Abstract
Understanding the transmission dynamics of generalist pathogens that infect multiple host species is essential for their effective control. Only by identifying those host populations that are critical to the permanent maintenance of the pathogen, as opposed to populations in which outbreaks are the result of ‘spillover’ infections, can control measures be appropriately directed. Rabies virus is capable of infecting a wide range of host species, but in many ecosystems, particular variants circulate among only a limited range of potential host populations. The Serengeti ecosystem (in northwestern Tanzania) supports a complex community of wild carnivores that are threatened by generalist pathogens that also circulate in domestic dog populations surrounding the park boundaries. While the combined assemblage of host species appears capable of permanently maintaining rabies in the ecosystem, little is known about the patterns of circulation within and between these host populations. Here we use molecular phylogenetics to test whether distinct virus–host associations occur in this species-rich carnivore community. Our analysis identifies a single major variant belonging to the group of southern Africa canid-associated viruses (Africa 1b) to be circulating within this ecosystem, and no evidence for species-specific grouping. A statistical parsimony analysis of nucleoprotein and glycoprotein gene sequence data is consistent with both within- and between-species transmission events. While likely differential sampling effort between host species precludes a definitive inference, the results are most consistent with dogs comprising the reservoir of rabies and emphasize the importance of applying control efforts in dog populations.
Keywords: rabies, evolution, statistical parsimony, Serengeti
1. Introduction
Rabies virus (RABV), a prototype member of the genus Lyssavirus, family Rhabdoviridae, is a multi-host pathogen capable of infecting a wide range of species. The paradigm of rabies epidemiology is the compartmentalization of the circulating virus by species and geographical area, leading to the evolution of distinct virus variants that establish sustained transmission networks in a single species, the reservoir host (Rupprecht et al. 1991). However, this paradigm largely applies to areas with relatively low species diversity, and it has been suggested that in some areas, particularly in species-rich communities, multiple variants of the virus circulate in different host species (East et al. 2001) or multiple host species independently maintain infection of a single variant (Thomson & Meredith 1993; Bingham et al. 1999a,b).
It is generally considered that, as a result of the fatal outcome of the disease, maintenance host populations can only maintain the virus if they have specific demographic and ecological characteristics. For instance, species that are terrestrial rabies reservoirs tend to have high birth rates, which allow rapid population recovery from rabies-induced mortality (Wandeler et al. 1994). Host–virus adaptation has also been proposed as a mechanism for increased efficiency of transmission in maintenance hosts, for example, through high rates of salivary virus excretion (Blancou 1988a). Conversely, transmission to non-adapted ‘spillover’ hosts typically results in short-lived chains of transmission. Occasionally, cross-species transfers may lead to sustained transmission when a virus variant gains access to a novel host species with favourable ecological, genetic and behavioural characteristics (e.g. the species jump from dogs, Canis familiaris, to the European red fox, Vulpes vulpes, in the twentieth century; Anderson et al. 1981; Bourhy et al. 1999).
Evidence from epidemiological studies coupled with the isolation of a typically canid-associated African variant (Africa 1b) from the domestic dog, African wild dog (Lycaon pictus), bat-eared fox (Otocyon megalotis) and white-tailed mongoose (Ichneumia albicauda; Cleaveland & Dye 1995; Kissi et al. 1995; East et al. 2001) have suggested that domestic dogs may be the sole maintenance host of rabies in the Serengeti ecosystem. However, these conclusions were drawn from a limited range of epidemiological data, and several alternative hypotheses have been proposed for the maintenance of rabies in multi-host communities in Africa (Thomson & Meredith 1993; Bingham et al. 1999a,b; East et al. 2001). The question is important because multiple variants in distinct hosts would prevent effective disease control by targeting a single host population.
An atypical pattern of infection proposed to account for rabies maintenance involves an infectious healthy carrier state where animals actively shed virus in the saliva for prolonged periods, but remain clinically normal. In rare instances, naturally infected healthy dogs have been documented to excrete virus in saliva (Fekadu 1972; Aghomo et al. 1989), and non-lethal rabies infection has been suggested to occur in spotted hyaenas (Crocuta crocuta) in the Serengeti (East et al. 2001). In East et al.'s study (2001), hyaenas were deduced to maintain an avirulent variant based on detection of viral RNA in saliva of healthy animals by reverse transcriptase polymerase chain reaction (RT-PCR). Sequence analysis of these PCR products indicated that the presumed hyaena variant was phylogenetically more closely related to European and Middle Eastern RABVs than to African isolates.
Bingham et al. (1999a,b) suggested that a single variant may be maintained by multiple canine species (i.e. dogs and jackals (Canis mesomelas and Canis adustus)) in southern Africa through independent cycles, although other studies have indicated that jackals are unlikely to support infection independently of dogs (Cleaveland & Dye 1995; Rhodes et al. 1998). Similarly, bat-eared foxes, which are also infected by this variant (von Teichman et al. 1995; Sabeta et al. 2003), have been implicated as maintenance hosts in the Western Cape (Thomson & Meredith 1993).
