Abstract
Tuberous sclerosis (TSC) is an autosomal dominant disease characterized by hamartoma formation in various organs and is caused by mutations targeting either the TSC1 or TSC2 genes. TSC1 and TSC2 proteins form a functionally interdependent dimeric complex. Phosphorylation of either TSC subunit by different kinases regulates the function of TSC and represents a major mechanism to integrate various signals into a centralized cell growth pathway. The majority of disease-associated mutations targeting either TSC1 or TSC2 results in a substantial decrease in protein level, suggesting that protein turnover also plays a critical role in TSC regulation. Here we report that TSC2 protein binds to FBW5, a DDB1-binding WD40 (DWD) protein, and is recruited by FBW5 to the DDB1–CUL4–ROC1 E3 ubiquitin ligase. Overexpression of FBW5 or CUL4A promotes TSC2 protein degradation, and this is abrogated by the coexpression of TSC1. Conversely, depletion of FBW5, DDB1, or CUL4A/B stabilizes TSC2. Ddb1 or Cul4 mutations in Drosophila result in Gigas/TSC2 protein accumulation and cause growth defects that can be partially rescued by Gigas/Tsc2 reduction. These results indicate that FBW5–DDB1–CUL4–ROC1 is an E3 ubiquitin ligase regulating TSC2 protein stability and TSC complex turnover.
[Keywords: TSC, DDB1, cullin, ubiquitination, cell growth]
The tuberous sclerosis 1 (TSC1) locus encodes a 130-kDa protein termed “hamartin,” and the TSC2 locus encodes a 180-kDa protein termed “tuberin” that contains a coiled-coil domain and a GAP (GTPase-activating protein) domain. Genetic studies in Drosophila melanogaster and biochemical analyses in mammalian cells have established the TSC1–TSC2 heterodimeric complex as a critical regulator in coupling various cellular conditions to cell growth through stimulation of GTP hydrolysis of RheB to antagonize the mTOR signaling pathway (Inoki et al. 2005). These studies have identified both TSC subunits as substrates of various kinases that mediate different cellular conditions such as nutrient availability, energy, hormones, and growth factor stimulation. TSC2 is a short-lived protein, and it is actively ubiquitinated (Chong-Kopera et al. 2006). Many disease-associated mutations in TSC1 and TSC2 result in a substantial decrease in the level of hamartin and tuberin, respectively (Inoki et al. 2002; Nellist et al. 2005), suggesting that protein turnover plays a critical role in TSC regulation. Identification of the ubiquitin ligase for TSC2 would shed light on the understanding of the regulation of TSC and cell growth pathways.
Results and Discussion
TSC2 binds to WD40 protein FBW5, DDB1, and CUL4
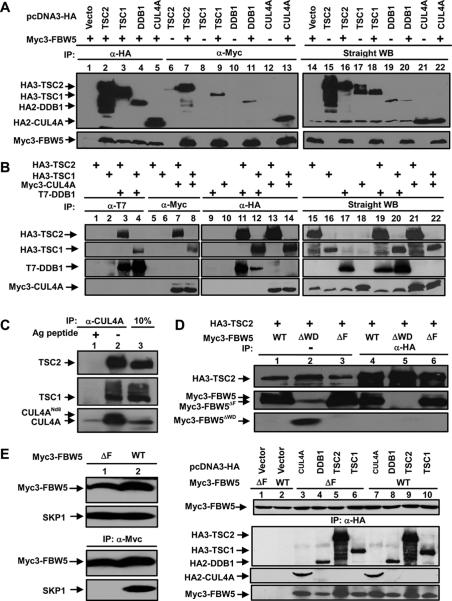
We used a yeast two-hybrid screen and coupled immunoprecipitation and immunoblotting (IP-Western) analyses to search for TSC2-interacting protein with potential ubiquitin ligase activity and identified two F-box proteins—FBW5 (Fig. 1A) and FBL6 (data not shown)—that bind with TSC2. Three additional control F-box proteins—SKP2, β-TrCP, and FBX5—did not bind with TSC2 in the same assay (data not shown), indicating specificity of the FBL6–TSC2 and FBW5–TSC2 interactions and also suggesting that the F-box motif is not sufficient for binding to TSC2. Consistent with TSC1–TSC2 dimerization, FBW5 also associates with TSC1 (Fig. 1A). Our subsequent genetic study in Drosophila (see below) led us to focus on determining the function and mechanism of the FBW5–TSC interaction. The functional significance of the FBL6–TSC interaction remains to be determined.
