SDS-PAGE were cut, trypsin digested and analyzed by mass spectrometry. The list of the identified proteins is reported in Table 1. Further details about the peptides identified for each protein are available in Table 1S [see Additional file 2].
Table 1.
Identification of proteins in the secretome of hMADS cells at early step of adipogenesis and osteogenesis.
| ID | Protein name | Function | Mw kDa | Loc | Day 0 | Adipo | Osteo | Ref |
| PROTEASES | ||||||||
| P00736 | Complement component 1, r subcomponent | serine protease | 80 | ex | Y | Y | [69] | |
| P09871 | Complement component 1, s subcomponent | serine protease | 77 | sp | Y | Y | ||
| P03956 | Matrix metallopeptidase 1 (interstitial collagenase) | collagen catabolism | 54 | sp | Y | |||
| P08253 | Matrix metallopeptidase 2, gelatinase A (MMP2) | collagen catabolism | 74 | sp | Y | Y | Y | |
| Q15113 | Procollagen C-endopeptidase enhancer | collagen catabolism | 48 | sp | Y | |||
| PROTEASE INHIBITORS | ||||||||
| P05121 | Serpin peptidase inhibitor, clade E, plasminogen activator inhibitor type 1(PAI-1) | fibrinolysis | 45 | sp | Y | Y | ||
| P36955 | Serpin peptidase inhibitor, clade F (PEDF) | inhibitor of angiogenesis | 46 | sp | Y | |||
| P05155 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | fibrinolysis | 55 | sp | Y | Y | ||
| P01033 | TIMP metallopeptidase inhibitor 1 | proteolysis inhibitor | 23 | sp | Y | Y | Y | |
| EXTRACELLULAR MATRIX COMPONENTS | ||||||||
| P50454 | Serpin H1 precursor (Collagen-binding protein) (Colligin) (47 kDa heat shock protein) (Proliferation-inducing gene 14 protein) | collagen-binding protein | 46 | sp | Y | |||
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein precursor | ECM component | 469 | sp | Y | Y | ||
| P02452 | Collagen, type I, alpha 1 | ECM component | 139 | sp | Y | Y | Y | |
| P08123 | Collagen, type I, alpha 2 | ECM component | 129 | sp | Y | Y | Y | |
| P02461 | Collagen, type III, alpha 1 | ECM component | 138 | sp | Y | Y | ||
| P12109 | Collagen, type VI, alpha 1 | ECM component | 108 | sp | Y | Y | Y | |
| P12110 | Collagen, type VI, alpha 2 | ECM component | 109 | sp | Y | Y | Y | |
| P12111 | Collagen, type VI, alpha 3 | ECM component | 343 | sp | Y | Y | Y | |
| P07585 | Decorin precursor (Bone proteoglycan II) | ECM component | 40 | sp | Y | |||
| P02751 | Fibronectin precursor | cell growth | 26 | sp | Y | Y | Y | |
| Q08380 | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) | growth regulation/cell-matrix | 65 | sp | Y | Y | ||
| Q16270 | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) | cell adhesion | 29 | sp | Y | |||
| P11047 | Laminin gamma 1 chain precursor (Laminin B2 chain) | ECM component | 178 | sp | Y | Y | ||
| P51884 | Lumican precursor (Keratin sulfate proteoglycan lumican) | ECM component | 38 | sp | Y | Y | Y | |
| P14543 | Nidogen 1 | ECM component | 136 | sp | Y | |||
| Q15063 | Periostin, osteoblast specific factor | cell adhesion | 93 | sp | Y | Y | ||
| Q02809 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 pecursor | collagen process | 83 | sp | Y | |||
| O00391 | Sulfhydryl oxidase 1 precursor, Quiescin Q6 (QSCN6) | growth regulation | 83 | sp | Y | |||
| P09486 | Secreted protein, acidic, cysteine-rich (osteonectin) | cell-matrix interaction | 35 | sp | Y | Y | Y | |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 precursor (BIGH3) | cell adhesion | 75 | sp | Y | Y | Y | |
| ANTI-INFLAMMATORY/ANTI-OXIDANT PROTEINS | ||||||||
| Q12841 | Follistatin-like 1 | immunity and defense | 35 | sp | Y | Y | Y | |
| P09211 | Glutathione S-transferase P | antioxidant | 23 | ex | Y | [70, 71] | ||
| P26022 | Pentraxin-related gene, rapidly induced by IL-1 beta (PTX3) | inflammatory response | 42 | sp | Y | Y | Y | |
| P30041 | PRDX6 peroxiredoxin 6 | antioxidant | 25 | ex | Y | [72] | ||
| Q16881 | TXNRD1 thioredoxin reductase 1 | antioxidant | 55 | ex | Y | [33] | ||
| METABOLIC ENZYMES | ||||||||
| Q04828* | Aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1) | progesterone conversion | 37 | in | Y | |||
| P06733 | Enolase 1 (ENO1) | glycolysis | 47 | ex | Y | Y | Y | [73, 74] |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | glycolysis | 36 | ex | Y | Y | Y | [31] |
| P40926 | Malate dehydrogenase 2, NAD (mitochondrial) | Krebs cycle/gluconeogenesis | 36 | ex | Y | Y | [75, 76] | |
| P00558 | Phosphoglycerate kinase 1 | glycolysis | 44 | ex | Y | [32] | ||
| P18669 | Phosphoglycerate mutase 1 | glycolysis | 29 | ex | Y | [70] | ||
| P14618 | Pyruvate kinase, muscle | glycolysis | 58 | ex | Y | Y | [77] | |
| P29401 | Transketolase | pentose phosphate-glycolysis | 68 | ex | Y | Y | [73] | |
| P60174 | Triosephosphate isomerase 1 | glycolysis | 27 | ex | Y | [76] | ||
| CYTOSKELETAL COMPONENTS | ||||||||
| P60709 | Actin, beta | cell growth | 42 | ex | Y | Y | Y | [70, 71] |
| P12814 | Actinin, alpha 1 | cell growth | 103 | ex | Y | Y | [73, 74] | |
| Q71U36 | Alpha-3 tubulin | cell growth | 50 | ex | Y | Y | [73] | |
| Q14019* | Coactosin-like protein | actin-binding protein | 16 | In | Y | |||
| P15924 | Desmoplakin | cell adhesion | 332 | ex | Y | [77] | ||
| Q16555* | Dihydropyrimidinase-like 2 | tubulin binding protein | 62 | In | Y | |||
| P14923* | Junction plakoglobin | cell adhesion | 82 | In | Y | |||
| P09382 | Lectin, galactoside-binding, soluble, 1 (galectin 1) | growth regulation | 15 | ex | Y | [21] | ||
| P26038 | Moesin | cell-matrix interaction | 68 | ex | Y | Y | Y | [77] |
| P07737 | Profilin 1 | cell growth | 15 | ex | Y | Y | [73] | |
| Q15293* | Reticulocalbin 1, EF-hand calcium binding domain | calcium-binding protein | 39 | In | Y | |||
| P08670 | Vimentin | cell growth | 54 | ex | Y | [71, 73] | ||
| P18206 | Vinculin | cell growth | 124 | ex | Y | Y | [73] | |
| O75083 | WD repeat-containing protein 1 isoform 1 variant | actin pol. control | 66 | ex | Y | Y | [76] | |
| HEAT SHOCK/PROTEIN FOLDING PROTEINS | ||||||||
| P11021 | 78 kDa glucose-regulated protein precursor (GRP78) | chaperone | 72 | sp | Y | Y | Y | |
| O43852 | Calumenin precursor (Crocalbin) | calcium-binding chaperone | 37 | sp | Y | Y | Y | |
| P27797 | Calreticulin | calcium-binding chaperone | 48 | sp | Y | Y | ||
| P50990* | Chaperonin containing TCP1, subunit 8 (theta) | chaperone | 60 | In | Y | |||
| P68104 | Elongation factor 1-alpha 1 (EF-1-alpha-1) | protein biosynthesis | 50 | ex | Y | [78] | ||
| P11142 | Heat shock 70 kDa protein 8 and homologs | chaperone | 71 | ex | Y | [28] | ||
| Heat shock 70 kDa protein 2 (P54652), Heat shock 70 kDa protein 1-like (P43931) | ||||||||
| P10809 | Heat shock 60 kDa protein 1 (chaperonin) | chaperone | 61 | ex | Y | [29] | ||
| P07900 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | chaperone | 85 | ex | Y | [77] | ||
| P30101 | PDIA3 protein disulfide-isomerase A3 precursor (EC 5.3.4.1) | protein folding | 57 | sp | Y | Y | Y | |
| P23284 | Peptidyl-prolyl cis-trans isomerase B precursor, (Rotamase) | protein folding | 23 | sp | Y | Y | Y | |