High species diversity of wild carnivores in the 27 000 km2 Serengeti ecosystem and the lack of fencing between wildlife-protected areas and human settlements provide an ideal interface for testing the paradigm of compartmentalization of RABVs in a multi-host community. Compartmentalization has never been tested in a system with coexisting species that have been implicated elsewhere as maintenance hosts of rabies, such as bat-eared foxes and jackals (Thomson & Meredith 1993; Bingham et al. 1999b).
With additional samples and epidemiological data available from the Serengeti, and the application of phylogenetic analyses, we are now in a position to examine these alternative hypotheses more rigorously. We genetically characterized RABVs isolated from a range of species from the Serengeti and the surrounding areas to determine the phylogeographic relationships among Serengeti viruses and RABVs recovered elsewhere (i.e. Europe, Middle East and Africa), and identify viral variants that might signify distinct virus–host associations. In a second analysis, we examined the genealogic relationships among Serengeti viruses to infer and identify transmission routes. We employed a parsimony-based network construction procedure (Templeton et al. 1992) that has proven useful in hypothesis testing of intra- and interspecific transmission of HIV and human and simian T-cell leukaemia/lymphoma virus type I (Crandall 1995, 1996). The application of this method to RABV sequence data illustrates how genetic analysis can reveal elusive aspects of virus transmission in a complex ecosystem.
2. Material and methods
(a) Study samples and rabies diagnosis
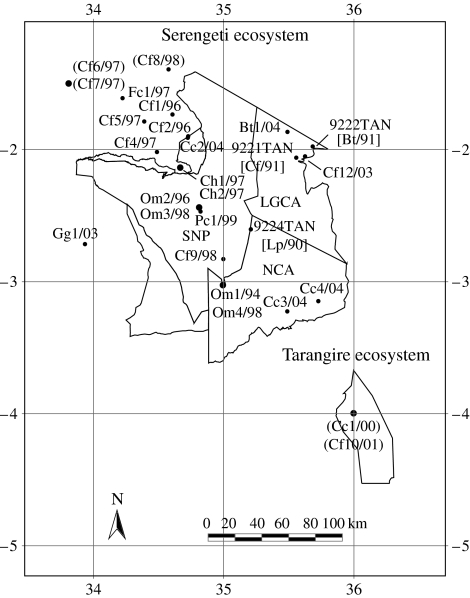
Twenty-four viruses obtained from a range of animal species in the Serengeti ecological region of northwestern Tanzania and Tarangire ecosystem (to the southeast of Serengeti) were included in this study (figure 1; for sample details see electronic supplementary material, S1). All the viruses were from animals diagnosed as rabies positive. For brains collected between 1994 and 2001, diagnostic tests and viral isolations were carried out at the Agence Française de Sécurité Sanitaire des Aliments (AFSSA), Malzéville, France using the fluorescent antibody test (FAT; Dean et al. 1996), inoculation of murine neuroblastoma cells and mouse inoculation (Barrat et al. 1988). Rabies diagnosis on more recent brain tissues was conducted at the Rabies Section of the Centers for Disease Control and Prevention (CDC), Atlanta, USA by FAT (Dean et al. 1996).
Figure 1.
Map of the Serengeti and Tarangire ecosystems showing the location where the field isolates originated (including three previously described viruses: 9221TAN; 9222TAN; and 9224TAN; Kissi et al. 1995). The precise sampling location of the isolates in round brackets is not known. The isolates are designated by a prefix indicating the species of origin (Bt, Bos taurus; Cc, Crocuta crocuta; Cf, Canis familiaris; Ch, Capra hircus; Fc, Felis catus; Gg, Genetta genetta; Om, Otocyon megalotis; Pc, Proteles cristatus), the isolate number and the year of collection. For isolates 9221TAN, 9222TAN and 9224TAN, the species of origin and the year of collection are indicated within square brackets (Lp, Lycaon pictus). SNP, Serengeti National Park; LGCA, Loliondo Game Control Area; NCA, Ngorongoro Conservation Area.
(b) RNA extraction, RT-PCR and nucleotide sequencing
Total RNA was extracted from infected brain material using the TRIzol method (Invitrogen, San Diego, CA) according to the manufacturer's recommendations. Reverse transcription of 11 isolates was performed at the Veterinary Laboratory Agency (VLA), Weybridge, Addlestone, Surrey, UK following methods of Heaton et al. (1997). RT-PCR of the other isolates and direct sequencing were performed at CDC using previously described methods (Sacramento et al. 1991) with primer sets for the regions encoding the nucleoprotein (N) and the central part of the ectodomain of the glycoprotein (G) published earlier (Smith 2002).
(c) Phylogenetic analyses
Sequence editing and translation to amino acid sequences were performed using BioEdit software v. 7.0.0 (Hall 1999). Multiple alignments were generated using the ClustalX package v. 1.83 (Jeanmougin et al. 1998), and sequence alignments were trimmed to include only complete non-stop codons. Prior to proceeding with phylogenetic analysis, we examined the alignments for the presence of recombination events using Worobey's informative sites test (Worobey 2001). No significant evidence of recombination was detected.