Figure 1.
FBW5 binds to both ubiquitin ligase DDB1–CUL4 and TSC1–TSC2. (A,B) 293T cells were transfected with plasmids expressing the indicated proteins and protein–protein association was examined by coupled IP-Western analysis. (C) Endogenous TSC2–CUL4A association was determined by IP-Western analysis. (D) The WD40 domain, but not the F-box, is required for bridging FBW5 to associate with TSC2. 293T cells were transfected with plasmids expressing HA3-TSC2 and Myc3-FBW5 or Myc3-FBW5 without either the F-box or WD40 domain. The cells were lysed by NP-40 buffer, and the lysates were applied to IP with anti-HA antibody followed by Western blot analysis with indicated antibodies. (E) The F-box is not required for bridging FBW5 to associate with TSC1/2 and DDB1–CUL4A. 293T cells transfected with a plasmid expressing myc-tagged wild-type or mutant FBW5 deleting the F-box (FBW5ΔF), followed by IP-Western analysis to examine protein complex formation.
Human FBW5 is a 60-kDa (566 residues) protein containing two recognizable domains; an N-terminally located F-box motif and seven WD40 repeats occupying most of the rest of the sequence. The F-box motif functions to bridge a substrate protein to the CUL1–ROC1 E3 ligase via the SKP1 linker (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997). Recently, we and others found that DDB1 binds to a subset of WD40 proteins referred to as DWD proteins (also known as DCAF, for Ddb1- and Cul4-associated factors; or CDW, for CUL4- and DDB1-associated WD40 repeat proteins), and bridges them to CUL4–ROC1 to constitute a potentially large and distinct family of DWD–DDB1–CUL4–ROC1 E3 ubiquitin ligases (Angers et al. 2006; He et al. 2006; Higa et al. 2006; Jin et al. 2006). FBW5 contains two recognizable DWD boxes between residues 436 and 451 and 484 and 499 (He et al. 2006), which led us to determine whether it can bind to and target TSC2 for ubiquitination by the DDB1–CUL4–ROC1 ligase.
We first examined the association between ectopically expressed TSC2 and DDB1–CUL4A complex in 293T cells. TSC2, as well as its partner TSC1, was readily detected in both DDB1 and CUL4A immunocomplexes (Fig. 1B). Both TSC1 and TSC2 proteins were also readily detected in CUL4A immunocomplexes from untransfected 293T or U2OS cells (shown for 293T cells in Fig. 1C), demonstrating association between endogenous TSC1–TSC2 and DDB1–CUL4A. The amount of TSC2 protein detected in CUL4A complexes is similar to or slightly greater than that detected by direct Western blotting using one-tenth of the cell extract, suggesting that at least 10% of the total TSC2 is associated with DDB1–CUL4A in 293T cells (Fig. 1C). We therefore conclude that the DDB1–CUL4 ligase plays a direct and important role in regulating the TSC complex in vivo.
To determine which domain—the F-box or the WD40 repeats—is required for FBW5 to bridge TSC1–TSC2 to DDB1–CUL4A, we generated FBW5 mutants, FBW5ΔF and FBW5ΔWD, with deletions of the F-box motif and WD40 domain, respectively. The FBW5ΔWD mutant completely lost its ability to bind with TSC2 (Fig. 1D). The FBW5ΔF mutant cannot bind to SKP1 but retains the ability to bridge TSC1 and TSC2 with DDB1–CUL4A (Fig. 1E). These results demonstrate that the WD40 repeats, but not the F-box, in FBW5 mediate TSC2 binding to DDB1, supporting the idea that a CUL4-, but not a CUL1-based E3 ligase regulates TSC2 protein stability (see below).