| Q15084 | Protein disulfide isomerase family A, member 6 | protein folding | 48 | ex | Y | Y | [71] | |
| P14625 | Endoplasmin precursor, (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1) | chaperone | 92 | sp | Y | |||
| OTHER PROTEINS | ||||||||
| P41250 | Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine – tRNA ligase) (GlyRS) | tRNA synthetase | 83 | sp | Y | |||
| Q9NTK5* | Putative GTP-binding protein 9 (putative) | GTP-binding protein | 45 | In | Y | |||
| P50395 | RAB GDP dissociation inhibitor beta | vesicular transport | 51 | sp | Y | |||
| P30153* | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, (Medium tumor antigen-associated 61 kDa protein) | serine/threonine phosphatase | 65 | In | Y |
Y: indicates the presence of the candidate in the sample; sp: signal peptide; ex: extracellular; in: intracellular; Mw: molecular weight; adipo:day 3 adipocytes; osteo: day 3 osteoblasts; Loc: localisation; ref: reference. * indicates only putatively
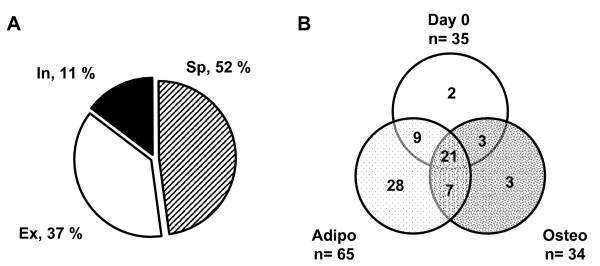
Seventy three proteins were classified according to their function and localization, determined by the Meta search engine Bioinformatic Harvester. As reported in Figure 2A and Table 1, a large amount of identified proteins (52 %) corresponded to "classical" secreted proteins characterized by a signal peptide. Among them, we found several components of the extracellular matrix (fibronectin, collagens etc...), proteases, serine protease inhibitors (serpins such as PAI-1, pigment epithelium derived factor (PEDF) and Plasma protease C1 inhibitor) and other proteins involved in collagen catabolism, protein folding/turnover and growth regulation. In differentiating osteoblasts at day 3, periostin, described previously to be associated with osteoblast differentiation [23] was identified, whereas its presence was not detected in secretion media of differentiating adipocytes. A high percentage of proteins (37 %), devoid of signal peptide, was also detected into the secretion medium. These proteins were labelled as extracellular since they have been previously described in the extracellular compartment. Some of these proteins were metabolic enzymes such as α-enolase (ENO1) and pyruvate kinase, cytoskeletal components or heat shock/chaperone proteins. Similar findings from literature have been reported (see references in Table 1) strengthening actually their presence in the extracellular medium of various cellular models as well as in the murine/human bloodstream. The remaining proteins (11 %) did not exhibit a signal peptide (in: intracellular) and, to the best of our knowledge, have not been so far described extracellularly and did not represent artefacts as cell lysis was very low [see Additional file 1]. These proteins are only putatively secreted and indicated by an asterisk in Table 1. As illustrated by the Venn diagram in Figure 2B, the secretion of 21 proteins resulted to be similar under the three culture conditions, while 28 proteins were specifically secreted under adipogenic condition compared to 3 proteins under osteogenic condition.
Figure 2.
Protein distribution of hMADS cell secretome. A. hMADS cell secretome is represented using a pie chart in three main groups: secreted proteins with signal peptide (sp, dashed area), secreted proteins without signal peptide (ex, white area) and intracellular proteins (in, dark area). The percentage of the proteins present in each group is reported in the scheme. One hundred per cent is referred to a total number of 73 identified proteins. B. Venn diagram of proteins expressed under the different culture conditions; n represents protein number in each condition.
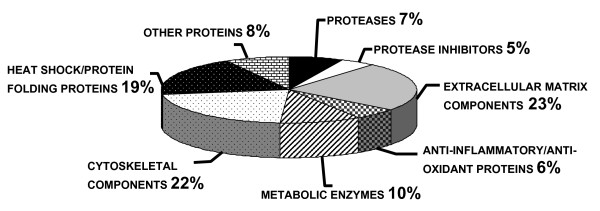
Finally, hMADS cell secretome was clustered in 8 main groups, as indicated in Table 1 and Figure 3. These clusters include: proteases, protease inhibitors, extracellular matrix (ECM) components, anti-inflammatory/anti-oxidant proteins, metabolic enzymes, cytoskeletal components, heat shock/protein folding proteins and other proteins. These clusters illustrated the diversity of hMADS cell secretome, thus suggesting that some of these proteins could be involved in the regulation of the adipocyte/osteoblast balance.
Figure 3.
Distribution into clusters of hMADS cell secretome. Identified proteins were distributed in 8 main clusters created to classify hMADS cell secreted proteins and are presented in the pie chart. For each cluster the percentage of proteins included is reported.
Validation of protein expression of selected candidates
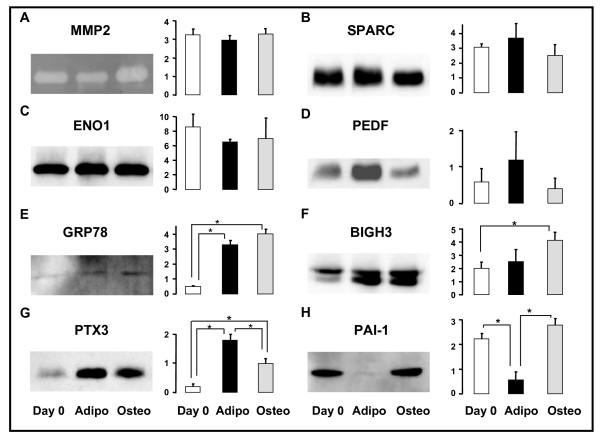
Mass spectrometry identification data were validated for 8 candidates, selecting them from different clusters, i.e. 1 protease, 2 protease inhibitors, 2 ECM components, 1 anti-inflammatory/anti-oxidant protein, 1 metabolic enzyme and 1 heat shock protein. Western blots or zymograms were employed to supplement the identification by mass spectrometry and to determine the expression profile among three different sets of conditions (day 0 and day 3 for adipogenesis or osteogenesis). For all the candidates, the results of immunoblotting/zymogram and a semi-quantification of the bands are reported in Figure 4.
Figure 4.
Secretion levels of 8 candidates released from hMADS cells during the commitment to adipocytes and osteoblasts. The activity of MMP2 has been evaluated by gelatin zymography (A). The expression of SPARC (B), ENO1(C), PEDF (D), GRP78 (E), BIGH3 (F), PTX3 (G) and PAI-1 (H) has been analyzed by Western blot. The bar graphs report the levels of expression of every single candidate as the mean of three independent experiments after 6 h of incubation. The values are indicated as arbitrary units. *: p < 0.05. Two μg of secreted proteins have been loaded for each gel.
Matrix metallopeptidase 2 (MMP2), secreted protein, acidic, cysteine-rich (SPARC) and ENO1 were secreted by hMADS cells under all conditions, confirming the pattern of detection obtained by SDS-PAGE-mass spectrometry analysis (Fig. 4A–C, to be compared with Table 1). However, no significant difference was observed under the three conditions for these candidates. Zymographic analysis of the secretion media revealed a prominent band of lysis at 66 kDa corresponding to the reported active form of MMP2 (Fig. 4A). Addition of EDTA, an inhibitor of gelatinase activity, led to the absence of lytic bands (data not shown), thus confirming the specificity of MMP activity. Pigment epithelium derived factor (PEDF) was highly expressed in day 3 adipocytes compared to day 0 and day 3 osteoblasts (Fig. 4D) in agreement with mass spectrometry identification.
For 78 kDa glucose-regulated protein precursor (GRP78), transforming growth factor-beta-induced protein (BIGH3), pentraxin 3 (PTX3) and PAI-1, we confirmed the pattern of detection reported in Table 1, with significant differences of expression between day 0, day 3 adipocytes and day 3 osteoblasts, as shown by Western blot and histogram semi-quantifications (Fig. 4E–H).