The evolutionary relationships among the Tanzanian isolates newly described in this article and selected representatives of African and European/Middle Eastern lineages of RABV (Kissi et al. 1995; Bourhy et al. 1999; Randall et al. 2004) were determined using Bayesian Markov chain Monte Carlo (MCMC) methods. The N gene was chosen because the N sequence of isolates is available for all four African lineages (Kissi et al. 1995). Bayesian reconstructions were conducted in MrBayes v. 3.0b4 (Ronquist & Huelsenbeck 2003). Two analyses were performed to check for any substantial sensitivity associated with fixing model parameters prior to analysis rather than estimating them as per MrBayes default settings. The first analysis specified the model of evolution and estimated parameters identified by the program ModelTest v. 3.7 (Posada & Crandall 1998) using Akaike Information Criterion (Sakamoto et al. 1986). The second analysis used the general time reversible (GTR) model with a proportion of invariable sites and a gamma-shaped distribution of rates across sites, (GTR+I+Γ; Yang et al. 1994) treating model parameters as unknown variables with uniform priors to be estimated in the analysis. Four MCMC chains with initial random starting trees without constraints were run for 1×107 generations with trees sampled every 100th generation, resulting in 1×105 sampled trees. To ensure that the chains had reached stationarity, log-likelihood values for sampling points were plotted against generation time, and the convergence diagnostic was examined. The first 25 000 trees were discarded as the burn-in phase, and the remaining trees were used to estimate consensus phylograms and Bayesian posterior probabilities. Posterior probability values of 0.95 or greater were considered significant. Graphical representations of the trees were generated with the program TreeView v. 1.6.6 (Page 1996).
In order to generate the highest possible degree of resolution for the Tanzanian sequence set, a phylogenetic tree was constructed using a Bayesian MCMC algorithm implemented in the BEAST program v. 1.4.1 (Drummond & Rambaut 2006) that permitted the year of virus isolation to be explicitly incorporated into the analysis. Analysis was performed assuming a constant viral population size and a relaxed molecular clock model (Drummond et al. 2002), which allows rates to vary over branches in an exponentially autocorrelated fashion. MCMC analysis chains were run for 1×107 generations with trees sampled every 1000th generation using the SRD06 substitution model (Shapiro et al. 2006). The pre-burn-in was set at 10 000 steps. BEAST output was assessed using the Tracer program (Drummond & Rambaut 2006).
Statistical parsimony (SP) networks were constructed to estimate the genealogical intra- and interspecific relationships among the Tanzanian N gene sequences included in the previously described analyses. An SP analysis was also performed on G gene data available for 15 Tanzanian isolates, a number of which were identical over a 398 bp region. Analyses were performed using the TCS software v. 1.20 (Clement et al. 2000), which implements the procedure of SP developed by Templeton et al. (1992), a population-based method for reconstructing historical relationships among gene sequences. The SP approach is based on the parsimony criterion as defined by Templeton et al. (1992) with a statistical procedure to evaluate the limits of the parsimony assumption, that is the probability that a nucleotide difference between two variant sequences is caused by a single mutational event (the parsimonious state) and not by multiple mutational events at a single site (the non-parsimonious state). An absolute distance matrix is calculated for all pairwise comparisons of sequences. The probability of parsimony is calculated for pairwise differences until the probability exceeds 95% using the model developed by Templeton et al. (1992; eqns (6)–(8)). The number of mutational differences just before this 95% cut-off point represents the maximum number of mutational steps between pairs of sequences justified by the parsimony criterion. The TCS program then connects the sequences into networks with the number of mutational steps connecting two sequences indicated by the lines connecting sequences.
3. Results
The majority-rule consensus tree of partial N gene sequences for RABVs from Tanzania compared with isolates recovered from other locations obtained after selecting and fitting an appropriate nucleotide substitution model (Posada & Crandall 1998) is shown in figure 2. The same topology was obtained when reconstruction was performed by Bayesian analysis with vague priors. The phylogeny revealed clear phylogeographic structure with all of the major clades supported by posterior probabilities greater than 0.95. All Tanzanian isolates grouped together and fell into the Africa 1b group of canid-associated viruses. Within the Tanzanian group (figure 2, inset), which included viruses isolated from domestic and wild species from the Serengeti and Tarangire ecosystems, there was little resolution as reflected in the low posterior probabilities (note that most of the nodes have no posterior probabilities associated with them as only nodes with values greater than or equal to 0.95 are labelled). However, a number of smaller groups (posterior probabilities≥0.95) were evident that corresponded to viruses recovered from outbreaks linked in time.
Figure 2.