FBW5 promotes TSC2 degradation
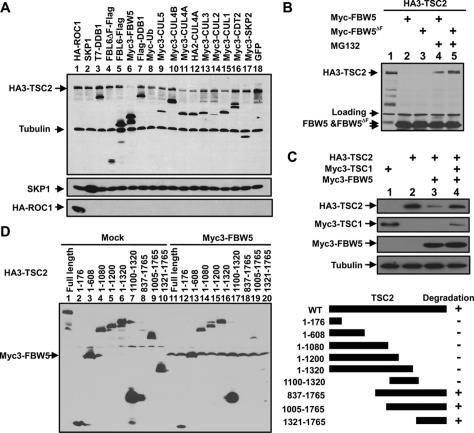
To determine whether FBW5–DDB1–CUL4 is an E3 ubiquitin ligase for TSC2 and promotes TSC2 degradation, we coexpressed TSC2 with a series of individual components of different E3 ubiquitin ligases. Overexpression of FBW5 substantially reduced the steady-state level of ectopically expressed TSC2 protein, while overexpression of another DWD protein, CDT2, or two F-box proteins, SKP2 and FBL6, did not exhibit any detectable effect on the level of TSC2 (Fig. 2A). In the same assay, overexpression of CUL4A also reduced the level of TSC2 protein. Overexpression of ROC1 or DDB1 did not show any effect on TSC2 levels, suggesting that a sufficient amount of both proteins is present in the cells to support the ligase activity toward TSC2. Overexpression of CUL1 also noticeably reduced the level of TSC2, albeit to a lesser extent than CUL4A overexpression. Consistent with the notion that FBW5ΔF is sufficient to bridge TSC2 to DDB1–CUL4A, overexpression of FBW5ΔF reduced TSC2 levels with a potency similarity to that of wild type (Fig. 2B). Degradation of TSC2 by FBW5 or FBW5ΔF was inhibited by the proteasome inhibitor MG132 (Fig. 2B), indicating that FBW5-promoted TSC2 degradation is proteasome-dependent. Coexpression of TSC1 protects TSC2 from degradation caused by FBW5 overexpression (Fig. 2C). This finding suggests that monomeric TSC2 is degraded by the FBW5–DDB1–CUL4–ROC1 E3 ligase and suggests a molecular explanation for why TSC2 function depends on the formation of a dimeric complex with TSC1.
Figure 2.
Ectopic expression of FBW5 decreases TSC2. (A) HA-tagged TSC2 was cotransfected with plasmids expressing various indicated proteins into 293T cells. The steady-state level of TSC2 protein was determined by direct immunoblotting. (B) FBW5 promotes TSC2 degradation in a proteasome-dependent manner. 293T cells were cotransfected with plasmids expressing indicated proteins and were either untreated or treated with proteasome inhibitor MG132. The steady-state level of TSC2 protein was determined by direct immunoblotting. (C) Coexpression of TSC1 protects TSC2 from degradation by FBW5. 293T cells were cotransfected with plasmids expressing indicated proteins, and the steady-state levels of TSC1, TSC2, and FBW5 were determined by direct immunoblotting. (D) A C-terminal domain of TSC2 is both required and sufficient for FBW5-promoted degradation. Various TSC2 deletion mutants were cotransfected with FBW5, and the steady-state level of TSC2 protein was determined by direct immunoblotting.
A series of deletion mutants of TSC2 was generated to map the region in TSC2 required for its degradation by FBW5–DDB1–CUL4A. All mutants containing amino acids from 1321 to 1765 were effectively degraded by the coexpression of FBW5, while all mutants missing this fragment were resistant to FBW5-promoted degradation (Fig. 2D). These results suggest that fragment 1321–1765 of TSC2, which contains the Rap-GAP domain and is highly conserved during evolution, is necessary and sufficient to mediate TSC2 degradation by the FBW5–DDB1–CUL4 ligase.