Analysis of the plasminogen system in hMADS cells differentiating towards adipocytes and osteoblasts
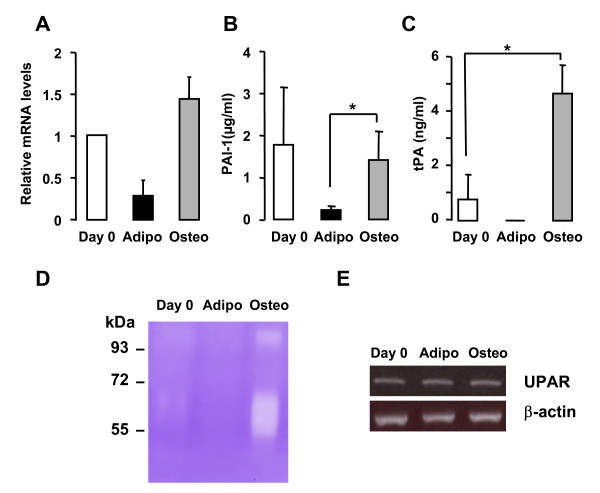
Given the pattern of expression at the early step of adipogenesis and osteogenesis of hMADS cells, PAI-1 appeared as one of the most promising candidates for investigating further the adipocyte/osteoblast balance. PAI-1 mRNA levels were similar to those of protein levels (Fig. 5A). Since PAI-1 is a key regulator of the conversion of plasminogen to plasmin, negatively acting on two plasminogen activators, namely uPA (urinary plasminogen activator) and tPA (tissue type plasminogen activator) [24], we decided to evaluate uPA and tPA levels in hMADS cell secretome. ELISA experiments were performed on secretion media from hMADS cells at day 0 and day 3 under adipogenic and osteogenic differentiating conditions after 6 h of incubation. As reported in Figure 5B, consistent with the immunoblotting data, PAI-1 levels strongly decreased in differentiating adipocytes as compared to day 0. By contrast, PAI-1 was secreted by day 3 osteoblasts, at albeit the same levels when compared to day 0. Concerning tPA measurement, we did not detect tPA in day 3 differentiating adipocytes, while a significant increase of tPA levels was observed in day 3 osteoblasts as compared to day 0 (Fig. 5C). By contrast, uPA remained undetectable in the secretion media of hMADS cells under the three conditions analyzed.
Figure 5.
Evaluation of the presence of the plasminogen system in hMADS cells. (A) PAI-1 mRNA levels have been determined in hMADS cells at day 0, day 3 adipocytes (adipo) and day 3 osteoblasts (osteo). (B) PAI-1 and (C) tPA protein levels have been measured by ELISA in the secretion media of hMADS cells at day 0, day 3 adipocytes (adipo) and day 3 osteoblasts (osteo) after 6 h of incubation. *: p < 0.05. (D) Zymographic analysis of plasminogen activators activity in hMADS cell conditioned media collected after 6 h of incubation. A representative casein-plasminogen zymogram out of three independent experiments is shown. (E) RT-PCR analysis of UPAR expression in hMADS cells at day 0, day 3 adipocytes and day 3 osteoblasts. β-actin expression is reported as internal control. PCR products have been separated on a 1% agarose gel.
Enzymatic activities of uPA and tPA in 6 h serum-free conditioned media of hMADS cells at d0 and day 3 of differentiating adipocytes and osteoblasts were then evaluated by casein/plasminogen zymographic analysis. As reported in Figure 5D, tPA was mostly secreted by day 3 osteoblasts, as observed by the lytic band at 63 kDa, while uPA was undetectable (expected band at 48 kDa), thus supporting ELISA results. The higher band at around 110 kDa may correspond to complexes between plasminogen activators and PAI-1 as classically reported [25]. Incubation of the gel with amiloride, which selectively abrogates uPA-dependent enzyme activities, did not affect tPA-dependent lytic bands (data not shown). Furthermore, we demonstrated that hMADS cells expressed uPA receptor (UPAR) mRNA as reported by semi-quantitative RT-PCR analysis (Fig. 5E). As these data actually suggest the involvement of plasminogen activation in the regulation of the fine balance between adipogenesis and osteogenesis, this hypothesis was next examined in a pathophysiological situation where such an imbalance occurs.
PAI-I expression in ovariectomized mice
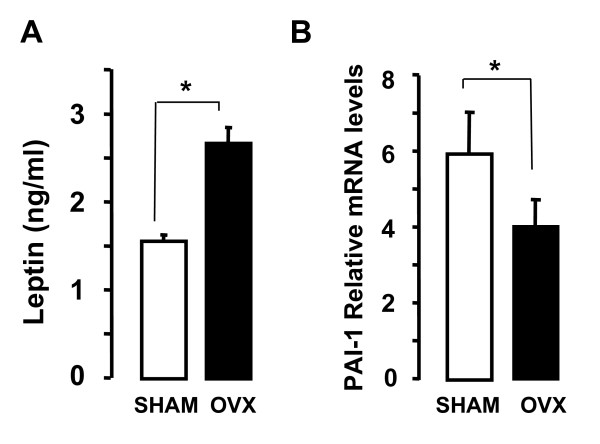
Disequilibrium of the adipocyte/osteoblast balance is associated with the development of osteoporosis after ovariectomy. It is well established that in the bone of ovariectomized (ovx) mice, adipocytes develop at expense of bone formation [26,27]. In order to evaluate whether osteoporosis affects PAI-1 in bone, we analyzed the expression of PAI-1 mRNA in the humerus of ovx compared to sham mice. Ten weeks after surgery, as previously described [26] mice increased fat mass as shown by leptin levels (Fig. 6A). As shown in Figure 6B, the levels of PAI-1 mRNA decreased significantly in the humerus of ovx compared to sham mice, thus pointing out at PAI-1 modulation in the adipocyte/osteoblast balance as a potential marker of osteoporosis.
Figure 6.
PAI-1 mRNA levels in the humerus of ovariectomized as compared to control mice. A. Plasma leptin levels have been determined by ELISA. B. PAI-1 mRNA expression has been evaluated by qRT-PCR in the humerus of ovx and sham mice and standardized with TBP expression levels. The data are expressed as the mean ± SD per group of 4 mice. *: p < 0.05.
Discussion
Characterization of hMADS cell secretome
The secretome of murine and human adipocytes has been characterized [19-22]. However, these studies primarily focused on the late events of adipogenesis. Concerning osteoblast secretome, a single detailed characterization has been performed so far by Xiao and collaborators, who analyzed the extracellular matrix vesicle proteome of mineralizing osteoblasts and identified proteins at late steps of osteogenesis of MC3T3-E1 cells [28].
Herein, proteomic analysis has been used for the first time to compare the secretome of hMADS cells differentiating into adipocytes and osteoblasts and to gain insights into the adipocyte/osteoblast balance. In order to delineate the initial secretory events of adipogenesis and osteogenesis, we performed a 6 h short term secretion which likely limits the number of secreted proteins and cell lysis. A total of 73 proteins was identified and classified into 8 clusters. The largest cluster corresponds to extracellular matrix proteins, thus confirming the secretome characteristic of our fraction. In addition, more than 50% of hMADS cell secretome is represented by proteins with signal peptide. Cytoskeletal proteins and heat shock and folding proteins are also largely represented with 14 and 12 proteins, respectively. Among the heat shock/folding proteins, two chaperones, e.g. HSP60 and HSP70 were found. These findings are reinforced by recent observations describing the presence of HSP60 and HSP70 in the bloodstream. HSP60 circulating levels in diabetic patients have been found associated to cardiovascular diseases [29], while elevated HSP70 circulating levels are related to systemic inflammatory reaction in patients with heart failure following acute myocardial infarction [30]. In the metabolic enzyme cluster, 9 enzymes were also found, including glyceraldehyde dehydrogenase (GAPDH) and phosphoglycerate kinase (PGK). According to literature, extracellular GAPDH, also found in rat serum, inhibits spreading of COS-7 cells [31], while PGK is secreted by tumor cells and its activity is regulated by hypoxia [32]. These enzymes might reflect increased metabolic requirements, but they could also exert independent and still undiscovered functions. Anti-oxidant proteins, like Thioredoxin reductase 1 (TXNRD1), represent another cluster of proteins secreted by hMADS cells. TXNRD1, that does not exhibit a signal peptide, has been described in rat as a circulating protein [33].