Majority-rule consensus tree of nucleoprotein gene sequences (1158 bp, 386 deduced amino acids, nucleotide positions 263–1420 on the SAD B19 genome; Conzelmann et al. 1990) for RABVs from Tanzania (Serengeti and Tarangire ecosystems) compared with isolates from other areas of Africa, Europe and the Middle East recovered with Bayesian phylogenetics under the GTR+invariant sites (I)+gamma shape (Γ) model of evolution (Yang et al. 1994; base frequencies=0.2911, 0.2132, 0.2396 and 0.2561; nucleotide substitution rates of the GTR rate matrix=1.4665, 6.5059, 0.7601, 0.1703 and 10.8510; I=0.3530; and Γ=0.7587). The tree is rooted with isolate 1500AFS defined as the out-group, representative of the lineage Africa 3 (Kissi et al. 1995). Numbers on branches indicate Bayesian bootstrap values and are shown next to key nodes only. For a detailed phylogenetic tree of the Tanzanian viruses, see inset (and methods described in main text). Only posterior probabilities greater than or equal to 0.95 are shown. The scales indicate branch length expressed as the expected number of substitutions per site. Isolates described in this study are designated by a prefix indicating the species from which virus was recovered (Bt, Bos taurus; Cc, Crocuta crocuta; Cf, Canis familiaris; Ch, Capra hircus; Fc, Felis catus; Gg, Genetta genetta; Om, Otocyon megalotis; Pc, Proteles cristatus), the isolate number and the year of collection. Strain names are given for published isolates (Kissi et al. 1995; Bourhy et al. 1999; Randall et al. 2004) and the species of origin is indicated within square brackets (Cp, Cynictis penicillata; Cs, Canis simensis; Hs, Homo sapiens; Lp, Lycaon pictus; Np, Nyctereutes procyonoides; Vv, Vulpes vulpes).
The Tanzanian isolates showed between 0.1 and 3.3% (average 1.6%) nucleotide and between 0.0 and 2.6% (average 0.7%) amino acid sequence divergence. Maximum nucleotide diversity was between a virus recovered from an African wild dog (9224TAN) in 1990, the oldest Serengeti isolate, and a virus recovered from a spotted hyaena in 2004 (nucleotide and amino acid divergences were 3.3 and 2.1%, respectively). The BEAST analysis generated an estimated rate of change for the molecular clock of 0.0013 nucleotide substitutions per year (95% CIs 0.0005–0.0021) for the gene, and dated the most recent common ancestor to the sampled sequences to be from 1976 (95% CIs 1953–1989).
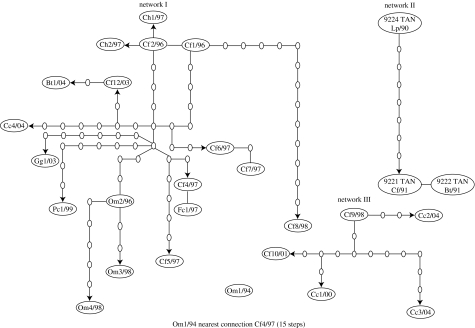
For the N gene dataset, parsimonious connections were justified (p≥0.95) among sequences differing by as many as 14 nucleotide substitutions. These sequences were connected into a single parsimony network (figure 3, network I and electronic supplementary material, S2), whereas other sequences formed independent networks (figure 3, networks II and III and electronic supplementary material, S3). Isolate Om1/94 could not be connected to any network.
Figure 3.
SP networks connecting the Tanzanian nucleoprotein gene sequences described in figure 2 (virus designations are the same as given in figure legend 2). Each branch represents a single mutational step (nucleotide substitution). The lengths of the connecting lines are not significant. Large ovals represent sequences, smaller ovals indicate nodes in the tree, which represent intermediate sequences not present in the sample. The arrows indicate temporal direction of evolutionary change.
For the G gene dataset, the SP procedure justified connections among sequences that differed by eight or fewer nucleotide substitutions. The resulting network is shown in figure 4 (see also electronic supplementary material, S4).
Figure 4.
Network of statistically supported relationships for the glycoprotein sequence data (398 bp, 132 deduced amino acids, nucleotide positions 3761–4158 on the SAD B19 genome; Conzelmann et al. 1990) available for 15 isolates described in figure 2 (virus designations are the same as given in figure legend 2) inferred using a SP approach. Asterisks indicate that identical genotypes were recovered from multiple animals.
4. Discussion
Our analysis strongly suggests that only a single Africa 1b virus variant circulates among Serengeti's domestic and wild mammal species, and cross-species transmission is a frequent event. These findings raise interesting questions about why communities with highly diverse species only support a single virus variant.
Overall, our phylogeny revealed site-specific rather than species-specific grouping, and the Tanzanian viruses clustered in a lineage associated primarily with domestic dogs throughout southern and eastern Africa (Kissi et al. 1995). No host-distinguishable variants were identified, and domestic dog isolates were present in all clusters. Divergences among viruses were low, consistent with previous analyses of Tanzanian viruses (Kissi et al. 1995; East et al. 2001) and southern Africa canid viruses (von Teichman et al. 1995; Sabeta et al. 2003; Johnson et al. 2004), suggesting that a single dog-introduced lineage can infect a range of hosts (e.g. dogs, jackals and bat-eared foxes). Although bat-eared fox viruses appear to be more distinct in South Africa (Sabeta et al. 2003), definitive virus–host associations have not yet been identified among canid species in this geographical area.