Depletion of FBW5, DDB1, or CUL4A/B stabilizes TSC2
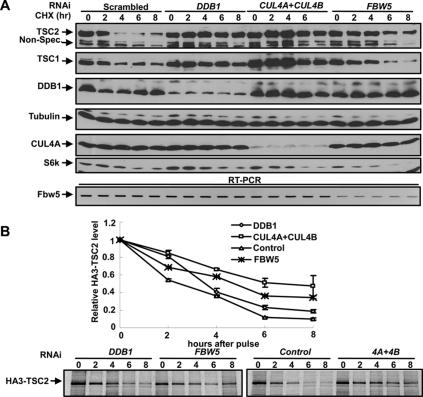
To further demonstrate the function of FBW5, DDB1, and CUL4A/B in targeting TSC2 protein degradation, we blocked new protein synthesis with cycloheximide treatment and determined the half-life of TSC2 protein. TSC2 is a short-lived protein with a half-life ∼2 h in control U2OS cells transfected with scrambled siRNA oligo (Fig. 3A). Silencing either DDB1 or the combination of CUL4A and CUL4B increased the half-life of TSC2 to >8 h (the duration of the experiment), and silencing FBW5 increased the half-life of TSC2 protein to ∼6 h (Fig. 3A). Silencing CUL4, DDB1, or FBW5 also consistently stabilized the half-life of ectopically expressed HA-TSC2, measured by 35S pulse-chase assay (Fig. 3B). These results suggest that FBW5–DDB1–CUL4 is an active E3 ubiquitin ligase that continuously ubiquitinates TSC2 and promotes its degradation.
Figure 3.
Silencing CUL4, DDB1, or Fbw5 stabilizes TSC2. (A) Silencing DDB1, FBW5, or a combination of CUL4A and CUL4B in mammalian cells increases the half-life of endogenous TSC2 protein. U2OS cells transfected with siRNAs targeting the indicated genes were treated with cycloheximide for different times. The steady-state level of endogenous TSC2 as well as TSC1 was determined by direct immunoblotting. (B) Silencing DDB1, FBW5, or a combination of CUL4A and CUL4B in mammalian cells increases the half-life of ectopic TSC2 protein. U2OS cells were transfected with siRNAs targeting the indicated genes 12 h after transfection of plasmid expressing HA-TSC2. The half-life of HA-STC2 was measured by 35S pulse-chase assay.
FBW5–DDB1–CUL4 promotes TSC2 in vivo and in vitro ubiquitination
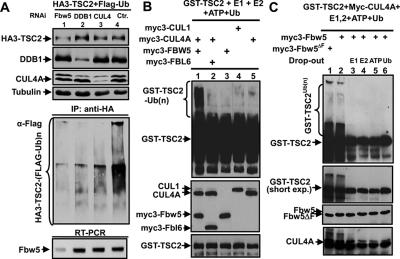
We next determined whether the FBW5–DDB1–CUL4–ROC1 ligase ubiquitinates TSC2. U2OS cells were cotransfected with plasmids expressing HA-tagged TSC2 and Flag-tagged ubiquitin, and 12 h later, the cells were transfected with siRNA oligo silencing DDB1, FBW5, or a combination of CUL4A/B. TSC2 in vivo ubiquitination in these cells was analyzed by measuring the amount of HA-TSC2–Flag-ubiquitin conjugates by IP-Western. TSC2 protein is actively ubiquitinated, and the level of TSC2 ubiquitination was substantially reduced by knocking down individually FBW5, DDB1, or CUL4A/B, with FBW5 knockdown exhibiting the most pronounced effect (Fig. 4A).
Figure 4.
In vivo and in vitro ubiquitination of TSC2 by FBW5–DDB1–CUL4 ligase. (A) CUL4, DDB1, and FBW5 are required for TSC2 ubiquitination in vivo. U2OS cells were cotransfected with plasmids expressing HA-TSC2 and Flag-ubiquitin and siRNA targeting CUL4A/B, DDB1, or FBW5. Cells were lysed 24 h after transfection, and TSC2 ubiquitination was analyzed by immunoprecipitating HA-TSC2 and blotted with an antibody recognizing Flag. (B) In vitro ubiquitination of TSC2 by FBW5 and CUL4A. 293T cells were cotransfected with plasmids expressing the indicated myc-tagged proteins. Individual protein complex was eluted by incubating with molar excess myc antigen peptide and mixed as indicated with substrate GST-TSC2 in the presence of E1, E2, ubiquitin, and ATP. Reaction mixtures were incubated for 30 min, terminated by boiling directly in SDS sample buffer, resolved by SDS-PAGE, and immunoblotted with indicated antibodies. (C) Drop-out assay was carried out as described in B to determine the specificity of in vitro TSC2 ubiquitination.