Eleven percent of secreted proteins from hMADS cells is however represented by intracellular proteins, presence of which has not been reported up to now outside the cells. A modest contamination by uncontrolled release of intracellular proteins cannot be excluded despite the fact that no cell lysis was detected. Although cell death can take place in culture and is unavoidably accompanied by a subsequent release of cytosolic components, it should be pointed out that cell viability was not affected [see Additional file 1]. Among non-artifactual explanations, it is worth pointing out that this phenomenon could implicate exosome secretion, as most of the identified proteins were reported to be present in exosomes. Exosomes are membrane vesicles that originate from the cell membrane and are released extracellularly [34]. Their occurrence has been also described in the bloodstream and in different tissues in vivo [35], suggesting their participation in physiological and/or pathological processes. To date, the exact function of exosomes in vivo is not completely understood. A large number of cell types, including reticulocytes, immune cells, platelets, epithelial cells, dendritic cells and others (see [34] and [36] for detailed reviews) appears to secrete exosomes either as a mechanism to extrude some proteins and/or to communicate between cells and/or to play immune functions. Typically, exosomes contain chaperones, cytoskeletal proteins (moesin, desmoplakin), elongation factors and several enzymes (ENO1), as described in literature through proteomic analysis of the secretion media from different sources as well as from analysis by flow cytometry and Western blot [34,37]. Therefore intracellular proteins recovered outside hMADS cells may be a mere reflection of events taking place within the cytosol at the time of exosome formation. Furthermore, 4 (Q14019, Q15293, P50990 and P30153) out of 8 candidates, known to be intracellular, have a score higher than a fixed threshold when analyzed using software predicting a non-classical protein secretion [38], it is likely that these proteins are secreted by non-traditional mechanisms and did not arise from cell lysis. Clearly, these proteins can only be considered as putatively secreted and their actual secretion should be assessed.
Selected candidates of hMADS cell secretome
Mass spectrometry data have been validated for 8 selected candidates. MMP2, SPARC and ENO1 displayed no significant differences in terms of quantification by immunoblotting or zymogram when comparing the three culture conditions.
MMP2, a zinc-dependent endopeptidase, involved in the degradation of the ECM, is actively secreted by adipocytes [39] and osteoblasts [40]. The presence of MMP2 in hMADS cells reinforces the concept that ECM remodelling represents a crucial event during the early steps of differentiation. As observed in this study, SPARC has been already described as a secreted molecule in both adipocytes [41] and osteoblasts [42]. SPARC is reported to be up-regulated in the adipose tissue of different models of murine obesity [41] and its circulating levels correlate with body mass index in humans [43]. A potential interest in further studying SPARC in the adipocyte/osteoblast balance relies upon the fact that bone marrow stromal cells from SPARC-null mice tend to form more adipocytes than cells from wild type mice [44]. ENO1 is mainly described for its role as an intracellular glycolytic enzyme, however its presence extracellularly has been already reported in the secretion medium of 3T3-L1 adipocytes [21], as well as in other cellular models [45]. ENO1 is a multifunctional protein and a putative plasminogen receptor, since it has been reported as a cell surface protein [46]. Therefore, a potential role of ENO1 in the plasminogen cascade cannot be excluded.
Concerning the five other candidates (PEDF, GRP78, BIGH3, PTX3 and PAI-1), relevant differences of expression between day 0 and day 3 of adipogenesis and osteogenesis have been found. PEDF has been reported to be preferentially secreted by 3T3-L1 preadipocytes as compared to fully differentiated adipocytes [20]. However, Zvonic et al. [22] have described an increase in PEDF expression during adipogenesis of primary cultures of human adipose-derived stem cells, in agreement with our observations in hMADS cells when comparing hMADS cells at day 0 and after complete adipocyte differentiation (data not shown). PEDF could represent a valuable candidate for the adipocyte/osteoblast balance given its expression also in differentiating hMADS osteoblasts. A putative role for PEDF as mediator of angiogenesis and in matrix remodelling of the bone has been recently postulated [47].
Several molecular chaperones have been found in the secretome of hMADS cells, including calumenin, calreticulin, protein disulfide isomerase 6, heat shock proteins and GRP78. As GRP78 controls intracellular protein transport [48], its up-regulation in hMADS differentiating adipocytes and osteoblasts as compared to day 0 could reflect an increased demand in protein folding or a protection against endoplasmic reticulum stress.
BIGH3 is reported to be down-regulated during the differentiation of murine osteoblasts and to play negative effects on the early stages of osteogenesis [49]. In hMADS cells, BIGH3 protein levels increase in differentiating osteoblasts as compared to day 0, however a decrease during osteoblast terminal differentiation was observed (data not shown). Interestingly, BIGH3 is also released by differentiating adipocytes at day 3. So far, no report has described the secretion of BIGH3 by adipocytes or adipose tissue, whereas BIGH3 is involved in distinct cellular functions, such as cell growth, tumorigenesis, wound healing, apoptosis and migration [50]. Thus a role for BIGH3 in adipogenesis remains to be established.
PTX3 is a member of the pentraxin family that we described to be secreted by adipocytes and to respond to inflammatory stimuli such as TNFα [51]. In differentiating hMADS cells, PTX3 appears up-regulated during both adipogenesis and osteogenesis. The presence of PTX3 in osteoblasts has never been reported; we speculate that, under conditions of stress or inflammation, PTX3 could represent a potential target of the adipocyte/osteoblast balance that might deserve further studies.
Finally, PAI-1 represents another member of the serpin family which is secreted by hMADS cells. PAI-1 is a pleiotropic molecule, exerting functional roles in wound healing, atherosclerosis, tumor angiogenesis, rheumatoid arthritis, fibrosis etc., besides its main role as regulator of fibrinolysis [52]. In addition, PAI-1 plays a role in metabolic disorders such as obesity and insulin resistance, representing a marker of metabolic syndrome [53]. A role for PAI-1 in bone remodeling has been postulated, since mice lacking PAI-1 are protected from trabecular bone loss after ovariectomy, suggesting a site-specific role for PAI-1 in bone turnover [54]. Altogether, these data indicate that PAI-1 should represent a reliable candidate for the study of the adipocyte/osteoblast balance. In hMADS cell secretome, PAI-1 signal disappears at day 3 of adipogenesis while remaining present in differentiating osteoblasts. Despite some demonstration that PAI-1 is produced by adipocytes during adipogenesis [53], it was recently shown that adipogenesis, as such, may not induce PAI-1; rather it enhances the potential of adipocytes to respond to PAI-1 inducers [55]. This may explain why PAI-1 synthesis dropped after differentiation of human adipocytes cultured in the absence of serum, known to contain several PAI-1 inducers [56,57]. Moreover overexpression of PAI-1 by adenovirus-mediated gene transfer inhibited adipocyte differentiation [58]. Conversely, preadipocytes from PAI-1-/- mice showed greater differentiation than those issued from wild type mice [58]. This suggests that not only PAI-1 is dispensable for adipocyte differentiation but could even be deleterious as it has been recently suggested that bone marrow-derived PAI-1 had an effect on the development of obesity through its effect in inflammation [59].
tPA is also preferentially secreted by hMADS differentiating osteoblasts, while no detectable levels of uPA have been detected, despite the presence of UPAR mRNA. Interestingly, PAI-1 and tPA serum levels have been reported as putative non-invasive diagnostic biomarkers of idiopathic osteonecrosis of the femoral head [60]. As for circulating PAI-1, a correlation with dysbaric osteonecrosis has been also described [61]. These data strongly suggest that the plasminogen/plasmin system could represent a potential target for further investigation of the adipocyte/osteoblast balance. Even if preliminary, our data concerning a decrease of PAI-1 mRNA levels in bone of ovx as compared to sham mice clearly indicate that PAI-1 expression is modulated under conditions altering the adipocyte/osteoblast balance, such as osteoporosis.
Conclusion
In conclusion, we have characterized the secretome of hMADS cells during the early events of adipogenesis and osteogenesis using a proteomic approach. hMADS cell secretome is represented by bona fide secreted proteins, proteins secreted through non-classical pathways and intracellular proteins, the presence of which in the extracellular medium being likely due to exosome secretion. Eight clusters have been identified, including proteases, protease inhibitors, ECM components, anti-inflammatory-antioxidant proteins, metabolic enzymes, cytoskeletal components, heat-shock/protein folding proteins and other proteins. Among the 8 selected proteins, PAI-1 constitutes the most promising candidate for further studies owing to the role of the plasminogen/plasmin system in adipogenesis and bone remodeling.
Methods
Materials
Cell culture media were purchased from Cambrex and FCS from Dutscher S.A. (Brumath, France). EGF was a product of Euromedex (Souffelweyersheim, France). Antibodies directed against ENO1, PEDF, BIGH3 and SPARC were supplied from Santa Cruz Biotechnology (Santa Cruz, CA), Chemicon International (Temecula, CA), R&D Systems (Minneapolis, MN) and Haematologic Technologies Inc. (Essex Junction, VT), respectively. PAI-1 antibody was a kind gift from Prof. PJ Declerck, (Leuven, Belgium). Rat anti-human PTX-3 antibody was kindly provided by Prof. A Mantovani [62]. Reverse transcriptase and trypsin were from Promega (Charbonnières-les-Bains, France). PVDF membranes were from Amersham Biosciences (Orsay, France). Complete protease inhibitor cocktail was from Roche Diagnostics (Meylan, France). All the other products were from Sigma-Aldrich (Saint Quentin Fallavier, France).