A number of viruses originating from the Serengeti and Tarangire ecosystems grouped together. One possible explanation for this is the seasonal migration of nomadic Maasai pastoralists and their dogs from Tarangire to the Crater Highlands each year.
The results of the Bayesian analyses suggest cross-species transmission of a single variant among a range of domestic and wild species, since viruses recovered from different hosts cluster together. The SP approach shows strong support for one Canidae-associated virus variant circulating within the Serengeti carnivore community. The estimation procedure applied to both the N and G gene sequences connects viruses recovered from a range of hosts into parsimony networks with domestic dog viruses present in all the networks. The sparse and necessarily opportunistic nature of the sampling process required of this sort of study introduces biases in the proportion of domestic and wild animal hosts represented in the dataset over space and time (which are not reflective of any obvious changes in the distribution or movement of host species). This sparse sampling process prohibits a definitive inference regarding the identity of the reservoir host, but the genealogic pattern repeatedly identified in these results is most consistent with the domestic dog comprising the reservoir of rabies.
Our findings suggest that, even in species-rich areas, the paradigm of maintenance of a single virus variant by a single host species holds true. Despite the abundance of other mammalian hosts, the domestic dog appears to act as the principal host of a typical canid variant. Similar characteristics of viruses isolated from a range of other species indicate that this variant is freely able to jump species boundaries, but the transmission networks suggest that wildlife species cannot establish stable infection cycles independently of dogs. The domestic dog population surrounding the Serengeti is rapidly expanding and is well suited to serve as a rabies reservoir, with high turnover rates generating large numbers of susceptible hosts. Several Serengeti species with attributes consistent with reservoir hosts (Wandeler et al. 1994) have been diagnosed with the disease (e.g. the bat-eared fox, the white-tailed mongoose, the small-spotted genet), and the limited sample sizes available for this study do not permit definitive rejection of these species as part of a reservoir system. However, with the possible exception of the bat-eared fox, the available evidence indicates that these species are all associated with sporadic, short-lived epidemics with no evidence for species-specific virus–host associations.
What are the factors preventing the establishment of sustained cycles in a new host in the ecosystem? First, no single Serengeti wild carnivore population may be large enough or reach high enough densities to support independent cycles of a host-adapted virus. Although the Serengeti is renowned for the abundance of its carnivore populations, the high diversity of species coexisting within the park may prevent any single species reaching high enough densities to maintain infection. For example, population densities of jackals in less diverse farmland in Zimbabwe far exceed those recorded in the Serengeti (Cleaveland & Dye 1995), and this is an explanation for the suggestion that dogs and jackals are both able to maintain rabies in Zimbabwe (Bingham et al. 1999a,b). Second, in general, there are no biogeographic barriers around the Serengeti to impede animal movement (as emphasized by the lack of genetic isolation of virus variants) that might promote localized viral evolution in specialized host niches (Bourhy et al. 1999). Third, while high species diversity might be expected to provide many opportunities for host-viral adaptation, such adaptation presumably requires successive generations of infection within the same species and may be inhibited by high levels of interference between generalist carnivores that afford frequent opportunities for cross-species transmission.
In contrast with our observations of a single species supporting the virus cycle in the ecosystem, East et al. (2001) suggested that healthy carrier hyaenas maintain a genetically distinct non-pathogenic variant on the basis of viral RNA detected in hyaena saliva by RT-PCR. East et al. (2001) provide evidence indicating that this variant is genetically most closely related to RABVs circulating in Europe and the Middle East, primarily among foxes, and distinct from hyaena viruses in our study (see electronic supplementary material, S5). Typically, fox RABVs cause rabies clinical signs and inevitable death in foxes (George et al. 1980) and are known to be pathogenic to a range of other species in which no evidence of survival has been documented (Blancou 1988b; Charlton et al. 1988). We consider the finding of this variant in healthy Serengeti hyaenas, without evidence for clinical disease, difficult to explain. In this study, diagnostic material was obtained from 41 hyaenas. Of these, four were confirmed rabies positive and Africa 1b RABVs were recovered. Clinical signs of rabies in hyaenas infected with this variant are quite typical, with signs of altered behaviour, increased aggression (attacking humans and animals), ataxia and death. Rabies morbidity and mortality in hyaenas have been reported elsewhere in Africa (Mills 1990; Swanepoel et al. 1993). There is no doubt that Serengeti hyaenas can die when infected with dog rabies and that rabid hyaenas pose a severe risk to humans and other mammals. With their intra- and interspecific kleptoparasitic behaviour (Kruuk 1972), wide-ranging ‘commuting’ outside the protected areas (Hofer & East 1995), scavenging in agricultural areas (Kruuk 1972) and predation on domestic dogs (Butler et al. 2004; S. Cleaveland, personal observation), hyaenas probably constitute a critical link in disease transmission between domestic and wild carnivore populations in the Serengeti and elsewhere (Cleaveland et al. 2000; Butler et al. 2004).
