Abstract
Snail family transcriptional repressors regulate epithelial mesenchymal transitions during physiological and pathological processes. A conserved SNAG repression domain present in all vertebrate Snail proteins is necessary for repressor complex assembly. Here, we identify the Ajuba family of LIM proteins as functional corepressors of the Snail family via an interaction with the SNAG domain. Ajuba LIM proteins interact with Snail in the nucleus on endogenous E-cadherin promoters and contribute to Snail-dependent repression of E-cadherin. Using Xenopus neural crest as a model of in vivo Snail- or Slug-induced EMT, we demonstrate that Ajuba LIM proteins contribute to neural crest development as Snail/Slug corepressors and are required for in vivo Snail/Slug function. Because Ajuba LIM proteins are also components of adherens junction and contribute to their assembly or stability, their functional interaction with Snail proteins in the nucleus suggests that Ajuba LIM proteins are important regulators of epithelia dynamics communicating surface events with nuclear responses.
Keywords: LIM proteins, Snail, Slug, Neural crest development, Epithelial Mesenchymal Transitions, E-cadherin, Xenopus
Introduction
Developmental processes such as gastrulation and neural crest delamination require epithelial cells to undergo a mesenchymal transition in order to invade and migrate (Thiery and Sleeman 2006). This epithelial to mesenchymal transition (EMT) occurs not only during development, but also during wound repair (Savagner et al. 2005; Thiery and Sleeman 2006), chronic inflammation and fibrosis (Kalluri and Neilson 2003), and tumor progression from localized epithelial adenomas to metastatic carcinomas (Thiery 2002). While many different environmental signals induce EMT, they all converge to activate transcription factors that effect an EMT “gene program.” This program requires transcriptional down-regulation of epithelial genes, such as E-cadherin, and up-regulation of mesenchymal genes, such as fibronectin, that allow epithelial cells to lose adherence to neighboring cells, migrate, and invade. A fundamental challenge then is to understand whether and how cell surface adhesive events communicate with nuclear processes to coordinate this dynamic transition.
Many transcription factors contribute to EMT through direct repression of epithelial cell surface adhesive receptor genes such as E-cadherin, claudins, and occludins. These include SIP1/ZEB2 (Comijn et al. 2001), E47 (Perez-Moreno et al. 2001), Twist (Yang et al. 2004), Snail (Batlle et al. 2000; Cano et al. 2000; Ikenouchi et al. 2003) and Slug (Hajra et al. 2002). Snail and Slug belong to the Snail family of zinc-finger transcriptional repressors and are central regulators of EMT in gastrulation (Carver et al. 2001; Hemavathy et al. 2004), neural crest induction, delamination, and migration (Nieto et al. 1994; LaBonne and Bronner-Fraser 2000; del Barrio and Nieto 2002; Aybar et al. 2003; Zhang et al. 2006), skin wound repair (Savagner et al. 2005), fibrosis (Barrallo-Gimeno and Nieto 2005), and tumor metastasis (Blanco et al. 2002; Barrallo-Gimeno and Nieto 2005; Moody et al. 2005). Specifically, the repressor functions of Snail and Slug are necessary for Xenopus neural crest induction and delamination (LaBonne and Bronner-Fraser 2000; Aybar et al. 2003). On a cellular level, repressor activities of Snail and Slug influence cell survival (Kajita et al. 2004; Vega et al. 2004), adhesion, and migration (Barrallo-Gimeno and Nieto 2005).
While Snail and Slug play integral roles in development and disease through gene repression, the precise mechanism of Snail-dependent repression remains unclear. Repression by Snail and Slug is sensitive to TSA, a histone deacetylase (HDAC) inhibitor (Hemavathy et al. 2000; Peinado et al. 2004), and Snail recruits an HDAC repressor complex to the promoter region (Peinado et al. 2004). How it assembles this complex at select promoters, however, is unknown. In Drosophila, Snail associates with a corepressor, CtBP, that is necessary to mediate its repressor activity (Nibu et al. 1998). Vertebrate Snail family members, however, do not contain a conserved CtBP binding domain. Instead, they mediate repression through an N-terminal SNAG (Snail-Gfi-1) domain that is necessary and sufficient for repression (Grimes et al. 1996; Nakayama et al. 1998; Hemavathy et al. 2000). The SNAG domain contributes to recruitment of HDAC proteins and assembly of a repressor complex (Peinado et al. 2004). Together, these data strongly support the presence of cellular co-repressor(s) that interact with the SNAG domain of vertebrate Snail, analogous to CtBP and Drosophila Snail.
In a screen to identify SNAG domain interacting co-repressors, we identified the Ajuba LIM protein family. Ajuba LIM proteins (Ajuba, LIMD1, WTIP) are closely related to Zyxin LIM proteins (Zyxin, LPP, Trip6). The Ajuba/Zyxin family is characterized by three homologous C-terminal LIM domains (LIM region) and a unique N-terminal region (preLIM region). These proteins localize to cell-cell or cell-matrix adhesion sites in epithelial and fibroblast cells, respectively, and influence cell adhesive complex formation and function (Marie et al. 2003; Srichai et al. 2004; Pratt et al. 2005; Hansen and Beckerle 2006; Hoffman et al. 2006). In addition, they shuttle to and from the nucleus, suggesting they have the potential to coordinate cell surface adhesive events with nuclear responses (Nix and Beckerle 1997; Kanungo et al. 2000; Sharp et al. 2004; Srichai et al. 2004).
We show that Ajuba LIM proteins, but not closely related Zyxin LIM proteins, specifically interact with Snail proteins in the nucleus and function as Snail corepressors to downregulate E-cadherin transcription. Ajuba LIM proteins are recruited to the endogenous E-cadherin promoter in a Snail-dependent manner. In vivo, Ajuba, LIMD1, and WTIP cooperate with Snail and Slug to mediate Xenopus neural crest development. Thus, in addition to regulating epithelial cell-cell adhesion, Ajuba LIM proteins contribute to epithelial-mesenchymal transitions as Snail co-repressors during neural crest development.
Results
Ajuba LIM proteins interact with Snail family proteins in the nucleus
The SNAG domain of vertebrate Snail proteins is necessary and sufficient for transcriptional repressor activity (Nakayama et al. 1998; Hemavathy et al. 2000), yet how it mediates assembly of a repressor complex remains unclear. Through a yeast two-hybrid protein-protein interactive screen, two Ajuba family LIM proteins (Ajuba and LIMD1) were identified as potential interactors with the SNAG domain. The biochemical interaction between Ajuba and a SNAG domain, as modeled by a synthetic SNAG domain-containing repressor, required a functional SNAG domain (Ayyanathan et al. 2007), but whether, and how, this was a biologically relevant interaction in cells and in vivo was not determined.
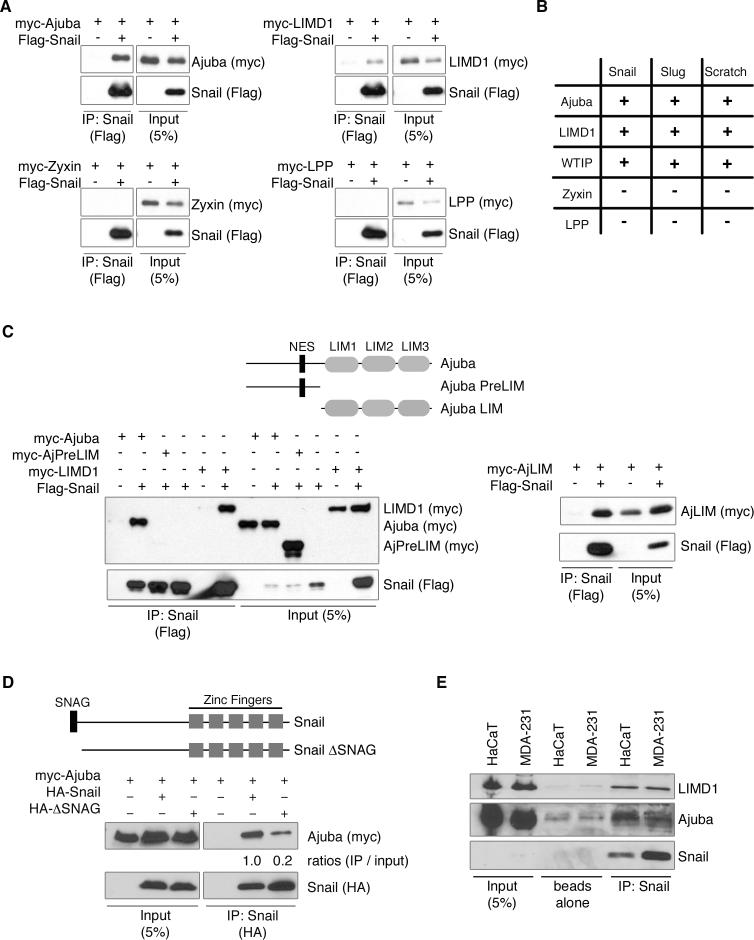
To determine whether Ajuba LIM proteins actually associate with SNAG domain containing proteins in vertebrate cells, HEK293 cells were cotransfected with epitope-tagged LIM and Snail proteins. Snail proteins were immunoprecipitated from total cell lysates and bound products Western blotted for the presence of LIM protein. Ajuba, LIMD1, and WTIP all interacted with Snail proteins Snail1, Slug (Snail2), and Scratch (Fig. 1A and B). Closely related LIM proteins, Zyxin and LPP, did not interact with Snail family members (Fig. 1A and B), however, indicating that Ajuba LIM proteins specifically interact with Snail proteins in vertebrate cells.