Cell culture
The establishment and characterization of the multipotency and self-renewal of hMADS cells have already been described [14-16]. In the experiments reported herein, hMADS-2 cells, established from the pubic region fat pad of a 5-year old male donor, were used at passages between 16 and 35 corresponding to 35 to 100 population doublings. Cells were seeded at a density of 5000 cells/cm2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, 2.5 ng/ml hFGF2, 60 μg/ml penicillin and 50 μg/ml streptomycin. The medium was changed every other day and hFGF2 was removed when cells reached confluence. At day 2 post-confluence (designated as day 0) adipogenic or osteogenic differentiation was induced, as described previously [14,17]. Briefly, for adipogenic differentiation hMADS cells were induced to differentiate in the presence of DMEM/Ham's F12 media supplemented with 0.85 μM insulin, 0.2 nM triiodothyronine, 10 μg/ml transferrin, 1 μM dexamethasone (DEX) and 500 μM isobutyl-methylxanthine (IBMX). Three days later, the medium was changed (DEX and IBMX omitted) and 100 nM Rosiglitazone was added up to day 9. For osteogenic differentiation, cells were induced to differentiate in α-MEM containing 1% FCS supplemented with 10 nM 1,25-dihydroxyvitamin D3, 100 nM DEX, 10 μM L-ascorbic acid phosphate, 10 mM β-glycerophosphate and 10 ng/ml EGF. For both differentiation protocols, the media were then changed every other day and cells were used at the indicated days.
Lactate dehydrogenase (LDH) activity has been performed as described previously [63]. Briefly, the activity has been measured in the culture media and compared to that obtained from cells treated with a detergent in order to release the whole intracellular LDH activity.
Animals
Eight weeks old C57Bl/6J mice were subjected either to bilateral ovariectomies (ovx) from the dorsal approach or to sham surgery during which the ovaries were exteriorized but replaced intact by the operator (Charles River, L'Arbresle, France). Mice were housed in local animal facility for a period of 10 weeks following surgery to await the development of osteoporosis. Animals were sacrificed with carbon dioxide according to the guidelines of the local animal care and experimentation committee. RNA from humerus was extracted using Totally RNA™ kit according to manufacturer's instructions (Ambion, Courtaboeuf, France).
Sample preparation
Conditioned media for each condition (24 ml, corresponding to 8 dishes of 100 mm diameter) were collected on ice, centrifuged and filtered to remove cell debris, if any, and supplemented with complete protease inhibitor cocktail. Twelve ml of the samples were concentrated by ultra-filtration (Millipore, Centricon, 5 kDa cut-off). Protein concentrations were determined using Bio-Rad Protein Assay reagent (Bio-Rad, Marnes-la-Coquette, France) and were: 0.12 ± 0.04, 0.15 ± 0.06; 0.12 ± 0.02 μg/μl for day 0, day 3 adipocyte and day 3 osteoblast respectively.
One-dimensional polyacrylamide gel electrophoresis
One-dimensional polyacrylamide gel electrophoresis (SDS-PAGE) was performed using Bio-Rad mini-Protean system. Gels were stained with Bio-Safe Coomassie Stain (Bio-Rad) for 2 h at room temperature and rinsed with water, according to manufacturer's instructions.
Trypsin digestion and Mass Spectrometry
After Coomassie staining, the whole lanes were manually excised into 10 bands which were processed for tryptic digestion according to a standard protocol. Briefly, after a washing step with 50% ethanol, proteins were reduced and alkylated with 10 mM DTT and 50 mM iodoacetamide, respectively. The spots were washed twice with 50% ethanol, shrunken with acetonitrile (ACN) and then subsequently digested with 100 ng of trypsin in 50 mM ammonium bicarbonate, 15% ACN. The peptides were extracted with 0.5% TFA, 50% ACN then 100% ACN. LC-MS/MS mass spectrometry was performed with a Surveyor system coupled with an LCQ DECA XP ion trap mass spectrometer (ThermoQuest, San Jose, CA). Peptides were separated on a C18 column (300 μm × 10 cm, Hypersil, ThermoQuest) at 5 μl/min with 1 to 40 % ACN gradient in 0.1 % formic acid. A 3 kV voltage was applied on the micro-ESI needle and the automatic acquisition was set as previously described [64].
Fragmentation spectra acquired by Xcalibur 1.3 (ThermoElectron Corp.) were searched with Sequest (Bio-Works version 3.3, Thermo Electron Corp.) against the Swiss-Prot protein database (235 673 entries, uniprot_sprot.fasta file downloaded on October 2006 from [65]). Sequest parameters were: i) peptide mass tolerance of 1.7 Da, ii) parent ion masses treated as monoisotopic, iii) fragmentation ion masses treated as monoisotopic with 1.0 Da mass tolerance, iv) trypsin (KR, strictly enzymatic specificity at the both ends) and two miss cleavages, v) a 57.0 Da static modification on cysteines accounted for alkylation and a 16.0-Da variable modification on methionine accounted for oxidation.
Sequest results were filtered with the following requirements: i) at least 2 peptides were required for protein identification, ii) peptides identified as first candidate, iii) Xcorr > 1.7, 2.2 and 3.3 for mono-, di- and tri-charged peptides, iv) P value up to 0.001 for peptide. The filters' relevance was checked with different tryptic digests as control (HSP90, albumin, lactoglobulin) and using the different SDS-PAGE molecular weight markers; filters automatically removed all the proteins from Bioworks results sheet with the exception of the exact identification. To address the database redundancy issue, only identifications with highest scoring and corresponding to human protein references were selected. Protein classification has been performed using annotation from Bioinformatic Harvester [66] and Swiss-Prot Protein database [67].
RNA extraction, RT-PCR and quantitative RT-PCR analysis (qRT-PCR)
Total RNA was isolated with TRI-Reagent kit (Euromedex, France) according to the manufacturer's instructions. Reverse transcriptase reactions and semi-quantitative PCR assays were performed as already described [14,51]. PCR products (25 cycles) were analyzed by 1.5% agarose gel electrophoresis visualized by ethidium-bromide staining.
qRT-PCR assays were run on an ABI Prism 7000 real-time PCR machine (PerkinElmer Life Sciences). Reactions were performed according to manufacturer's instructions using SYBR green master mix (Eurogentec, France). The expression of selected genes was normalized to the expression of the TATA-binding protein (TBP) encoding gene. Gene expression was quantified using the comparative-delta Ct method.
Primer sequences and annealing temperatures are reported in Table 2S [see Additional file 3].
Immunoblotting
Equal amounts of proteins (2 μg) were denatured, reduced and separated on 12% polyacrylamide-SDS gels. Proteins were transferred in 25 mM Tris, 192 mM glycine and 20% ethanol onto PVDF membrane. Blots were blocked for 30 min with TBS (10 mM Tris-HCl pH 7.5 and 150 mM NaCl) plus 0.1% Tween 20 (TTBS buffer) containing 5% dried milk powder (blocking buffer) and then hybridized in the same buffer with specific primary antibodies (0.2 μg/ml) at 4°C. After overnight incubation, the blots were washed twice in TTBS and incubated 1 hr at room temperature in blocking buffer using the appropriate secondary antibodies (Sigma). After 5 washes in TTBS, immunoreactive proteins were visualized using the ECL chemiluminescence's detection kit (Amersham) according to the manufacturer's instructions.
Determination of MMP activity by gelatin zymography
Proteins with gelatinolytic activity were identified by electrophoresis in 10% SDS-PAGE containing 1 mg/ml gelatin. Two μg of secretion media were directly loaded on gels under nonreducing conditions. After electrophoresis, proteins were renatured by washing twice with 2.5% Triton X-100 in 50 mM Tris-HCl pH 7.4. The gels were then incubated at 37°C for 16 h in an activation buffer (50 mM Tris-HCl pH 7.4, 5 mM CaCl2 and 0.2 M NaCl) and stained with Coomassie Brilliant Blue R-250 (in 40% methanol and 10% acetic acid). Destaining was performed in the same buffer in the absence of the dye. Migration of proteins was compared with that of a prestained molecular weight marker. EDTA (15 mM) was added to the incubation buffer in parallel gels, in order to inhibit cation-dependent enzymatic activities.