Viral generalist pathogens pose a grave threat to biodiversity and human health (Cleaveland et al. 2001). The impact of rabies on African wild canids can be substantial, as documented following rabies outbreaks in the African wild dog and the Ethiopian wolf (Canis simensis: Gascoyne et al. 1993; Randall et al. 2004; Haydon et al. 2006). The disease also inflicts a considerable public health burden in many parts of the world (Knobel et al. 2005). Our study is consistent with the view that, in the Serengeti, domestic dogs maintain a single major virus variant belonging to the Africa 1b group with spillover cases occurring in other species, and does not provide evidence for the co-circulation of multiple variants associated with distinct hosts. Efforts directed at controlling infection in dogs through mass vaccination can therefore be expected to eliminate rabies in all other species with benefits for both human health and wildlife conservation.
Acknowledgments
This work was supported by the joint National Institutes of Health/National Science Foundation Ecology of Infectious Diseases Program under grant no. NSF/DEB0225453. T.L. was supported by the Royal (Dick) School of Veterinary Studies, University of Edinburgh and the University of Edinburgh Development Trust (visits to CDC); S.C. by the Wellcome Trust and the Department for International Development Animal Health Programme; and A.R.F. by a grant from the UK Department for Environment, Food and Rural Affairs (Defra, project SEV3500). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agencies.
We thank Tanzania Government ministries, the Tanzania National Parks, the Tanzania Wildlife Research Institute, the Ngorongoro Conservation Area Authority, the Tanzania Commission for Science and Technology and the National Institute for Medical Research for permission to undertake research; the Tanzania National Parks Veterinary Unit, the Viral Transmission Dynamics Project, the Serengeti Lion and Cheetah Projects, the Frankfurt Zoological Society, the livestock officers of the Ministry of Water and Livestock Development in the Mara, Shinyanga and Arusha Regions, Mathias Magoto, Paul Tiringa and Barbara Schachennuann-Suter for assistance with sample collection; Katie Hampson for valuable comments on the manuscript; the staff of the Rabies Section of the CDC, especially Lillian A. Orciari for expert technical assistance; Denise Marston (VLA) for molecular sequencing; and two anonymous referees for constructive comments on an earlier version of the manuscript.
Use of trade names and commercial sources are for identification only and do not imply endorsement by the US Department of Health and Human Services.
Ethical clearance for this work was obtained from the National Institute for Medical Research, Ministry of Health, Dar es Salaam, Tanzania.
Footnotes
Data deposition. The sequences of RABVs produced in this study have been deposited in the GenBank database (accession nos. DQ900547–DQ900579).
Supplementary Material
Details of rabies virus sequences produced in this study
Nucleotide and amino acid substitutions for network I in figure 3
Nucleotide and amino acid substitutions for networks II and III in figure 3
Nucleotide and amino acid substitutions for the network in figure 4
Phylogenetic tree comparing viruses from Tanzanian rabid hyaenas obtained in this study and previously published sequences from healthy Serengeti hyaenas
References
- Aghomo H.O, Oduye O.O, Tomori O, Ibe M. Isolation of rabies virus from clinically healthy and previously unvaccinated dogs. Bull. Anim. Health Prod. Afr. 1989;37:131–135. [Google Scholar]
- Anderson R.M, Jackson H.C, May R.M, Smith A.M. Population dynamics of fox rabies in Europe. Nature. 1981;289:765–771. doi: 10.1038/289765a0. doi:10.1038/289765a0 [DOI] [PubMed] [Google Scholar]
- Barrat J, Barrat M.J, Picard M, Aubert M.F.A. Diagnostic de la rage sur culture cellulaire, comparaison des résultats de l'inoculation au neuroblastome murin et de l'inoculation à la souris. Comp. Immunol. Microbiol. Infect. Dis. 1988;11:207–214. doi: 10.1016/0147-9571(88)90039-2. doi:10.1016/0147-9571(88)90039-2 [DOI] [PubMed] [Google Scholar]
- Bingham J, Foggin C.M, Wandeler A.I, Hill F.W.G. The epidemiology of rabies in Zimbabwe. 1. Rabies in dogs (Canis familiaris) Onderstepoort J. Vet. Res. 1999a;66:1–10. [PubMed] [Google Scholar]
- Bingham J, Foggin C.M, Wandeler A.I, Hill F.W.G. The epidemiology of rabies in Zimbabwe 2. Rabies in jackals (Canis adustus and Canis mesomelas) Onderstepoort J. Vet. Res. 1999b;66:11–23. [PubMed] [Google Scholar]