To demonstrate that the FBW5–DDB1–CUL4–ROC1 ligase directly promotes TSC2 ubiquitination in vitro, we immunoprecipitated CUL4A complexes from transfected 293T cells and used it as a source of E3 ligase. Incubation of purified GST-TSC2 substrate with CUL4A and FBW5 immunocomplexes resulted in robust TSC2 polyubiquitination in the presence of E1, E2-UBC5, ubiquitin, and ATP (Fig. 4B, lane 1). Omitting either CUL4A (Fig. 4B, lane 3) or FBW5 (Fig. 4B, lane 5) or substitution of FBW5 with FBL6 (Fig. 4B, lane 2) abolished GST-TSC2 polyubiquitination. To further confirm GST-TSC2 ubiquitination by the FBW5–DDB1–CUL4A–ROC1 ligase, we performed dropped out experiments (Fig. 4C). Omitting individually E1 (Fig. 4C, lane 3), E2-UBC5 (Fig. 4C, lane 4), ATP (Fig. 4C, lane 5), and ubiquitin (Fig. 4C, lane 6) each eliminated GST-TSC2 polyubiquitination. Consistent with the finding that the F-box motif is not required for mediating TSC2 ubiquitination by DDB1–CUL4A, FBW5ΔF was fully capable of substituting for wild-type FBW5 in promoting TSC2 ubiquitination (Fig. 4C, lane 2). Together, these results demonstrate that FBW5 recruits TSC2 to the DDB1–CUL4A–ROC1 ligase for polyubiquitination in vitro.
Drosophila Cul4 or Ddb1 mutants accumulate Gigas/TSC2
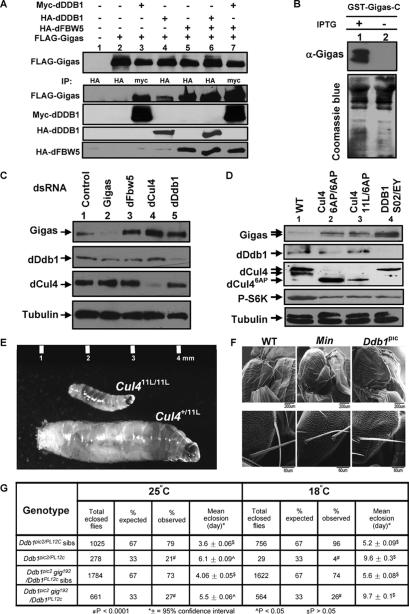
To seek further in vivo evidence for FBW5–DDB1–CUL4-mediated TSC2 degradation, we turned to D. melanogaster in which both the TSC-mediated cell growth control pathway and the DDB1–CUL4 E3 ligase are conserved. Drosophila also contains a gene, CG9144, encoding an F-box and WD40 protein that exhibits 40% identity and 58% similarity to human FBW5, and thus represents an FBW5 homolog. We first ectopically expressed these three fly proteins in cultured S2 cells and examined their binding by coimmunoprecipitation assay. As shown in Figure 5A, Flag-Gigas/TSC2 was readily detected in Myc-dDDB1 (lane 3), HA-dDDB1 (lane 4), and HA-dFBW5 immunocomplexes (lane 5), and Myc-dDDB1 complexes contained detectable Flag-Gigas/TSC2 and HA-dFBW5 (lane 7). These results demonstrate that like its mammalian homologs, Drosophila TSC2/Gigas also bind directly to FBW5 and DDB1.
Figure 5.
Accumulation of Gigas/Tsc2 protein and growth defects in Drosophila Cul4 and Ddb1 mutants. (A) Drosophila S2 cells were transfected with plasmids expressing the indicated proteins, and protein–protein association was examined by coupled IP-Western analysis. (B) Characterization of an anti-Gigas antibody. Escherichia coli BL21 cells were transformed with a plasmid expressing a C-terminal portion of the Gigas protein, and were cultured in LB media with or without IPTG to induce the expression of GST-Gigas. Cells were lysed in SDS buffer followed by Western analysis. (C) S2 cells were treated with dsRNA targeting Gigas, dFbw5, dCul4, and dDdb1 and control dsRNA for 2 d. The cells were lysed in RIPA buffer followed by Western analysis with the indicated antibodies. (D) Gigas accumulates in Drosophila Cul4 and Ddb1 mutants. Total cell lysates were prepared from Drosophila Cul4 and Ddb1 mutant first instar larvae, and the steady-state levels of Cul4, Ddb1, Giags, tubulin, and phospho-S6K proteins were determined by direct Western blotting. (E) Size comparison of growth-arrested Cul411L/11L homozygous mutant larvae with a normal Cul411L/+ heterozygous sibling third instar larvae. (F) Drosophila hypomorphic Ddb1 mutants display small and missing macrochaete. Scanning electron micrographs of wild-type, Minute, and Ddb1pic2/PL12C mutant adults. (G) Growth defects of Cul4 and Ddb1 mutants are rescued by heterozygosity of gig (TSC2). Total eclosed flies and mean eclosion time of Ddb1pic2/Ddb1PL12c mutants and their siblings and Ddb1pic2gig192/Ddb1PL12c mutants and their siblings were measured at 18°C and 25°C.