Determination of uPA and tPA activities by casein-plasminogen zymography
Two μg of conditioned media were analyzed on 10% polyacrylamide gels containing 1 mg/ml alpha-casein and 10 μg/ml plasminogen (Sigma), under nonreducing conditions. After the electrophoretic run, gels were rinsed once in 100 mM glycine buffer with 2.5% Triton-X 100 for 45 min and then incubated overnight at 37°C in 100 mM glycine buffer, 15 mM EDTA, pH 8.0. Identical gels were incubated in the above buffers containing 500 μM amiloride, in order to discriminate uPA from tPA bands. Gels were then stained with a 0.25% Coomassie Blue/10% acetic acid/40% methanol solution for 1 hr, followed by destaining in a 40% methanol/10% acetic acid mixture. Caseinolytic activity resulting from plasminogen activation was visualized by white lytic bands on a blue background after conversion of plasminogen to plasmin by uPA and tPA. Molecular weights were calculated from the position of prestained markers that were subjected to electrophoresis in parallel lanes. No lytic bands were observed in plasminogen-free gels.
ELISA
Supernatant PAI-1 antigen was assayed using ELISA's specific for human PAI-1, as previously described [68]. PAI-1 ELISA detects latent and active forms of human PAI-1 and PAI-1 complexes. The assay is insensitive to PAI-2. The amount of uPA and tPA antigen was measured with the Immunobind uPA ELISA kit (American Diagnostic, Greenwich, CT) and the commercially available kit Asserachrom tPA (Diagnostica Stago, France) according to the instructions of the manufacturer. Inactive and active forms of plasminogen activators are all recognized by the ELISA kit. Plasma concentration of leptin was measured by ELISA using EIA kit (SPI-Bio, France).
Statistical analysis
Data are expressed as mean values ± standard deviation (SD). qRT-PCR and ELISA data and quantifications of immunoblot/zymogram experiments were analyzed using Student's t-test. Statistical significance was assumed at p level <0.05.
Abbreviations
BIGH3, transforming growth factor-beta-induced protein; ECM, extracellular matrix; EGF, epidermal growth factor; ENO1, α-enolase; ex, extracellular; GRP78, 78 kDa glucose-regulated protein precursor; hFGF2, human fibroblast growth factor 2; hMADS cells, human multipotent adipose-derived stem cells; in, intracellular; LC-MS/MS, liquid chromatography-tandem mass spectrometry; LDH, lactate dehydrogenase; MMP2, Matrix metallopeptidase 2; ovx, ovariectomized; PAI-1, plasminogen activator inhibitor type 1; PEDF, pigment epithelium derived factor; PTX3, pentraxin 3; sp, signal peptide; SPARC, secreted protein, acidic, cysteine-rich; tPA, tissue type plasminogen activator; uPA, urinary plasminogen activator; UPAR, receptor for urokinase-type plasminogen activator
Authors' contributions
CC did most of the work, including cell culture, sample media preparation, monodimensional gels, immunoblotting, RNA extraction and qRT-PCR experiments, zymograms and contributed to manuscript preparation. OC contributed to cell culture experiments, sample media and RNA preparation and provided tissues from ovx and sham mice. LN and MS conducted mass spectrometry experiments, peptides identification and database searches; LN also contributed to manuscript preparation. MP performed ELISA assays. GA provided useful suggestions and contributed to manuscript preparation. MCA contributed with reagents, provided useful suggestions and participated to manuscript preparation. CD contributed to data interpretation and manuscript preparation. EA contributed to the experimental design, analysis, interpretation, and manuscript preparation. All authors have read and approved the final manuscript.
Supplementary Material
Figure 1S: LDH activity in media. LDL activity released in the medium after 6 hours of exposition to serum-free medium (white columns) compared to LDH release by complete cell lysis (black columns) from cells at day 0, day 3 adipodipocyte or osteoblast differentiation.
Table 1S: List of peptides identified by mass spectrometry for the 73 proteins found in hMADS cell secretome. The identified proteins are indicated in bold with their corresponding peptides listed under the protein annotation. The different values correspond to parameters produced by Bioworks 3.3 after the data filtering.
Table 2S: List of primer sequences for gene expression analysis by qRT-PCR and RT-PCR.
Acknowledgments
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique (CNRS, Programme Protéomique et génie des protéines), and by a grant from: «Equipe FRM, soutenue par la Fondation pour la Recherche Médicale». Prof. Alberto Mantovani is acknowledged for generous gift of anti-PTX3 antibody. We are grateful to Dr. Nathalie Billon, Dr. Joaquin Lopez-Soriano and Dr. Delphine Bastelica for critical review of the manuscript and for help with ELISA experiments. CC is a recipient of a fellowship from Fondation pour la Recherche Médicale.
Contributor Information
Chiara Chiellini, Email: Chiara.Chiellini@gmail.com.
Olivia Cochet, Email: cochet@unice.fr.
Luc Negroni, Email: negroni@unice.fr.
Michel Samson, Email: samson@unice.fr.
Marjorie Poggi, Email: Marjorie.Poggi@medecine.univ-mrs.fr.
Gérard Ailhaud, Email: ailhaud@unice.fr.
Marie-Christine Alessi, Email: Marie-Christine.Alessi@medecine.univ-mrs.fr.
Christian Dani, Email: dani@unice.fr.
Ez-Zoubir Amri, Email: amri@unice.fr.
References
- Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocr Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- Ailhaud G. Adipose tissue as a secretory organ: from adipogenesis to the metabolic syndrome. C R Biol. 2006;329:570–7; discussion 653-5. doi: 10.1016/j.crvi.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Dani C. [Stem cells from human adipose tissue: a new tool for pharmacological studies and for clinical applications] J Soc Biol. 2006;200:45–50. doi: 10.1051/jbio:2006006. [DOI] [PubMed] [Google Scholar]
- Ahima RS. Adipose tissue as an endocrine organ. Obesity (Silver Spring) 2006;14 Suppl 5:242S–249S. doi: 10.1038/oby.2006.317. [DOI] [PubMed] [Google Scholar]
- Simha V, Garg A. Lipodystrophy: lessons in lipid and energy metabolism. Curr Opin Lipidol. 2006;17:162–169. doi: 10.1097/01.mol.0000217898.52197.18. [DOI] [PubMed] [Google Scholar]
- Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol. 2006;2:35–43. doi: 10.1038/ncprheum0070. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- Thomas T, Gori F, Khosla S, Jensen MD, Burguera B, Riggs BL. Leptin acts on human marrow stromal cells to enhance differentiation to osteoblasts and to inhibit differentiation to adipocytes. Endocrinology. 1999;140:1630–1638. doi: 10.1210/en.140.4.1630. [DOI] [PubMed] [Google Scholar]
- Martin A, de Vittoris R, David V, Moraes R, Begeot M, Lafage-Proust MH, Alexandre C, Vico L, Thomas T. Leptin modulates both resorption and formation while preventing disuse-induced bone loss in tail-suspended female rats. Endocrinology. 2005;146:3652–3659. doi: 10.1210/en.2004-1509. [DOI] [PubMed] [Google Scholar]
- Oshima K, Nampei A, Matsuda M, Iwaki M, Fukuhara A, Hashimoto J, Yoshikawa H, Shimomura I. Adiponectin increases bone mass by suppressing osteoclast and activating osteoblast. Biochem Biophys Res Commun. 2005;331:520–526. doi: 10.1016/j.bbrc.2005.03.210. [DOI] [PubMed] [Google Scholar]
- Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Elabd C, Delteil F, Astier J, Vernochet C, Saint-Marc P, Guesnet J, Guezennec A, Amri EZ, Dani C, Ailhaud G. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem Biophys Res Commun. 2004;315:255–263. doi: 10.1016/j.bbrc.2004.01.053. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Pisani D, Dechesne CA, Turc-Carel C, Kurzenne JY, Wdziekonski B, Villageois A, Bagnis C, Breittmayer JP, Groux H, Ailhaud G, Dani C. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. J Exp Med. 2005;201:1397–1405. doi: 10.1084/jem.20042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragosi LE, Ailhaud G, Dani C. Autocrine fibroblast growth factor 2 signaling is critical for self-renewal of human multipotent adipose-derived stem cells. Stem Cells. 2006;24:2412–2419. doi: 10.1634/stemcells.2006-0006. [DOI] [PubMed] [Google Scholar]