Figure 1. Ajuba LIM proteins interact with Snail transcriptional repressors.
A. Myc-tagged LIM proteins and Flag-Snail were cotransfected into HEK293 cells. Snail was immunoprecipitated (anti-Flag) and bound products Western blotted for LIM protein (anti-myc) and Snail (anti-Flag). Control Western blot of lysate is on right panel of each set. B. Table of interactions between Snail proteins and LIM proteins, as determined by co-immunoprecipitation, as described in A. C. Top: Schematic of Ajuba constructs used. NES – nuclear export sequence. Bottom: Co-immunoprecipitation experiments as in A. D. Top: Schematic of Snail constructs used. Bottom: Myc-tagged Ajuba and HA-tagged Snail constructs were cotransfected into HEK293 cells. Snail was immunoprecipitated (anti-HA) and bound products Western blotted for Ajuba (anti-myc) and Snail (anti-HA). Control Western blot of lysate is shown on left. The amount of Ajuba immunoprecipitated relative to input was quantified by densitometry and controlled for the amount of Snail immunoprecipitated. (Ajuba immunoprecipitated with full-length Snail was arbitrarily set to 1). E. Endogenous Snail was immunoprecipitated from lysates of HaCaT or MDA-231 cells and bound products Western blotted for the presence of Ajuba, LIMD1 and Snail. Controls include pulldown with Protein G beads alone and lysate input.
Next, we determined the region of Ajuba necessary for binding Snail, and the region of Snail necessary for binding Ajuba in cells. HEK293 cells were co-transfected with epitope-tagged Ajuba, N-terminal preLIM region of Ajuba, or C-terminal LIM region of Ajuba (Fig. 1C) and Snail. Cells were lysed, Snail immunoprecipitated, and bound products Western blotted for the presence of Ajuba isoforms. The LIM region of Ajuba, but not the preLIM region, interacted with Snail (Fig. 1C). Conversely, HEK293 cells were co-transfected with epitope-tagged Snail or Snail lacking the conserved 7 amino acid SNAG domain (Snail.ΔSNAG) and Ajuba (Fig. 1D). While loss of the SNAG domain decreased the interaction by 80%, Snail.ΔSNAG still bound to Ajuba, suggesting that other regions of Snail may also contribute to the interaction (Fig. 1D). Further determination of Snail region(s) that contribute to the association with Ajuba was complicated by instability of N-terminal and C-terminal deletions, as has been previously observed (Peinado et al. 2004). Nonetheless, this analysis demonstrated that the major domain in Snail that mediated its interaction with Ajuba in cells was the SNAG domain.
To confirm that Ajuba and Snail associated in cells containing endogenous levels of each protein we used two epithelial cell lines that express endogenous Snail: HaCaT human keratinocytes and MDA-231 human breast cancer cells. Since Snail is degraded following phosphorylation by GSK3β (Zhou et al. 2004), cells were treated with MG132, a proteasome inhibitor, and LiCl, a GSK3β inhibitor, for five hours to stabilize endogenous Snail (Zhou et al. 2004). Snail was then immunoprecipitated from total cell lysates and bound products Western blotted for the presence of Ajuba and LIMD1. Ajuba and LIMD1 co-immunoprecipitated with Snail in both cell types (Fig. 1E), indicating that the interaction occurred in cells with endogenous levels of each protein.
After demonstrating that Ajuba and Snail associate in total cell lysates, we sought to determine whether these proteins also co-localize in cells. Both Ajuba and Snail shuttle to and from the nucleus (Kanungo et al. 2000; Dominguez et al. 2003; Zhou et al. 2004). When GFP-Snail or RFP-Ajuba was expressed alone in MCF-7 human breast epithelial cells, GFP-Snail was predominantly nuclear (95% of cells) while RFP-Ajuba was predominantly cytosolic (85−90% of cells). In 10−15% of the cells a low level of nuclear staining for Ajuba, in addition to cytosolic staining, was observed. MCF-7 cells were then co-transfected with RFP-Ajuba in combination with GFP, GFP-Snail, or YFP-Snail.ΔSNAG. The percent of cells with nuclear GFP-Snail remained the same in the presence or absence of Ajuba. Snail expression, however, caused a greater than three-fold increase in the percent of cells that contained nuclear Ajuba (Fig. 2A and B), whereas Snail.ΔSNAG did not influence the subcellular localization of Ajuba (Fig. 2B). Snail 8 Ser-Ala (Sn8SA) is a mutant form of Snail that cannot be phosphorylated by nuclear GSK3β and, thus, is stabilized and constitutively localized to the nucleus (Zhou et al. 2004). When GFP-Sn8SA and RFP-Ajuba were co-expressed, a greater percentage of cells (76%) contained nuclear Ajuba (Fig. 2B). Since Sn8SA does not exit the nucleus, this result suggested that Snail was likely trapping Ajuba in the nucleus.
Figure 2. Ajuba interacts with Snail in the nucleus.
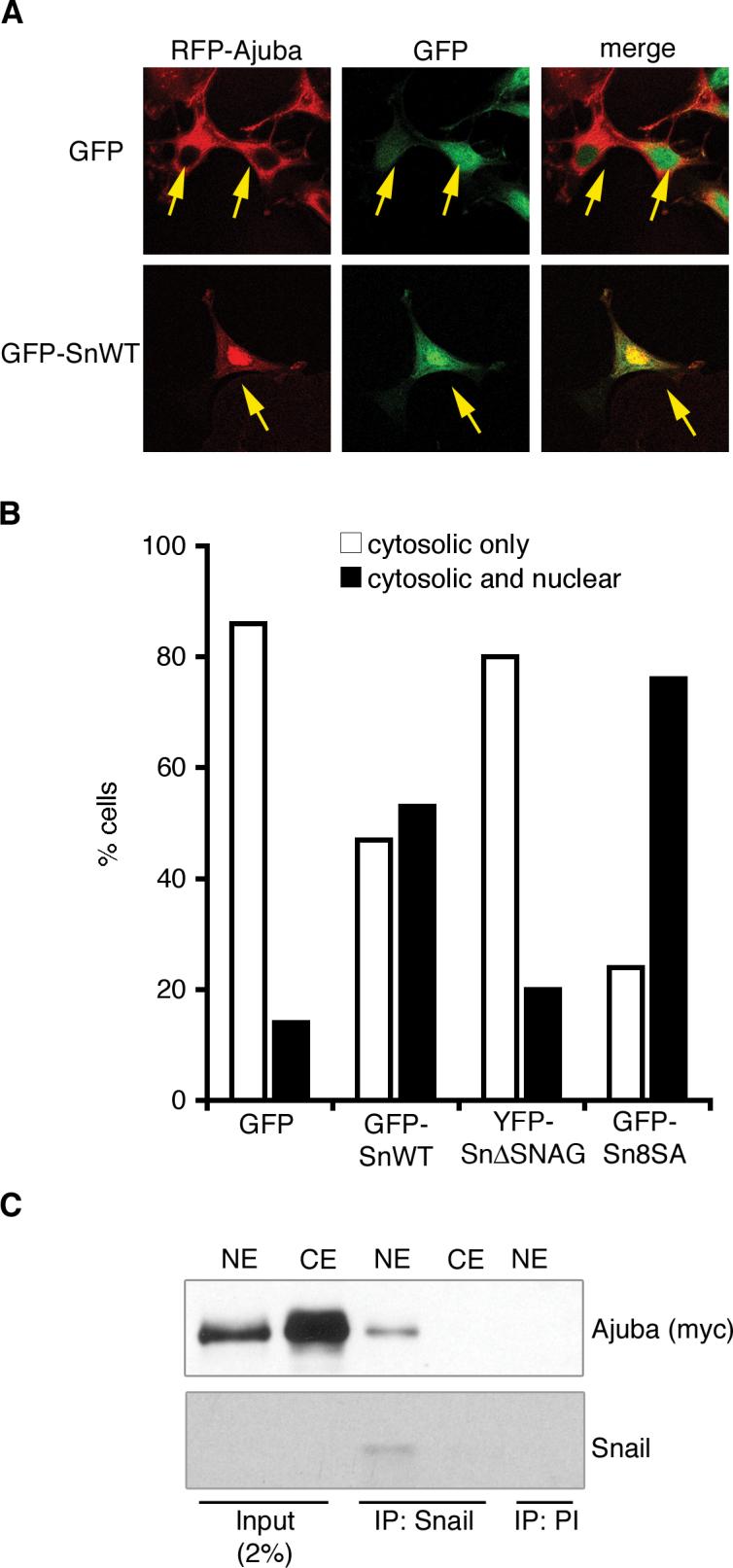
A. Confocal immunofluorescence analysis of MCF-7 cells co-transfected with RFP-Ajuba and GFP or GFP-Snail. Arrows indicate cotransfected cells. B. Quantification of immunofluorescence results. Shown is the percent of cells in which RFP-Ajuba is localized to the cytosol (white columns) or to the cytosol and the nucleus (black columns) in the presence of GFP, GFP-Snail, YFP-Snail.ΔSNAG or GFP-Snail8SA. For each sample, at least 100 cells were counted. The experiment was repeated three times with similar results. Shown is one representative experiment. C. Endogenous Snail was immunoprecipitated from nuclear extracts (NE) or cytosolic extracts (CE) of HaCaT cells stably expressing myc-Ajuba. Bound products were Western blotted for the presence of Ajuba (anti-myc) and Snail. Control immunoprecipitation was performed with rabbit preimmune sera (PI). Western blot of input controls is shown on the left.