We then generated and characterized an antibody to the Drosophila TSC2 protein, Gigas, and determined the steady-state level of Gigas/TSC2 protein in cultured S2 cells after silencing dFbw5, dCul4, and dDdb1. The specificity of this newly generated Gigas/TSC2 antibody was verified by its recognition of bacterially produced GST-Gigas fusion protein (Fig. 5B) and by the detection of endogenous Gigas/TSC2 that was reduced in cells transfected with dsRNA silencing Gigas (Fig. 5C). Silencing of dFbw5, dCul4, or dDdb1 all led to accumulation of Gigas/TSC2 (Fig. 5C). We next generated and characterized a series of mutations in the single Cul4 and Ddb1 genes (S. Zacharek, Y. Xiong, and R. Ruronio, unpubl.). Cul46AP and Cul411L were generated by P-element-mediated excision mutagenesis and encode predicted proteins containing C-terminal deletions of 82 and 65 amino acids, respectively. Cul411L produces no protein and is therefore null, while Cul46AP produces a stable truncated protein (Fig. 5D). Ddb1S026316 and Ddb1EY01408 are P-element insertion-null alleles that produce no detectable protein (Fig. 5D). The steady-state level of Gigas/TSC2 was substantially increased in both dCul46AP/6AP (Fig. 5D, lane 2) and dCul411L/6AP (Fig. 5D, lane 3) mutant larvae as well as in dDdb1S02/EY mutant larvae (Fig. 5D, lane 4). Together, these results demonstrate, in both cultured cells and animals, that the FBW5–DDB1–CUL4 E3 ligase is functionally conserved in Drosophila in promoting TSC2 degradation. We noted that while CUL4 and DDB1 promote TSC2 degradation in both mammalian and Drosophila cells, loss of function of either CUL4 or DDB1 results in an increase of steady-state levels of Gigas/TSC2 in fly cells, but not in mammalian cells, raising an intriguing possibility that there may exist an additional negative feedback loop in human cells, possibly via a post-transcriptional control, to maintain the TSC2 protein level.
Loss of Drosophila Ddb1 results in growth defects that are alleviated by a reduction of Tsc2/Gigas
Notably, both Cul4- and Ddb1-null mutations cause growth arrest in the first (Cul4) or second (Ddb1) instar larvae, with the larvae capable of surviving for >10 d on rich medium without growing (Fig. 5E). These results reveal a previously unrecognized function of Ddb1 and Cul4 in cell growth control. We next isolated and characterized viable Ddb1 hypomorphic alleles that cause growth defects and determined by complementation analysis that a previously defined locus termed piccolo (pic) was allelic to Ddb1. Semilethal piccolo mutants were originally characterized based on shared irregularities in bristle, wing, and tergite growth (Rushlow and Chovnick 1984; Clark and Chovnick 1985, 1986). This type of growth defect is reminiscent of phenotypes observed in hypomorphic myc mutants and Minute mutants, which are heterozygous for null alleles of ribosomal protein genes (Fig. 5F). pic2 is an X-ray-induced hypomorphic allele of Ddb1 encoding a G21 → D substitution that results in severely reduced Ddb1 protein levels (S. Zacharek, Y. Xiong, and R. Ruronio, unpubl.). We generated another hypomorphic allele of Ddb1 (Ddb1PL12c) by excision of the Ddb1EY01408 P element located in the 5′ untranslated region. Ddb1pic2/PL12c mutants develop into viable adults, but these flies show a significant developmental delay and eclose on average 2 and 4 d later than control siblings at 25°C and 18°C, respectively (Fig. 5G). In addition, these flies have cuticle defects including missing bristles and small bristles that resemble Min mutants (the piccolo phenotype) (Fig. 5F). Reduction of Gigas/Tsc2 gene dose by half substantially suppressed the late eclosion phenotype and the semilethality of Ddb1pic2/PL12c mutants (Fig. 5G), providing genetic support that Tsc2 is a downstream target of Ddb1. The bristle defects caused by Ddb1 mutation, on the other hand, were not significantly alleviated by the Tsc2/Gigas heterozygosity. This suggests that Tsc2 accumulation contributes to some of the growth defects caused by Ddb1 mutations and that Ddb1–Cul4 E3 ligases may regulate additional substrates that are involved in cell growth control.