- Blancou J. Ecology and epidemiology of fox rabies. Rev. Infect. Dis. 1988a;10:S606–S609. doi: 10.1093/clinids/10.supplement_4.s606. [DOI] [PubMed] [Google Scholar]
- Blancou J. Epizootiology of rabies: Eurasia and Africa. In: Campbell J.B, Charlton K.M, editors. Rabies. Kluwer Academic Publishers; Boston, MA: 1988b. pp. 243–265. [Google Scholar]
- Bourhy H, Kissi B, Audry L, Smreczak M, Sadkowska-Todys M, Kulonen K, Tordo N, Zmudzinski J.F, Holmes E.C. Ecology and evolution of rabies virus in Europe. J. Gen. Virol. 1999;80:2545–2557. doi: 10.1099/0022-1317-80-10-2545. [DOI] [PubMed] [Google Scholar]
- Butler J.R.A, du Toit J.T, Bingham J. Free-ranging domestic dogs (Canis familiaris) as predators and prey in rural Zimbabwe: threats of competition and disease to large wild carnivores. Biol. Conserv. 2004;115:369–378. doi:10.1016/S0006-3207(03)00152-6 [Google Scholar]
- Charlton K.M, Webster W.A, Casey G.A, Rupprecht C.E. Skunk rabies. Rev. Infect. Dis. 1988;10:S626–S628. doi: 10.1093/clinids/10.supplement_4.s626. [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Dye C. Maintenance of a microparasite infecting several host species: rabies in the Serengeti. Parasitology. 1995;111:S33–S47. doi: 10.1017/s0031182000075806. [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Appel M.G.A, Chalmers W.S.K, Chillingworth C, Kaare M, Dye C. Serological and demographic evidence for domestic dogs as a source of canine distemper virus infection for Serengeti wildlife. Vet. Microbiol. 2000;72:217–227. doi: 10.1016/s0378-1135(99)00207-2. doi:10.1016/S0378-1135(99)00207-2 [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson M.K, Taylor L.H. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. doi:10.1098/rstb.2001.0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Conzelmann K.K, Cox J.H, Schneider L.G, Thiel H.J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. doi:10.1016/0042-6822(90)90433-R [DOI] [PubMed] [Google Scholar]
- Crandall K.A. Intraspecific phylogenetics: support for dental transmission of human immunodeficiency virus. J. Virol. 1995;69:2351–2356. doi: 10.1128/jvi.69.4.2351-2356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall K.A. Multiple interspecies transmissions of human and simian T-cell leukemia/lymphoma virus type I sequences. Mol. Biol. Evol. 1996;13:115–131. doi: 10.1093/oxfordjournals.molbev.a025550. [DOI] [PubMed] [Google Scholar]
- Dean D.J, Abelseth M.K, Atanasiu P. The fluorescent antibody test. In: Meslin F.-X, Kaplan M.M, Koprowski H, editors. Laboratory techniques in rabies. 4th edn. World Health Organization; Geneva, Switzerland: 1996. pp. 88–95. [Google Scholar]
- Drummond A.J, Nicholls G.K, Rodrigo A.G, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2006 BEAST v. 1.4. See http://www.beast.bio.ed.ac.uk
- East M.L, Hofer H, Cox J.H, Wulle U, Wiik H, Pitra C. Regular exposure to rabies virus and lack of symptomatic disease in Serengeti spotted hyenas. Proc. Natl Acad. Sci. USA. 2001;98:15 026–15 031. doi: 10.1073/pnas.261411898. doi:10.1073/pnas.261411898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekadu M. Atypical rabies in dogs in Ethiopia. Ethiop. Med. J. 1972;10:79–86. [PubMed] [Google Scholar]
- Gascoyne S.C, Laurenson M.K, Lelo S, Borner M. Rabies in African wild dogs (Lycaon pictus) in the Serengeti region, Tanzania. J. Wildl. Dis. 1993;29:396–402. doi: 10.7589/0090-3558-29.3.396. [DOI] [PubMed] [Google Scholar]
- George J.P, George J, Blancou J, Aubert M.F.A. Description clinique de la rage du renard. Etude expérimentale. Revue Méd. Vét. 1980;131:153–160. [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Haydon D.T, et al. Low-coverage vaccination strategies for the conservation of endangered species. Nature. 2006;443:692–695. doi: 10.1038/nature05177. doi:10.1038/nature05177 [DOI] [PubMed] [Google Scholar]
- Heaton P.R, Johnstone P, McElhinney L.M, Cowley R, O'Sullivan E, Whitby J.E. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. J. Clin. Microbiol. 1997;35:2762–2766. doi: 10.1128/jcm.35.11.2762-2766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer H, East M. Population dynamics, population size and the commuting system of Serengeti spotted hyenas. In: Sinclair A.R.E, Arcese P, editors. Serengeti II: dynamics, management and conservation of an ecosystem. University of Chicago Press; Chicago, IL: 1995. pp. 332–363. [Google Scholar]