In another approach to assess whether Ajuba and Snail interact in the nucleus, HaCaT cells stably expressing myc-Ajuba were fractionated into nuclear and cytosolic extracts. Endogenous Snail was immunoprecipitated from both fractions, and bound products Western blotted for the presence of Ajuba. Snail was only detected in the nuclear extract and, therein, Snail associated with Ajuba (Fig. 2C). This, in combination with immunofluorescence data, indicated that Ajuba accumulated in the nucleus upon expression of Snail, in a SNAG-dependent manner, and that Ajuba interacted with Snail in the nucleus.
Ajuba acts as a corepressor to Snail
A central function of Snail during EMT is transcriptional repression of epithelial adhesive receptors, such as E-cadherin (Batlle et al. 2000; Cano et al. 2000). Since Ajuba interacted with the SNAG-repressor domain of Snail, in the nucleus, we asked whether Ajuba affects the ability of Snail to repress transcription. Transient reporter assays were performed using a human E-cadherin promoter that contains all three Snail-binding E-boxes driving luciferase expression. MCF-7 cells were transfected with the reporter construct along with Snail and Ajuba alone or in combination. Both Snail and Ajuba alone were able to repress transcription from the E-cadherin promoter (Snail had a greater effect than Ajuba), but when co-expressed there was increased repression over either alone, suggesting that Ajuba cooperated with Snail to repress E-cadherin transcription (Fig. 3A). Mutation of all three E boxes in the E-cadherin promoter completely abolished Snail repression. The capacity of Ajuba to repress transcription was also significantly reduced upon mutation of the E-boxes, but some repression remained (data not shown). The ability of Ajuba alone to repress transcription could be due to trace levels of endogenous Snail in MCF-7 cells (data not shown), or Ajuba may also contribute to repression in a Snail-independent manner as has been observed for the interaction between other Ajuba LIM proteins and other nuclear transcriptional regulators (Sharp et al. 2004).
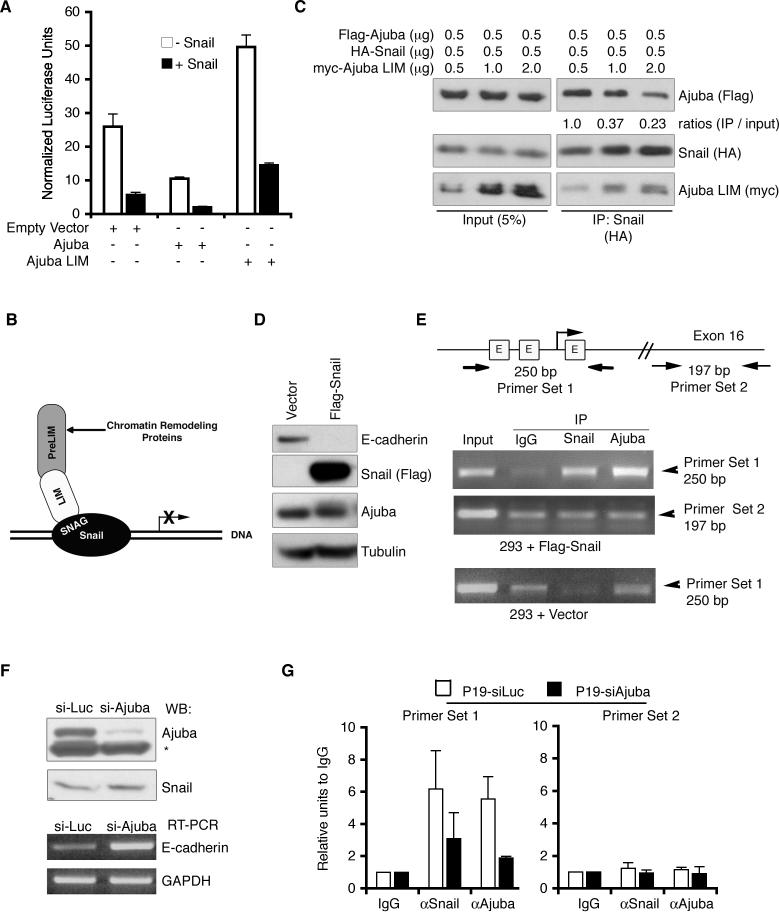
Figure 3. Ajuba is a corepressor of Snail.
A. Transient luciferase reporter assay using luciferase driven by the E-cadherin promoter. Constructs, as indicated, were expressed in MCF-7 cells and luciferase activity determined and normalized to β-gal activity (from co-transfected CMV-β–gal construct). Experiments were performed in triplicate. Shown are mean normalized luciferase values +/− standard deviations. B. Schematic of Ajuba interacting with Snail bound at a promoter to enhance Snail-dependent repression. C. HEK293 cells were co-transfected with Flag-Ajuba, HA-Snail, and myc-Ajuba LIM region as indicated. Snail was immunoprecipitated from lysates (anti-HA) and bound products Western blotted for full-length Ajuba (anti-Flag), Snail (anti-HA), and Ajuba LIM region (anti-myc). The amount of Ajuba immunoprecipitated relative to input was quantified by densitometry and controlled for the amount of Snail immunoprecipitated. The value for lane 4 was arbitrarily set to equal 1. D. Lysates of HEK293 cells stably transfected with empty vector or Flag-Snail were immunoblotted for E-cadherin, Flag, Ajuba, and Tubulin (as loading control). E. ChIPs were performed in HEK293 cells stably transfected with empty vector or Flag-Snail using antibodies to Snail and Ajuba. IgG was used as a control. PCRs were performed using primers flanking the three E-boxes (labeled E in schematic) in the human E-cadherin promoter (primer set 1) or flanking a region of Exon 16 (primer set 2). F. P19 cells were stably transfected with siRNA constructs targeting luciferase (Luc) or Ajuba. Top panel: lysates were immunoblotted for presence of Ajuba or Snail (* marks non-specific band). Bottom panel: RT-PCR was performed to detect E-cadherin levels. GAPDH is a loading control. G. ChIPs were performed in P19-siLuc (white bars) and P19-siAjuba (black bars) cells using antibodies to Snail and Ajuba. IgG was used as a control. Quantitative PCR was performed using primers flanking the E-boxes of the mouse E-cadherin promoter (primer set 1) or flanking a region of Exon 15 (primer set 2).