In this study, we provide five lines of evidence indicating that the FBW5–DDB1–CUL4–ROC1 ubiquitin ligase promotes the ubiquitination and degradation of the TSC2 tumor suppressor. First, CUL4, DDB1, and FBW5 associate with TSC1 and TSC2. Second, overexpression of FBW5 and CUL4A decreases the ectopically expressed TSC2 in cultured mammalian cells. Third, depletion of CUL4, DDB1, or FBW5 increases the half-life of TSC2 in cultured mammalian cells by RNAi and the steady-state levels in cultured Drosophila cells. Fourth, CUL4, DDB1, and FBW5 are required for the ubiquitination of TSC2 in vivo and in vitro. Finally, Drosophila Ddb1 hypomorphic alleles accumulate Gigas/Tsc2 protein and cause growth defects that can be rescued by reduction of Gigas/Tsc2 gene dose by half. It has been previously reported that the HERC1 ubiquitin ligase may contribute to TSC2 degradation (Chong-Kopera et al. 2006). While both HERC1 and FBW5–DDB1–CUL4–ROC1 E3 ligases promote TSC2 degradation in mammalian cells, there is no recognizable HERC1 homolog in Drosophila. This indicates that the FBW5–DDB1–CUL4–ROC1 E3 ligase was first evolved to regulate TSC2, and HERC1 was evolved later and used by the mammalian cells to provide an additional regulation of TSC2.
The TSC1–TSC2 complex plays a key role in mediating the PI3K–mTOR pathway of cell growth control (Engelman et al. 2006). Recent studies have demonstrated that TSC activity is regulated by as many as five other kinases: AKT, GSK3, AMPK, RSK, ERK, and MAPKAPK2 upon various signals including mitogenic stimulation, energy level, and amino acid availability (Crino et al. 2006). The physiological condition(s) signaling FBW5 to promote TSC2 protein turnover by DDB1–CUL4 ligase remains to be elucidated. We speculate that FBW5–DDB1–CUL4–ROC1 may function to regulate the homeostasis of TSC complexes instead of mediating a specific cellular growth condition. TSC2 is a short-lived protein undergoing constant turnover during normal cell growth conditions (t1/2 < 2 h) (Fig. 3). By maintaining a dynamic regulation of TSC complexes, cells can ensure continued responsiveness of TSC complexes to different growth conditions and signaling pathways. One way to achieve this is to limit the lifetime of TSC2 subunit and thus of TSC1–TSC2 complex, thereby avoiding locking TSC complexes in a specific state by an individual signaling growth condition.
Materials and methods
Plasmids, cell culture, and cell transfection
Plasmids expressing GFP, SKP1, SKP2, CUL1, CUL2, CUL3, CUL4A, CUL5, ROC1, CAND1, and DDB1 were described previously (Michel and Xiong 1998; Hu et al. 2004). Plasmids expressing CDT2 and myc3-ubiquitin were obtained from collaborator Hui Zhang (Yale University, CT). Plasmids expressing Flag-ubiquitin and TSC1, TSC2, and TSC2 mutants were obtained from Kun-Liang Guan (University of Michigan at Ann Arbor, MI). Plasmids expressing FBW5 and FBL6 were cloned from a HeLa cDNA library. Mutations were introduced by site-directed mutagenesis using the QuickChange Kit (Stratagene) and were verified by DNA sequencing. All human cells were cultured in DMEM containing 10% FBS in a 37°C incubator with 5% CO2. Cell transfections were performed using a calcium-phosphate buffer.