- Jeanmougin F, Thompson J.D, Gouy M, Higgins D.G, Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. doi:10.1016/S0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- Johnson N, Letshwenyo M, Baipoledi E.K, Thobokwe G, Fooks A.R. Molecular epidemiology of rabies in Botswana: a comparison between antibody typing and nucleotide sequence phylogeny. Vet. Microbiol. 2004;101:31–38. doi: 10.1016/j.vetmic.2004.03.007. doi:10.1016/j.vetmic.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Kissi B, Tordo N, Bourhy H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology. 1995;209:526–537. doi: 10.1006/viro.1995.1285. doi:10.1006/viro.1995.1285 [DOI] [PubMed] [Google Scholar]
- Knobel D.L, Cleaveland S, Coleman P.G, Fèvre E.M, Meltzer M.I, Miranda M.E.G, Shaw A, Zinsstag J, Meslin F.-X. Re-evaluating the burden of rabies in Africa and Asia. Bull. World Health Organ. 2005;83:360–368. [PMC free article] [PubMed] [Google Scholar]
- Kruuk H, editor. The spotted hyaena. University of Chicago Press; Chicago, IL: 1972. [Google Scholar]
- Mills M.G.L, editor. Kalahari hyaenas. Unwin Hyman; London, UK: 1990. [Google Scholar]
- Page R.D.M. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. doi:10.1093/bioinformatics/12.4.357 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Randall D.A, et al. Rabies in endangered Ethiopian wolves. Emerg. Infect. Dis. 2004;10:2214–2217. doi: 10.3201/eid1012.040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes C.J, Atkinson R.P.D, Anderson R.M, Macdonald D.W. Rabies in Zimbabwe: reservoir dogs and the implications for disease control. Phil. Trans. R. Soc. B. 1998;353:999–1010. doi: 10.1098/rstb.1998.0263. doi:10.1098/rstb.1998.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rupprecht C.E, Dietzschold B, Wunner W.H, Koprowski H. Antigenic relationships of Lyssaviruses. In: Baer G.M, editor. The natural history of rabies. 2nd edn. CRC Press; Boca Raton, FL: 1991. pp. 69–100. [Google Scholar]
- Sabeta C.T, Bingham J, Nel L.H. Molecular epidemiology of canid rabies in Zimbabwe and South Africa. Virus Res. 2003;91:203–211. doi: 10.1016/s0168-1702(02)00272-1. doi:10.1016/S0168-1702(02)00272-1 [DOI] [PubMed] [Google Scholar]
- Sacramento D, Bourhy H, Tordo N. PCR technique as an alternative method for diagnosis and molecular epidemiology of rabies virus. Mol. Cell. Probes. 1991;5:229–240. doi: 10.1016/0890-8508(91)90045-l. doi:10.1016/0890-8508(91)90045-L [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Ishiguro M, Kitagawa G, editors. Akaike information criterion statistics. Springer; New York, NY: 1986. [Google Scholar]
- Shapiro B, Rambaut A, Drummond A.J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. doi:10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- Smith J.S. Molecular epidemiology. In: Jackson A.C, Wunner W.H, editors. Rabies. Academic Press; New York, NY: 2002. pp. 79–112. [Google Scholar]
- Swanepoel R, Barnard B.J.H, Meredith C.D, Bishop G.C, Bruckner G.K, Foggin C.M, Hübschle O.J.B. Rabies in southern Africa. Onderstepoort J. Vet. Res. 1993;60:325–346. [PubMed] [Google Scholar]
- Templeton A.R, Crandall K.A, Sing C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson G.R, Meredith C.D. Rabies in bat-eared foxes in South Africa. Onderstepoort J. Vet. Res. 1993;60:399–403. [PubMed] [Google Scholar]
- von Teichman B.F, Thomson G.R, Meredith C.D, Nel L.H. Molecular epidemiology of rabies virus in South Africa: evidence for two distinct virus groups. J. Gen. Virol. 1995;76:73–82. doi: 10.1099/0022-1317-76-1-73. [DOI] [PubMed] [Google Scholar]
- Wandeler A.I, Nadin-Davis S.A, Tinline R.R, Rupprecht C.E. Rabies epidemiology: some ecological and evolutionary perspectives. In: Rupprecht C.E, Dietzschold B, Koprowski H, editors. Lyssaviruses. Springer; Berlin, Germany: 1994. pp. 297–324. [DOI] [PubMed] [Google Scholar]
- Worobey M. A novel approach to detecting and measuring recombination: new insights into evolution in viruses, bacteria, and mitochondria. Mol. Biol. Evol. 2001;18:1425–1434. doi: 10.1093/oxfordjournals.molbev.a003928. [DOI] [PubMed] [Google Scholar]
- Yang Z, Goldman N, Friday A. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol. Biol. Evol. 1994;11:316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of rabies virus sequences produced in this study
Nucleotide and amino acid substitutions for network I in figure 3
Nucleotide and amino acid substitutions for networks II and III in figure 3
Nucleotide and amino acid substitutions for the network in figure 4
Phylogenetic tree comparing viruses from Tanzanian rabid hyaenas obtained in this study and previously published sequences from healthy Serengeti hyaenas