The LIM and preLIM regions of Ajuba often function to bring together distinct targets for a common cellular purpose (Marie et al. 2003; Pratt et al. 2005). Since Ajuba appears to act as a Snail corepressor, via a LIM region interaction with Snail (Fig. 1C), this leaves the preLIM region available to potentially coordinate assembly of a chromatin repressor complex (see Fig. 3B). If true, then expression of the LIM region alone might interfere with the interaction between endogenous full-length Ajuba and Snail, and thus inhibit Snail-dependent transcriptional repression. To test this, HEK293 cells were cotransfected with fixed and equal amounts of Ajuba and Snail, and increasing amounts of Ajuba LIM region. Snail was immunoprecipitated, and bound products Western blotted for the presence of full-length Ajuba. As the amount of Ajuba LIM region transfected increased, the interaction of Snail with full-length Ajuba decreased (Fig. 3C), suggesting that the LIM region indeed blocked the Snail-Ajuba interaction. When MCF-7 cells were transfected with the LIM region in the presence or absence of Snail, E-cadherin transcription was increased (i.e., “de-repressed”) compared to control cells (Fig. 3A). These results suggested that it was the interaction of full-length LIM proteins with Snail that contributes to their ability to repress Snail-dependent transcription.
If Ajuba functions as a Snail co-repressor, it should be present at promoters of Snail repressed genes in a Snail-dependent manner. To test this, Ajuba chromatin immunoprecipitations (ChIP) were performed in the presence or absence of Snail, using the endogenous E-cadherin promoter as the target sequence. Expression of Flag-Snail in HEK293 human epithelial cells resulted in decreased E-cadherin levels, indicating that Snail was active (Fig. 3D). Nuclear chromatin preparations from these and control cells were immunoprecipitated with IgG (control) or antibodies to Snail or Ajuba and PCR performed for the E-cadherin promoter using primers that flanked the three E-boxes to which Snail binds. Snail was present on the E-cadherin promoter in cells overexpressing Flag-Snail as shown by Snail ChIP (Fig. 3E). Importantly, compared to the IgG control, endogenous Ajuba was specifically detected on the E-cadherin promoter, but only in cells expressing functional Snail (Fig. 3E). As a ChIP control, PCR was performed with primers that amplify a region of Exon 16 of the human E-cadherin gene, well outside the promoter region. Neither Snail nor Ajuba bound this region when compared to the nonspecific control IgG ChIP (Fig. 3E). This result demonstrated that Ajuba was present in the nucleus on the promoter of a gene physiologically repressed by Snail, but only in the presence of Snail.
To determine whether Ajuba was required for Snail-mediated E-cadherin transcriptional repression, Ajuba levels were stably reduced in P19 mouse embryonal carcinoma cells using siRNA. Ajuba knockdown in P19 cells resulted in increased levels of E-cadherin mRNA and protein levels, without any change in Snail protein level (Fig. 3F and data not shown). When ChIP experiments were performed on control siRNA and Ajuba siRNA P19 cell lines, the amount of both Snail and Ajuba detected on the mouse E-cadherin promoter was decreased in Ajuba RNAi P19 cells (Fig. 3G). Thus reduction of one of three mouse Ajuba LIM proteins affected Snail-mediated repression, indicating that Ajuba LIM proteins likely contribute to Snail-mediated repression of E-cadherin transcription in cells.
Gain- and loss-of-function of Ajuba LIM proteins phenocopies that of Snail proteins in Xenopus neural crest development
Snail and Slug have well-characterized roles as transcriptional repressors in Xenopus neural crest development, a developmental EMT process (LaBonne and Bronner-Fraser 1998; Carl et al. 1999; LaBonne and Bronner-Fraser 2000; Aybar et al. 2003). We utilized this model system to determine whether the interaction between Ajuba LIM proteins and Snail proteins is biologically relevant in vivo. We confirmed that mouse Ajuba LIM proteins co-immunoprecipitated with XSnail (Fig. 6A, and data not shown). One cell of the two-cell stage Xenopus embryo was injected with mAjuba, mLIMD1 or mWTIP mRNA. This introduces the mRNA into one bilateral half of the embryo while the other half serves as an internal control. Embryos were co-injected with β-gal mRNA to track the injected regions. The embryos were fixed and X-Gal stained at stage 18/19 and in situ hybridization was performed for markers of neural crest: Slug, Twist, and FoxD3. Overexpression of mAjuba, mLIMD1, or mWTIP, like overexpression of XSnail and XSlug, resulted in marked expansion of neural crest area on the injected side of the embryos as detected by all three markers (Fig. 4A and 4B and data not shown). Therefore, mAjuba LIM proteins phenocopied the effects of XSnail and XSlug to increase neural crest area.
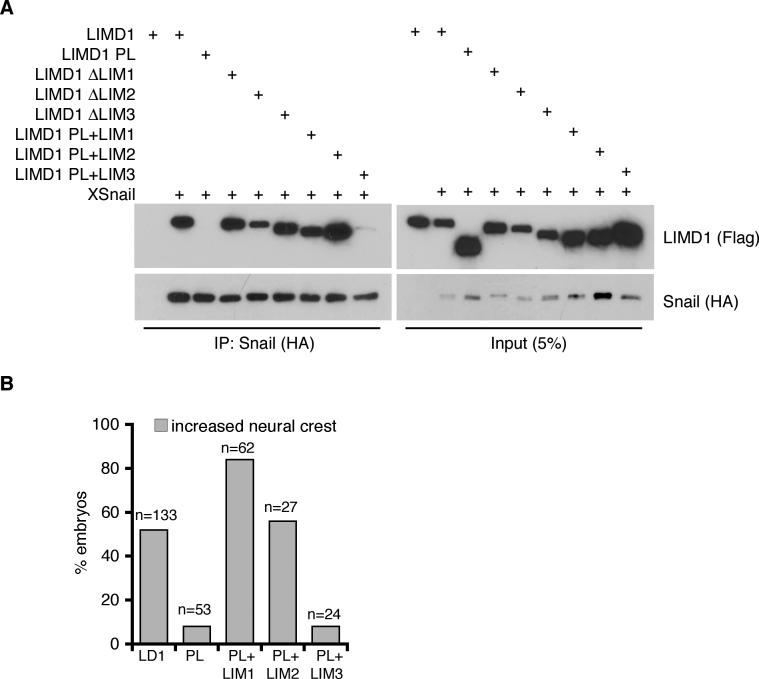
Figure 6. Biochemical and functional mapping of LIM domain-Snail interaction.
A. HEK293 cells were cotransfected with Flag-tagged mLIMD1 constructs and HA-XSnail as shown. Snail was immunoprecipitated from lysates (anti-HA) and bound products Western blotted for the presence of LIMD1 isoforms (Flag) and Snail (HA). Control Western blot of lysate is on right. B. Graph displaying the percent of embryos with increased neural crest on injected side by Slug in situ hybridization following injection of mLIMD1 (LD1), mLIMD1 PreLIM (PL), PL+LIM1, PL+LIM2, or PL+LIM3 as shown. The total number of embryos injected is shown over each column (n).
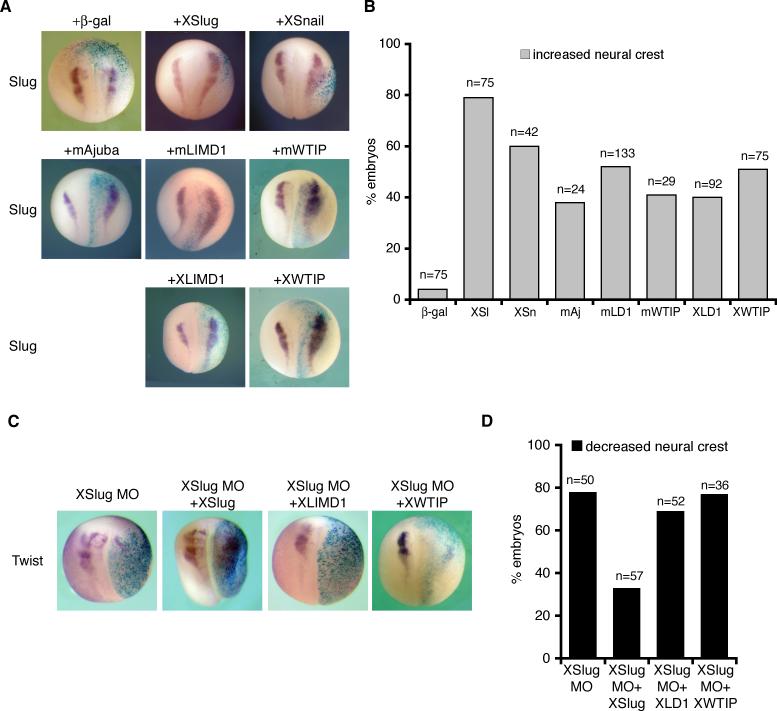
Figure 4. Ajuba LIM protein expression in Xenopus embryos enhances neural crest development in a Slug-dependent manner.