- Elabd C, Chiellini C, Massoudi A, Cochet O, Zaragosi LE, Trojani C, Michiels JF, Weiss P, Carle G, Rochet N, Dechesne CA, Ailhaud G, Dani C, Amri EZ. Human adipose tissue-derived multipotent stem cells differentiate in vitro and in vivo into osteocyte-like cells. Biochem Biophys Res Commun. 2007;361:342–348. doi: 10.1016/j.bbrc.2007.06.180. [DOI] [PubMed] [Google Scholar]
- Chen X, Cushman SW, Pannell LK, Hess S. Quantitative proteomic analysis of the secretory proteins from rat adipose cells using a 2D liquid chromatography-MS/MS approach. J Proteome Res. 2005;4:570–577. doi: 10.1021/pr049772a. [DOI] [PubMed] [Google Scholar]
- Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, Hoek A, Wolffenbuttel BH, Roelofsen H, Vonk RJ. Characterization of the Human Visceral Adipose Tissue Secretome. Mol Cell Proteomics. 2007;6:589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- Kratchmarova I, Kalume DE, Blagoev B, Scherer PE, Podtelejnikov AV, Molina H, Bickel PE, Andersen JS, Fernandez MM, Bunkenborg J, Roepstorff P, Kristiansen K, Lodish HF, Mann M, Pandey A. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol Cell Proteomics. 2002;1:213–222. doi: 10.1074/mcp.M200006-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang P, Mariman E, Keijer J, Bouwman F, Noben JP, Robben J, Renes J. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell Mol Life Sci. 2004;61:2405–2417. doi: 10.1007/s00018-004-4256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonic S, Lefevre M, Kilroy G, Floyd ZE, DeLany JP, Kheterpal I, Gravois A, Dow R, White A, Wu X, Gimble JM. Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6:18–28. doi: 10.1074/mcp.M600217-MCP200. [DOI] [PubMed] [Google Scholar]
- Litvin J, Selim AH, Montgomery MO, Lehmann K, Rico MC, Devlin H, Bednarik DP, Safadi FF. Expression and function of periostin-isoforms in bone. J Cell Biochem. 2004;92:1044–1061. doi: 10.1002/jcb.20115. [DOI] [PubMed] [Google Scholar]
- Vassalli JD, Sappino AP, Belin D. The plasminogen activator/plasmin system. J Clin Invest. 1991;88:1067–1072. doi: 10.1172/JCI115405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulisse S, Baldini E, Toller M, Marchioni E, Giacomelli L, De Antoni E, Ferretti E, Marzullo A, Graziano FM, Trimboli P, Biordi L, Curcio F, Gulino A, Ambesi-Impiombato FS, D'Armiento M. Differential expression of the components of the plasminogen activating system in human thyroid tumour derived cell lines and papillary carcinomas. Eur J Cancer. 2006;42:2631–2638. doi: 10.1016/j.ejca.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Jansson JO, Moverare-Skrtic S, Berndtsson A, Wernstedt I, Carlsten H, Ohlsson C. Leukemia inhibitory factor reduces body fat mass in ovariectomized mice. Eur J Endocrinol. 2006;154:349–354. doi: 10.1530/eje.1.02082. [DOI] [PubMed] [Google Scholar]
- Wronski TJ, Lowry PL, Walsh CC, Ignaszewski LA. Skeletal alterations in ovariectomized rats. Calcif Tissue Int. 1985;37:324–328. doi: 10.1007/BF02554882. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Camalier CE, Nagashima K, Chan KC, Lucas DA, de la Cruz MJ, Gignac M, Lockett S, Issaq HJ, Veenstra TD, Conrads TP, Beck GR., Jr. Analysis of the extracellular matrix vesicle proteome in mineralizing osteoblasts. J Cell Physiol. 2007;210:325–335. doi: 10.1002/jcp.20826. [DOI] [PubMed] [Google Scholar]
- Shamaei-Tousi A, Stephens JW, Bin R, Cooper JA, Steptoe A, Coates AR, Henderson B, Humphries SE. Association between plasma levels of heat shock protein 60 and cardiovascular disease in patients with diabetes mellitus. Eur Heart J. 2006;27:1565–1570. doi: 10.1093/eurheartj/ehl081. [DOI] [PubMed] [Google Scholar]
- Satoh M, Shimoda Y, Akatsu T, Ishikawa Y, Minami Y, Nakamura M. Elevated circulating levels of heat shock protein 70 are related to systemic inflammatory reaction through monocyte Toll signal in patients with heart failure after acute myocardial infarction. Eur J Heart Fail. 2006;8:810–815. doi: 10.1016/j.ejheart.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Yamaji R, Chatani E, Harada N, Sugimoto K, Inui H, Nakano Y. Glyceraldehyde-3-phosphate dehydrogenase in the extracellular space inhibits cell spreading. Biochim Biophys Acta. 2005;1726:261–271. doi: 10.1016/j.bbagen.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Daly EB, Wind T, Jiang XM, Sun L, Hogg PJ. Secretion of phosphoglycerate kinase from tumour cells is controlled by oxygen-sensing hydroxylases. Biochim Biophys Acta. 2004;1691:17–22. doi: 10.1016/j.bbamcr.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Oberley TD, Verwiebe E, Zhong W, Kang SW, Rhee SG. Localization of the thioredoxin system in normal rat kidney. Free Radic Biol Med. 2001;30:412–424. doi: 10.1016/S0891-5849(00)00486-X. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem (Tokyo) 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Mignot G, Roux S, Thery C, Segura E, Zitvogel L. Prospects for exosomes in immunotherapy of cancer. J Cell Mol Med. 2006;10:376–388. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RM. Exosomes biological significance: A concise review. Blood Cells Mol Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Segura E, Amigorena S, Thery C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells Mol Dis. 2005;35:89–93. doi: 10.1016/j.bcmd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- CBS SecretomeP 2.0 Server http://www.cbs.dtu.dk/services/SecretomeP/
- Bouloumie A, Sengenes C, Portolan G, Galitzky J, Lafontan M. Adipocyte produces matrix metalloproteinases 2 and 9: involvement in adipose differentiation. Diabetes. 2001;50:2080–2086. doi: 10.2337/diabetes.50.9.2080. [DOI] [PubMed] [Google Scholar]
- Parikka V, Vaananen A, Risteli J, Salo T, Sorsa T, Vaananen HK, Lehenkari P. Human mesenchymal stem cell derived osteoblasts degrade organic bone matrix in vitro by matrix metalloproteinases. Matrix Biol. 2005;24:438–447. doi: 10.1016/j.matbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Tartare-Deckert S, Chavey C, Monthouel MN, Gautier N, Van Obberghen E. The matricellular protein SPARC/osteonectin as a newly identified factor up-regulated in obesity. J Biol Chem. 2001;276:22231–22237. doi: 10.1074/jbc.M010634200. [DOI] [PubMed] [Google Scholar]
- Kasugai S, Todescan R, Jr., Nagata T, Yao KL, Butler WT, Sodek J. Expression of bone matrix proteins associated with mineralized tissue formation by adult rat bone marrow cells in vitro: inductive effects of dexamethasone on the osteoblastic phenotype. J Cell Physiol. 1991;147:111–120. doi: 10.1002/jcp.1041470115. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nagaretani H, Funahashi T, Nishizawa H, Maeda N, Kishida K, Kuriyama H, Shimomura I, Maeda K, Hotta K, Ouchi N, Kihara S, Nakamura T, Yamashita S, Matsuzawa Y. The expression of SPARC in adipose tissue and its increased plasma concentration in patients with coronary artery disease. Obes Res. 2001;9:388–393. doi: 10.1038/oby.2001.50. [DOI] [PubMed] [Google Scholar]
- Delany AM, Kalajzic I, Bradshaw AD, Sage EH, Canalis E. Osteonectin-null mutation compromises osteoblast formation, maturation, and survival. Endocrinology. 2003;144:2588–2596. doi: 10.1210/en.2002-221044. [DOI] [PubMed] [Google Scholar]
- Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- Lopez-Alemany R, Suelves M, Diaz-Ramos A, Vidal B, Munoz-Canoves P. Alpha-enolase plasminogen receptor in myogenesis. Front Biosci. 2005;10:30–36. doi: 10.2741/1503. [DOI] [PubMed] [Google Scholar]