A. X. laevis embryos co-injected with β-gal and either XSlug, XSnail, mAjuba, mLIMD1, mWTIP, XLIMD1, or XWTIP capped mRNAs were fixed at stage 18 and in situ hybridization performed for XSlug. B. Graph displaying the percent of embryos with increased neural crest on the injected side (by Slug in situ hybridization). The total number of embryos injected is shown over each column (n). C. X. laevis embryos were co-injected with β-gal and the XSlug MO alone or in combination with XLIMD1, XWTIP, or XSlug mRNA, and in situ hybridization for Twist performed. D. Graph displaying the percent of embryos with decreased neural crest on the injected side (by Twist in situ hybridization). The total number of embryos injected is shown over each column (n).
We identified and cloned two Xenopus orthologs of Ajuba LIM proteins – XLIMD1 (GenBANK Accession DQ913740) and XWTIP (GenBANK Accession EU257484) (supplemental Figs. 1-3). The endogenous expression patterns of XLIMD1 and XWTIP during Xenopus development were determined and largely overlapped with that of XSnail and XSlug in premigratory and migratory neural crest (supplemental Fig. 4), consistent with a physiological role in the development of these tissues. We confirmed that XLIMD1 co-immunoprecipitated with XSnail (data not shown). Embryos were then injected with XLIMD1 or XWTIP mRNA. As with overexpression of the mouse proteins, expression of XLIMD1 or XWTIP also resulted in increased neural crest on the injected side (Fig. 4A and B).
To determine if the capacity of Xenopus Ajuba LIM proteins to influence neural crest development was dependent on Snail or Slug, we made use of an antisense morpholino (MO) to block translation of Slug in Xenopus (Zhang et al. 2006). XSlug MO inhibited neural crest development, and this effect was rescued by co-expression with XSlug mRNA (Fig. 4C and D). Neither XLIMD1 nor XWTIP co-expression was sufficient to enhance neural crest development in the absence of XSlug, however (Fig. 4C and D). Therefore, overexpression of Xenopus Ajuba LIM proteins resulted in an expansion of neural crest territory in a manner dependent on XSlug expression.
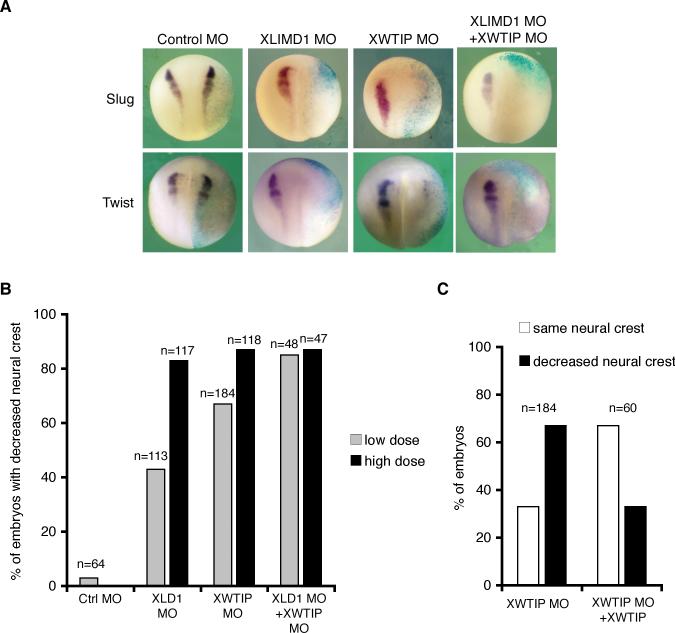
In another approach, we designed morpholinos (MOs) to XLIMD1 and XWTIP. Injection of either MO alone reduced or eliminated neural crest marker staining on the injected half of the embryo in a dose-dependent manner (Fig. 5A and B). XLIMD1 and XWTIP MOs were specific as when co-injected with mRNA for either XLIMD1 or XWTIP mRNA neural crest loss was rescued to an extent comparable to the XSlug mRNA rescue of XSlug MO (Fig. 5C). In sum, overexpression or knockdown of Ajuba LIM proteins phenocopied similar manipulations of Snail and Slug in Xenopus neural crest development.
Figure 5. Depletion of XLIMD1 or XWTIP blocks neural crest development in Xenopus.
A. X. laevis embryos were co-injected with β-gal and control MO, XLIMD1 MO (20ng), XWTIP MO (10ng), or a combination of XLIMD1 and XWTIP MOs. Embryos were fixed at stage 18−19, and in situ hybridization for XSlug or XTwist performed. B. Graph displaying percent of embryos with decreased neural crest on injected side by Slug or Twist in situ hybridization following injection of low dose (gray columns; 10ng XLIMD1 MO, 5ng XWTIP MO) or high dose (black columns; 20ng XLIMD1 MO, 10ng XWTIP MO) of morpholinos. The total number of embryos injected is shown over each column (n). C. X. laevis embryos were co-injected with β-gal, the XWTIP MO (5ng) and XWTIP capped mRNA as shown. Black columns indicate the percent of embryos with decreased neural crest on the injected side (by Slug or Twist in situ) and white columns indicate the percent of embryos where neural crest was the same on the injected and uninjected sides. The total number of embryos injected is shown over each set of columns (n).
The ability of Ajuba LIM proteins to interact with Snail is required for their capacity to enhance neural crest development
A number of nuclear roles for Ajuba family LIM proteins have been identified and include regulation of cell proliferation (Kanungo et al. 2000), cell cycle progression (Hirota et al. 2003; Sharp et al. 2004), and differentiation (Kanungo et al. 2000). Often, the LIM region, through either single or multiple LIM domains, mediates these effects. We sought to identify whether specific LIM domain(s) mediated the interaction with Snail family members, and correlate this with Snail/Slug biological responses (e.g., neural crest development). Coimmunoprecipitation experiments in HEK293 cells with Flag-tagged mLIMD1 LIM domain mutants and HA-tagged Snail revealed that LIM1 and LIM2, but not LIM3, mediated the interaction with Snail (Fig. 6A). To determine if these biochemical associations correlated with the ability to enhance Xenopus neural crest development, mRNA of full length LIMD1 (positive control), preLIM region (negative control), or of the preLIM region in combination with either LIM1, LIM2, or LIM3 was overexpressed in Xenopus embryos and neural crest development assessed. Mutants containing LIM1 or LIM2 resulted in neural crest expansion as well as, or better than, full length LIMD1 (Fig. 6B), while neither the preLIM region alone (no LIM domains) or preLIM + LIM3 affected neural crest development (Fig. 6B). Thus, the biochemical mapping of LIM domain interaction with Snail directly correlated with the ability to expand Xenopus neural crest.
Ajuba LIM proteins affect neural crest through modulation of cell survival and border territory identity, but not proliferation
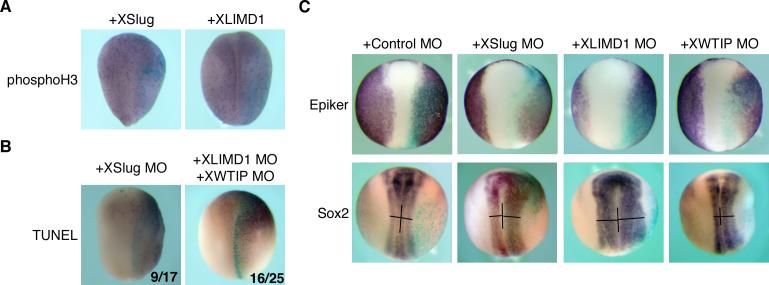
Snail and Slug have been described to affect cell fate determination, proliferation, survival, and migration in different tissues or cellular contexts (Barrallo-Gimeno and Nieto 2005). Therefore, we sought to determine whether Ajuba LIM proteins and Snail proteins affected neural crest development through the same pathways. Previously, in mouse embryonal carcinoma cells, overexpression of Ajuba resulted in increased proliferation. Structure-function studies revealed that LIM domains 1 or 2 affected cellular proliferation, while expression of LIM domain 3 affected cellular differentiation (Kanungo et al. 2000). When Xenopus embryos were injected with XLIMD1 or XWTIP and stained for phosphohistone-H3 (marker of proliferation) no significant difference was observed (Fig. 7A). Although different from results in P19 cells, this result was similar to the effects of XSlug. XSlug did not affect proliferation in Xenopus (Fig. 7A) and cellular proliferation is not required for Snail or Slug-mediated increase in neural crest (Aybar et al. 2003). To assess whether Ajuba LIM proteins influenced survival of Xenopus cells, in vivo, as Slug does (Tribulo et al. 2004; Zhang et al. 2006), embryos were injected with either XSlug MO or XLIMD1 + XWTIP MOs. Both conditions resulted in increased numbers of apoptotic cells on the injected side, as detected by TUNEL staining (Fig. 7B), suggesting that Ajuba LIM proteins, like Snail/Slug, function as anti-apoptotic factors during neural crest development.
Figure 7. Ajuba LIM proteins affect cell survival and border territories without affecting proliferation.
A. X. laevis embryos were injected with XSlug or XLIMD1 mRNA, fixed at stage 16 and immunohistochemistry performed for phosphohistone-H3. B. X. laevis embryos were injected with XSlug MO or combination of XLIMD1 +XWTIP MOs. Embryos were fixed at stage 16 and TUNEL staining performed. The number of embryos displaying increase TUNEL staining on the injected side over the total number of embryos analyzed is shown in the bottom right corner. C. X. laevis embryos were injected with MOs as shown, fixed at stage 16 and in situ hybridization performed for Epiker (epidermal) and Sox2 (neural).
Snail- or Slug-mediated increase in neural crest occurs with a concomitant decrease in neural plate area (LaBonne and Bronner-Fraser 2000; Aybar et al. 2003). We asked whether Ajuba LIM protein's effect upon neural crest, like that of Snail and Slug, influenced epidermal and neural fates at the border of these territories. The expression patterns of Sox2, a marker of the neural plate, and Epidermal keratin (Epiker), a marker of the non-neuronal epidermis were determined. At neurula stages, when we observed a dramatic decrease in neural crest upon XLIMD1 or XWTIP MO injection, we observed a consistent increase in Sox2 expression and disruption of Epiker expression at the border of these territories on the injected side of the embryo (Fig. 7C). This was similar to that observed following XSlug MO injection (Fig. 7C) in which reduction of neural crest development resulted in compensatory changes in epidermal and neural fates. In sum, the results mirrored those of Snail and Slug in that both border cell fates and cell survival were altered without affecting cell proliferation, consistent with Ajuba LIM proteins affecting neural crest development through a Snail/Slug-dependent pathway.
Ajuba LIM proteins are required as corepressors for Snail-induced neural crest development
To determine whether loss of LIM protein function specifically affected Snail/Slug-regulated neural crest development, we co-injected XSnail or XSlug mRNA along with XLIMD1 + XWTIP MOs and compared these embryos to embryos injected with XSnail or XSlug mRNA alone. When XSlug or XSnail mRNA was injected into embryos depleted of Ajuba LIM proteins there was no expansion of neural crest, in contrast to when these mRNAs were injected alone (see Fig. 4A and B). Rather, coinjection of XLIMD1+XWTIP MOs with XSlug or XSnail mRNA resulted in persistent inhibition of neural crest development (Fig. 8A and B and data not shown), suggesting that Ajuba LIM proteins are required for Snail/Slug function as transcriptional repressors.
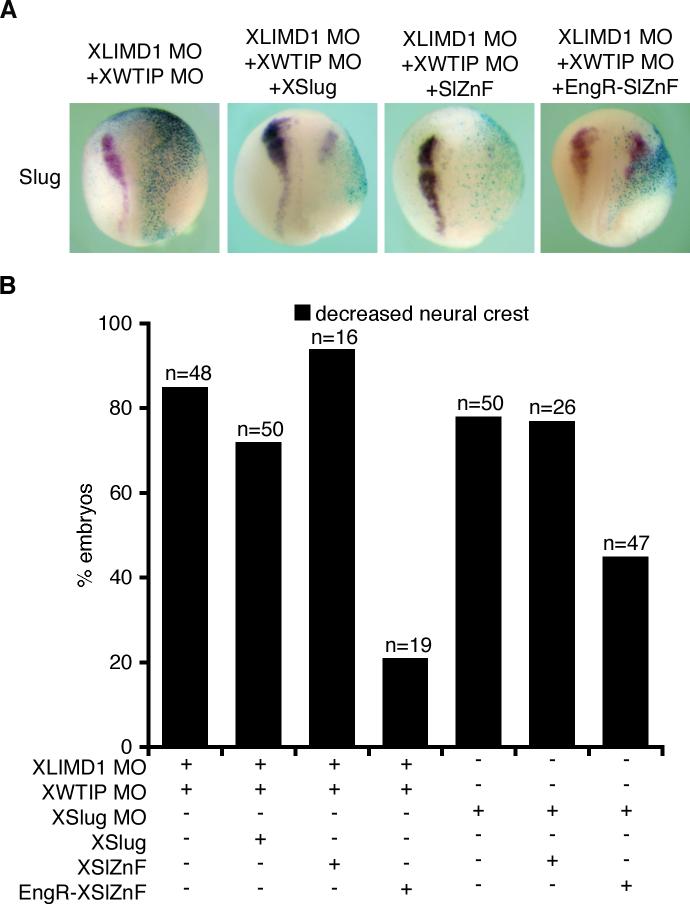
Figure 8. Depletion of both XLIMD1 and XWTIP blocks Slug repressor activity during neural crest development in Xenopus.
A. X. laevis embryos were co-injected with β-gal, XLIMD1 MO (5ng) and XWTIP MO (10ng) alone or in combination with XSlug, SlZnF, or EngR.SlZnF capped mRNA. Embryos were fixed at stage 18−19 and in situ hybridization performed for XSlug. B. Graph displaying the percent of embryos with decreased neural crest on the injected side by Slug in situs (following injection of MOs and RNAs as shown). The total number of embryos injected is shown over each column (n).
If the loss of Ajuba LIM proteins primarily blocks Xenopus neural crest development because of a loss of Snail/Slug repressor activity, then one might predict that XLIMD1 and XWTIP depletion would be rescued by a dominant Slug-Engrailed repressor (EngR.SlZnF) (LaBonne and Bronner-Fraser 2000). This mutant contains the DNA-binding Zinc finger region of Slug, but replaces the N-terminal repressor domain (including the Ajuba LIM protein-interacting SNAG domain) with the Drosophila Engrailed repressor domain. Embryos were co-injected with XSlug MO and either the Slug Zinc finger region alone (SlZnF) or the EngR.SlZnF mRNA. The XSlug MO block in neural crest development was rescued by co-expression of EngR.SlZnF, but not SlZnF, as expected by previous reports (Fig. 8B and (LaBonne and Bronner-Fraser 2000)). Next, embryos were co-injected with XLIMD1+XWTIP MOs and either SlZnF or EngR.SlZnF mRNA. The block in neural crest development that resulted from XLIMD1 and XWTIP depletion was not affected by SlZnF expression but was dramatically rescued by EngR.SlZnF (Fig 8A and B). This result is consistent with a primary role for Ajuba LIM proteins in neural crest development as Snail/Slug co-repressors.
Discussion
Transcriptional repression by Snail family members contributes to many biological and cellular processes, including Xenopus neural crest development, by affecting cellular differentiation, survival, and migration (Carl et al. 1999; LaBonne and Bronner-Fraser 2000; Aybar et al. 2003; Tribulo et al. 2004; Zhang et al. 2006). An N-terminal SNAG domain is both necessary and sufficient for Snail/Slug-mediated repression, but the precise mechanisms regulating repression in different tissues and organisms remain unclear. Here, we show that Ajuba family LIM proteins, Ajuba, LIMD1, and WTIP, specifically function as Snail/Slug corepressors in mammalian cells as well as in vivo to regulate Xenopus neural crest development. In support of this conclusion; 1) Ajuba LIM proteins interact predominately with the SNAG domain of Snail/Slug in cells and accumulate in the nucleus in a SNAG-dependent manner. 2) Ajuba LIM proteins contribute to Snail-dependent repression of E-cadherin transcription in cells. 3) Ajuba LIM proteins are present on endogenous promoters of Snail-regulated genes (e.g. E-cadherin), but only in the presence of Snail. 4) Expression of Ajuba LIM proteins in Xenopus embryos mimics expression of Snail/Slug in that they both enhance neural crest development, and this effect of Ajuba LIM proteins requires Slug expression. 6) Only isoforms of LIMD1 capable of interacting with Snail cause increased neural crest in Xenopus. 7) Depletion of Ajuba LIM proteins in Xenopus embryos phenocopies depletion of Slug with both resulting in a block or inhibition of neural crest development. 8) The effect of Ajuba LIM proteins upon Xenopus neural crest development occurs via mechanisms similar to those regulated by Snail/Slug. Upon loss of either, there is a compensatory increase in Sox2 (neural) expression in border territories and a decrease in cell survival, with no change in proliferation. 9) The block in Xenopus neural crest development upon depletion of Ajuba LIM proteins was not rescued by concurrent Slug expression but can be rescued by the dominant repressor protein, EngR.SlZnF.