- Tombran-Tink J, Barnstable CJ. Osteoblasts and osteoclasts express PEDF, VEGF-A isoforms, and VEGF receptors: possible mediators of angiogenesis and matrix remodeling in the bone. Biochem Biophys Res Commun. 2004;316:573–579. doi: 10.1016/j.bbrc.2004.02.076. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Thapa N, Kang KB, Kim IS. Beta ig-h3 mediates osteoblast adhesion and inhibits differentiation. Bone. 2005;36:232–242. doi: 10.1016/j.bone.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Morand S, Buchillier V, Maurer F, Bonny C, Arsenijevic Y, Munier FL, Schorderet DF. Induction of apoptosis in human corneal and HeLa cells by mutated BIGH3. Invest Ophthalmol Vis Sci. 2003;44:2973–2979. doi: 10.1167/iovs.02-0661. [DOI] [PubMed] [Google Scholar]
- Abderrahim-Ferkoune A, Bezy O, Chiellini C, Maffei M, Grimaldi P, Bonino F, Moustaid-Moussa N, Pasqualini F, Mantovani A, Ailhaud G, Amri EZ. Characterization of the long pentraxin PTX3 as a TNFalpha-induced secreted protein of adipose cells. J Lipid Res. 2003;44:994–1000. doi: 10.1194/jlr.M200382-JLR200. [DOI] [PubMed] [Google Scholar]
- Lijnen HR. Pleiotropic functions of plasminogen activator inhibitor-1. J Thromb Haemost. 2005;3:35–45. doi: 10.1111/j.1538-7836.2004.00827.x. [DOI] [PubMed] [Google Scholar]
- Alessi MC, Juhan-Vague I. PAI-1 and the metabolic syndrome: links, causes, and consequences. Arterioscler Thromb Vasc Biol. 2006;26:2200–2207. doi: 10.1161/01.ATV.0000242905.41404.68. [DOI] [PubMed] [Google Scholar]
- Daci E, Verstuyf A, Moermans K, Bouillon R, Carmeliet G. Mice lacking the plasminogen activator inhibitor 1 are protected from trabecular bone loss induced by estrogen deficiency. J Bone Miner Res. 2000;15:1510–1516. doi: 10.1359/jbmr.2000.15.8.1510. [DOI] [PubMed] [Google Scholar]
- Venugopal J, Hanashiro K, Nagamine Y. Regulation of PAI-1 gene expression during adipogenesis. J Cell Biochem. 2007;101: 369 –3380. doi: 10.1002/jcb.21173. [DOI] [PubMed] [Google Scholar]
- Wang B, Jenkins JR, Trayhurn P. Expression and secretion of inflammation-related adipokines by human adipocytes differentiated in culture: integrated response to TNF-alpha. Am J Physiol Endocrinol Metab. 2005;288:E731–40. doi: 10.1152/ajpendo.00475.2004. [DOI] [PubMed] [Google Scholar]
- Bastelica D, Morange P, Berthet B, Borghi H, Lacroix O, Grino M, Juhan-Vague I, Alessi MC. Stromal cells are the main plasminogen activator inhibitor-1-producing cells in human fat: evidence of differences between visceral and subcutaneous deposits. Arterioscler Thromb Vasc Biol. 2002;22:173–178. doi: 10.1161/hq0102.101552. [DOI] [PubMed] [Google Scholar]
- Liang X, Kanjanabuch T, Mao SL, Hao CM, Tang YW, Declerck PJ, Hasty AH, Wasserman DH, Fogo AB, Ma LJ. Plasminogen activator inhibitor-1 modulates adipocyte differentiation. Am J Physiol Endocrinol Metab. 2006;290:E103–E113. doi: 10.1152/ajpendo.00605.2004. [DOI] [PubMed] [Google Scholar]
- De Taeye BM, Novitskaya T, Gleaves L, Covington JW, Vaughan DE. Bone marrow plasminogen activator inhibitor-1 influences the development of obesity. J Biol Chem. 2006;281:32796–32805. doi: 10.1074/jbc.M606214200. [DOI] [PubMed] [Google Scholar]
- Tan X, Cai D, Wu Y, Liu B, Rong L, Chen Z, Zhao Q. Comparative analysis of serum proteomes: discovery of proteins associated with osteonecrosis of the femoral head. Transl Res. 2006;148:114–119. doi: 10.1016/j.trsl.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Miyanishi K, Kamo Y, Ihara H, Naka T, Hirakawa M, Sugioka Y. Risk factors for dysbaric osteonecrosis. Rheumatology (Oxford) 2006;45:855–858. doi: 10.1093/rheumatology/kel013. [DOI] [PubMed] [Google Scholar]
- Bottazzi B, Vouret-Craviari V, Bastone A, De Gioia L, Matteucci C, Peri G, Spreafico F, Pausa M, D'Ettorre C, Gianazza E, Tagliabue A, Salmona M, Tedesco F, Introna M, Mantovani A. Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J Biol Chem. 1997;272:32817–32823. doi: 10.1074/jbc.272.52.32817. [DOI] [PubMed] [Google Scholar]
- Grimaldi P, Negrel R, Ailhaud G. Induction of the triglyceride pathway enzymes and of lipolytic enzymes during differentiation in a 'preadipocyte' cell line. Eur J Biochem. 1978;84:369–376. doi: 10.1111/j.1432-1033.1978.tb12177.x. [DOI] [PubMed] [Google Scholar]
- Valot B, Negroni L, Zivy M, Gianinazzi S, Dumas-Gaudot E. A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics. 2006;6 Suppl 1:S145–55. doi: 10.1002/pmic.200500403. [DOI] [PubMed] [Google Scholar]
- Swiss-Prot Protein database ftp://ftp.expasy.org/databases/swiss-prot/release/
- Harvester http://www.harvester.embl.de/
- Swiss-Prot http://www.expasy.org/sprot/
- Declerck PJ, Alessi MC, Verstreken M, Kruithof EK, Juhan-Vague I, Collen D. Measurement of plasminogen activator inhibitor 1 in biologic fluids with a murine monoclonal antibody-based enzyme-linked immunosorbent assay. Blood. 1988;71:220–225. [PubMed] [Google Scholar]
- Bortoluzzi S, Scannapieco P, Cestaro A, Danieli GA, Schiaffino S. Computational reconstruction of the human skeletal muscle secretome. Proteins. 2006;62:776–792. doi: 10.1002/prot.20803. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Chen SX, Huang Y, Luo WJ, Jiang HH, Hu QH, Zhang PF, Yi H. Proteomics-based identification of secreted protein dihydrodiol dehydrogenase as a novel serum markers of non-small cell lung cancer. Lung Cancer. 2006;54:87–94. doi: 10.1016/j.lungcan.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Dupont A, Tokarski C, Dekeyzer O, Guihot AL, Amouyel P, Rolando C, Pinet F. Two-dimensional maps and databases of the human macrophage proteome and secretome. Proteomics. 2004;4:1761–1778. doi: 10.1002/pmic.200300691. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Akimoto Y, Suzuki T, Kitamura S, Ohta S. Identification of prostatic-secreted proteins in mice by mass spectrometric analysis and evaluation of lobe-specific and androgen-dependent mRNA expression. J Endocrinol. 2006;190:793–803. doi: 10.1677/joe.1.06733. [DOI] [PubMed] [Google Scholar]
- Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- Dupont A, Corseaux D, Dekeyzer O, Drobecq H, Guihot AL, Susen S, Vincentelli A, Amouyel P, Jude B, Pinet F. The proteome and secretome of human arterial smooth muscle cells. Proteomics. 2005;5:585–596. doi: 10.1002/pmic.200400965. [DOI] [PubMed] [Google Scholar]
- Volmer MW, Stuhler K, Zapatka M, Schoneck A, Klein-Scory S, Schmiegel W, Meyer HE, Schwarte-Waldhoff I. Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics. 2005;5:2587–2601. doi: 10.1002/pmic.200401188. [DOI] [PubMed] [Google Scholar]
- Lim JW, Bodnar A. Proteome analysis of conditioned medium from mouse embryonic fibroblast feeder layers which support the growth of human embryonic stem cells. Proteomics. 2002;2:1187–1203. doi: 10.1002/1615-9861(200209)2:9<1187::AID-PROT1187>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Wakabayashi K, Matsuoka T. Proteome analysis of secreted proteins during osteoclast differentiation using two different methods: two-dimensional electrophoresis and isotope-coded affinity tags analysis with two-dimensional chromatography. Proteomics. 2003;3:616–626. doi: 10.1002/pmic.200300410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S: LDH activity in media. LDL activity released in the medium after 6 hours of exposition to serum-free medium (white columns) compared to LDH release by complete cell lysis (black columns) from cells at day 0, day 3 adipodipocyte or osteoblast differentiation.
Table 1S: List of peptides identified by mass spectrometry for the 73 proteins found in hMADS cell secretome. The identified proteins are indicated in bold with their corresponding peptides listed under the protein annotation. The different values correspond to parameters produced by Bioworks 3.3 after the data filtering.
Table 2S: List of primer sequences for gene expression analysis by qRT-PCR and RT-PCR.