Although Ajuba LIM proteins play a number of roles in cells that could potentially affect neural crest development, our evidence strongly supports a major role as Snail/Slug corepressors. Throughout, gain- and loss-of function experiments of Ajuba LIM proteins consistently phenocopy Snail and Slug. It is possible that Ajuba LIM proteins directly or indirectly (through affecting a non-Snail/Slug-mediated neural crest pathway) affect the expression of Snail or Slug, and this could have resulted in similar phenotypes. Mapping experiments, however, revealed a direct correlation between the ability of LIMD1 to interact with Snail and its capacity to enhance neural crest development when overexpressed. Moreover, if influencing expression of Snail/Slug is a role for Ajuba LIM proteins, we would expect expression of Snail or Slug to rescue the loss of Ajuba LIM proteins expression in Xenopus. They do not rescue that loss, but the EngR.SlZnF fusion protein, which eliminates a dependence on SNAG interactions, does. While we cannot rule out the possibility that other functions of Ajuba LIM proteins may also contribute to neural crest development, these results demonstrate that the Snail/Slug corepressor function of Ajuba LIM proteins is likely their primary role in this process.
We propose a model in which Ajuba LIM proteins bind nuclear Snail on specific gene promoters through an interaction of the LIM region (LIM1 and/or 2) with the SNAG domain of Snail. LIM domains do not directly bind DNA, but LIM proteins have been shown to regulate gene expression through direct LIM domain interaction with known transcription factors (Zhao et al. 1999; Sharp et al. 2004; Srichai et al. 2004; Guo et al. 2006). In most cases, the LIM region directs the interaction with the DNA binding protein. Accumulated data on Ajuba function have shown that the LIM and preLIM regions often interact with distinct targets to bring together proteins that contribute to common cellular functions (Marie et al. 2003; Pratt et al. 2005). We hypothesize that the preLIM regions are important for corepressor activity, perhaps through interactions with repressor complex proteins (see Fig. 3B). This model is based, in part, on evidence that overexpression of the LIM region, which mediates the Snail interaction, blocks or inhibits association of full-length Ajuba and Snail in cells, inhibits Snail-dependent repression of E-cadherin transcription in transient assays, and inhibits neural crest development in vivo (unpublished data). It will be critical to define the specific role(s) of the preLIM region in Snail/Slug-mediated repression.
All three Ajuba LIM proteins interact with Snail family members, suggesting that they serve redundant functions. The potential for compensation between Ajuba LIM proteins is apparent in mice as we do not observe overt developmental pathologies in Ajuba−/−, Limd1−/− or Ajuba−/−;Limd1−/− mice ((Marie et al. 2003; Feng et al. 2007) and unpublished data). In Xenopus, however, we observe a dose-dependent response to depletion of either XLIMD1 or XWTIP. While the precise reason why depletion of only one family member gives a significant phenotype in Xenopus neural crest is not known, it may be that this system is more sensitive to reduction in total amount of Ajuba LIM proteins. Indeed, we observe a greater effect on neural crest development when both XLIMD1 and XWTIP are depleted. In addition, Slug overexpression rescued loss of XLIMD1 alone, but not the loss of both proteins (Fig. 8 and unpublished data). The discrepancy between the murine and Xenopus neural crest phenotypes following loss of Ajuba LIM proteins is most likely explained by the fact that neither Snail nor Slug is required for early neural crest development in mice (Murray and Gridley 2006).
The precise hierarchy of transcription factors controlling Xenopus neural crest development is complicated by multiple feedback loops and cross talk. Ajuba LIM proteins, as corepressors to both Snail and Slug, likely impact at multiple points during this process. Through a temporal analysis of neural crest markers, we found that Ajuba LIM proteins are required early in neural crest development. Loss of either XLIMD1 or XWTIP results in disruption of Pax3 and Snail staining as early as stage 12 (supplemental Table 1). In developing neural crest, Snail expression has been observed as early as stage 11−12, preceding Slug expression (Linker et al. 2000), suggesting that this early function of Ajuba LIM proteins may be due to its interaction with Snail instead of Slug. However, we also observed that the Slug depletion caused a similar loss of early neural crest markers (supplemental Table 1). This suggests that Slug may function earlier than previously appreciated and may contribute in a feedback loop to maintain expression of Snail, as well as other neural crest markers. Further work will be necessary to fully understand the temporal relationships between Ajuba LIM proteins, Snail, Slug, and other factors that regulate neural crest development, as well as to distinguish between the Snail and Slug corepressor functions of the Ajuba LIM proteins during neural crest development.
We describe a nuclear role for Ajuba LIM proteins as corepressors of Snail that contribute to E-cadherin repression. Ajuba LIM proteins are also components of epithelial cell-cell junctions (Marie et al. 2003; Srichai et al. 2004). Ajuba influences the formation and/or stability of adherens junctions, possibly by coupling the E-cadherin adhesive complex to the actin cytoskeleton (Marie et al. 2003). Because of distinct roles for separate cellular pools, Ajuba may play an integral role in communicating between cell surface adhesive complexes and the nucleus to provide precise regulation of epithelial dynamics. While we do not have evidence that Ajuba LIM proteins alone initiate EMT, we observe Ajuba accumulation in the nucleus at Snail-regulated promoters in the presence of Snail. Ajuba LIM proteins, as Snail co-repressors, could then contribute to a feed forward loop to maintain the mesenchymal phenotype. By contributing to E-cadherin repression, Ajuba may also indirectly allow more Ajuba to be available for entry into the nucleus. Consistent with this, Ajuba is released from adherens junctions upon expression of Snail and subsequent transcriptional downregulation of E-cadherin (Jamora et al. 2005). We have also observed the release of Ajuba from epithelial junctions in HaCaT epithelial cells upon treatment with TGF-β, which induces EMT in these cells (unpublished data).
Whether Ajuba LIM proteins also play a role in mesenchymal to epithelial transitions (MET) is an interesting possibility. MET occurs physiologically during mammalian nephrogenesis (Kanwar et al. 2004) and pathologically as metastatic cells reform epithelial-like tumors at metastatic sites (Thiery and Sleeman 2006). Recently, Snail was shown to be capable of repressing its own transcription (Peiro et al. 2006), perhaps providing a mechanism to initiate MET. Ajuba, as a Snail corepressor, may enhance Snail-mediated repression of Snail transcription. As Snail expression decreases, Ajuba would exit the nucleus and E-cadherin transcription would resume. Ajuba could then be recruited to newly forming adherens junctions and contribute to epithelia formation.
Materials and Methods
Cell Culture, Transfection, Antibodies, and Plasmids
HEK293T, MCF-7, HaCaT, MDA-231, and P19 cells were grown in DMEM supplemented with 10% FBS, 2mM L-Glutamine, 100 U/ml penicillin, and 100μg/ml streptomycin. Stable lines of HaCaT cells expressing myc-Ajuba were selected and grown in G418, HEK293T cells expressing Snail were selected and grown in Zeocin. For details of the siRNA retroviral constructs and infection of P19 cells, see supplementary methods. All transfections were performed with Trans-IT LT-1 (Mirus) according to the manufacturer's protocol. Antibodies used were: Ajuba (Cell Signaling, and (Pratt et al. 2005)), LIMD1 (Feng et al. 2007), Snail (Santa Cruz), E-cadherin (Cell Signaling), and HA (Sigma). HRP conjugated antibodies to Flag and Myc tags were from Sigma. For all plasmid information, see supplemental methods.
Immunoprecipitation, Immunoblots, and Immunofluorescence
For IPs, cells were lysed in IP buffer (10mM Tris pH7.4, 150mM NaCl, 1mM EDTA 10mM NaF, 10% glycerol, 1% NP-40, protease inhibitors) and extracts clarified. Lysates were pre-cleared with protein A or G beads alone for 1 hour, then incubated overnight with antibody. Protein A or G beads were added for 1 hour, washed 4 times with IP buffer, and bound proteins eluted into SDS-PAGE sample buffer. For Flag IPs, M2 conjugated beads (Sigma) were used and bound proteins eluted by competition with Flag peptide. SDS-PAGE and immunoblots were performed using standard protocols. For nuclear and cytosolic extracts, HaCaT cells were lysed in hypotonic buffer, cell membranes disrupted by dounce homogenization, and nuclei pelleted by centrifugation. Supernatant was used as cytosolic extract. Nuclei were extracted in hypertonic lysis buffer and centrifuged again. Extracts were adjusted to isotonic and IPs performed. Immunofluorescence was performed as described (Marie et al. 2003). Images were taken on a Zeiss confocal microscope using LSM 550 software.
Luciferase Assay
MCF-7 cells were transiently transfected with an 0.12μg E-cadherin-luciferase reporter construct (Hajra et al. 2002), 0.12μg CMV-β-galactosidase and combinations of empty vector, mAjuba, mAjuba LIM, and mSnail to equal 1.2μg total DNA. Cells were lysed in Cell Culture Lysis Reagent (Promega), and lysate used in both a luminescent β-galactosidase detection assay (BD Biosciences) and luciferase assay. All samples were read on a luminometer and values shown were obtained by normalizing luciferase values to β-galactosidase activity.
Chromatin Immunoprecipitation and RT-PCR
For ChIPs, cells were grown to 70−90% confluency, fixed in 1% formaldehyde, and harvested, and ChIPs performed using the EZ-CHIP kit (Upstate) according to the manufacturer's protocol. PCR or qPCR was performed on immunoprecipitated DNA. For RT-PCR, RNA was isolated using RNeasy kit (Qiagen) and cDNA synthesized with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. All primer sequences are available in supplementary methods.
Xenopus embryos, injections, in situ hybridizations, TUNEL and phosphohistone-H3 staining
X. laevis albino embryos were obtained through in vitro fertilization and raised as described (Kroll et al. 1998). Embryos were staged following Nieuwkoop and Faber (Nieuwkoop and Faber 1967). For injections, capped mRNA was transcribed in vitro (mMessage mMachine kit, Ambion). RNAs (250−500pg) were coinjected into one blastomere of two-cell stage embryos with β-galactosidase mRNA (50pg). Translation blocking MOs to XSlug (Zhang et al. 2006), XLIMD1, and XWTIP as well as a control MO were purchased from Gene Tools, Inc (see supplementary methods for sequences). Embryos were raised until indicated stages, fixed 1 hour in MEMFA, X-gal stained and analyzed by in situ hybridizations as described (Harland 1991). Probes were generated by in vitro transcription with digoxigenin-11-UTP (Roche) and were detected with alkaline phosphatase (AP)-conjugated anti-digoxigenin antibodies (Roche) with Nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP; Roche). For probe information, see supplemental methods. Whole mount TUNEL staining was performed as described (Tribulo et al. 2004). Rabbit polyclonal anti-phosphohistone-H3 antibody (Upstate) was used for immunohistochemistry as described (Bellmeyer et al. 2003). All images are shown as dorsal views with the anterior end oriented up. X-Gal staining (indicating the injected region of the embryos) has been oriented to the right.
Acknowledgments
EML is an HHMI predoctoral fellow. This work was supported by grants NIH GM66815-01, the March of Dimes (#1-FY06-374), and the American Cancer Society (RSG-06-148-01-DDC) to KLK; NIH Core CA10815, CA92088, CA095561, DAMD17-96-1-6141, 17-02-1-0631, the Irving A. Hansen Memorial Foundation, the Susan G. Komen Breast Cancer Foundation, and the Emerald Foundation to FJR; and NIH CA75315, CA106496, GM080673 and the Washington University/Pfizer Biomedical Research Program to GDL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130(3):483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K, Peng H, Hou Z, Fredericks WJ, Goyal RK, Langer EM, Longmore GD, Rauscher FJ., 3rd The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res. 2007;67(19):9097–9106. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4(6):827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21(20):3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense slug RNA. Dev Biol. 1999;213(1):101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21(23):8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- del Barrio MG, Nieto MA. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129(7):1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C, Garcia de Herreros A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol. 2003;23(14):5078–5089. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhao H, Luderer HF, Epple H, Faccio R, Ross FP, Teitelbaum SL, Longmore GD. The LIM protein, Limd1, regulates AP-1 activation through an interaction with Traf6 to influence osteoclast development. J Biol Chem. 2007;282(1):39–48. doi: 10.1074/jbc.M607399200. [DOI] [PubMed] [Google Scholar]
- Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16(11):6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Sallis RE, Greenall A, Petit MM, Jansen E, Young L, Van de Ven WJ, Sharrocks AD. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol Cell Biol. 2006;26(12):4529–4538. doi: 10.1128/MCB.01667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62(6):1613–1618. [PubMed] [Google Scholar]
- Hansen MD, Beckerle MC. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem. 2006;281(23):16178–16188. doi: 10.1074/jbc.M512771200. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT. Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol. 2000;20(14):5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Hu X, Ashraf SI, Small SJ, Ip YT. The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Dev Biol. 2004;269(2):411–420. doi: 10.1016/j.ydbio.2004.01.029. [DOI] [PubMed] [Google Scholar]
- Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114(5):585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172(5):771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116(Pt 10):1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- Jamora C, Lee P, Kocieniewski P, Azhar M, Hosokawa R, Chai Y, Fuchs E. A signaling pathway involving TGF-beta2 and snail in hair follicle morphogenesis. PLoS Biol. 2005;3(1):e11. doi: 10.1371/journal.pbio.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24(17):7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112(12):1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell. 2000;11(10):3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar YS, Wada J, Lin S, Danesh FR, Chugh SS, Yang Q, Banerjee T, Lomasney JW. Update of extracellular matrix, its receptors, and cell adhesion molecules in mammalian nephrogenesis. Am J Physiol Renal Physiol. 2004;286(2):F202–215. doi: 10.1152/ajprenal.00157.2003. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125(16):3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125(13):2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221(1):195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Linker C, Bronner-Fraser M, Mayor R. Relationship between gene expression domains of Xsnail, Xslug, and Xtwist and cell movement in the prospective neural crest of Xenopus. Dev Biol. 2000;224(2):215–225. doi: 10.1006/dbio.2000.9723. [DOI] [PubMed] [Google Scholar]
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, Moss SJ, Troyanovsky S, Attwell D, Longmore GD, Braga VM. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278(2):1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci U S A. 2006;103(27):10300–10304. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Scott IC, Cross JC. The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev Biol. 1998;199(1):150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. Embo J. 1998;17(23):7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264(5160):835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis. North-Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138(5):1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro S, Escriva M, Puig I, Barbera MJ, Dave N, Herranz N, Larriba MJ, Takkunen M, Franci C, Munoz A, Virtanen I, Baulida J, Garcia de Herreros A. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006;34(7):2077–2084. doi: 10.1093/nar/gkl141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276(29):27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Pratt SJ, Epple H, Ward M, Feng Y, Braga VM, Longmore GD. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J Cell Biol. 2005;168(5):813–824. doi: 10.1083/jcb.200406083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P, Kusewitt DF, Carver EA, Magnino F, Choi C, Gridley T, Hudson LG. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J Cell Physiol. 2005;202(3):858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- Sharp TV, Munoz F, Bourboulia D, Presneau N, Darai E, Wang HW, Cannon M, Butcher DN, Nicholson AG, Klein G, Imreh S, Boshoff C. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc Natl Acad Sci U S A. 2004;101(47):16531–16536. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srichai MB, Konieczkowski M, Padiyar A, Konieczkowski DJ, Mukherjee A, Hayden PS, Kamat S, El-Meanawy MA, Khan S, Mundel P, Lee SB, Bruggeman LA, Schelling JR, Sedor JR. A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J Biol Chem. 2004;279(14):14398–14408. doi: 10.1074/jbc.M314155200. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Sanchez SS, Mayor R. A balance between the anti-apoptotic activity of Slug and the apoptotic activity of msx1 is required for the proper development of the neural crest. Dev Biol. 2004;275(2):325–342. doi: 10.1016/j.ydbio.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18(10):1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zhang C, Carl TF, Trudeau ED, Simmet T, Klymkowsky MW. An NF-kappaB and slug regulatory loop active in early vertebrate mesoderm. PLoS ONE. 2006;1:e106. doi: 10.1371/journal.pone.0000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MK, Wang Y, Murphy K, Yi J, Beckerle MC, Gilmore TD. LIM domain-containing protein trip6 can act as a coactivator for the v-Rel transcription factor. Gene Expr. 1999;8(4):207–217. [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.