Antibodies, proteins, and immunological procedures
Antibodies to hemagglutinin (HA) (12CA5; Boehringer-Mannheim), Myc (9E10; NeoMarker), T7 (Novagen), and Flag (M2; Sigma) were purchased commercially. Antibodies to CUL4A, DDB1, ROC1, CAND1, TSC1, and TSC2 were described previously (Shumway et al. 2003; Hu et al. 2004; Zacharek et al. 2005). Polyclonal antibody to Gigas was raised against the C-terminal epitope of Gigas protein (DMDDQRGDFIKYT). Procedures for protein purification, immunoprecipitation, and immunoblotting have been described (Ohta et al. 1999; Furukawa et al. 2003; Hu et al. 2004).
RNAi
All siRNA oligonucleotides were synthesized with 3′ dTdT overhangs by Dharmacon in a purified and annealed duplex form. The sequences targeting each human gene were as follows: DDB1, 5′-CCUGUUGAUUGC CAAAAAC-3′; CUL4A, 5′-GAACUUCCGAGACAGACCU-3′; Cul4B, 5′-AAGCCUAAAUUACCAGAAA-3′; Fbw5, 5′-CCAAGGAGGGC UUGCGGCACUUUCU-3′. OPTI-MEM medium (500 μL) was mixed with Lipofectamine 2000 reagent (10 μL) for 5 min and then incubated with 10 μL (20 mM) of siRNA for 20 min at room temperature. DMEM-10%-FBS medium (1.5 mL) was added to the mixture, and the entire 2.5 mL was added to HeLa cells cultured on a 60-mm plate at 30%–40% confluency. The cells were transfected once with siRNAs and analyzed 48–72 h after transfection.
The RNAi in Drosophila S2 cells followed the protocol developed by the Jack Dixon laboratory (http://cmm.ucsd.edu/Lab_Pages/Dixon/protocols.php). The Ambion Megascript T7 kits were used to generate the dsRNAs targeting the first 500 bases of dCul4, dDdb1, and dFbw5 genes. Eighty percent confluent S2 cells cultured in 35-mm dishes were added with 5 μg of dsRNAs and incubated for 2 d. The dsRNA-treated S2 cells were lysed in RIPA buffer and applied to Western blot analysis.
35S pulse-chase assay
Forty-eight hours after siRNA transfection, the medium was aspirated off, and the cells were washed once with prewarmed PBS. Three milliliters of prewarmed labeling medium (DMEM, without Met and Cys, with 10% dialyzed FBS; ICN, Inc.) were added into each P100 dish. The cells were incubated for 30 min at 37°C to deplete the intracellular pools of sulfur-containing amino acids. 35S-Met (0.5 mCi) was added into each P100 dish, and the cells were incubated for 4 h. Labeling medium containing 35S-Met was then removed, and the cells were washed once with prewarmed PBS followed by the addition of DMEM medium with 10% FBS. The cells were lysed every 2 h and applied for IP using anti-HA antibodies. The immunocomplexes were resolved by SDS-PAGE. The gel was dried for 1 h at 80°C and developed by autoradiography. Each time point was repeated three times.
Ubiquitination assays
For in vivo ubiquitination assay, the U2OS cells transfected with plasmids expressing HA3-TSC2 and Flag-ubiquitin were further transfected with siRNAs targeting Fbw5, DDB1, or CUL4A + CUL4B, or scrambled siRNA 12 h later. The cells were lysed with SDS lysis buffer (50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 2% Triton X-100, 1% SDS) 48 h after siRNA transfection, and then the lysates were diluted with 10× NP-40 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NaCl, 50 mM NaF). The diluted lysates were applied to immunoprecipitation with anti-HA antibody and immunoblotting with anti-Flag antibody.
For the in vitro ubiquitination assay, the procedures for ubiquitin labeling were as described (Liu et al. 2002; Furukawa et al. 2003; Hu et al. 2004). Briefly, to purify substrate, GST-TSC2 was ectopically expressed in 293T cells, extracted in RIPA lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS), and pulled down with glutathione Sepharose 4B beads (Amersham Biosciences) followed by 10 mM glutathione elution. To purify CUL4A, CUL1, FBL6, and FBW5 ligases, Myc3-CUL4A, Myc3-CUL1, Myc3-FBL6, and Myc3-FBW5 immunocomplexes were immunoprecipitated using anti-Myc antibody from transfected 293T cells, immobilized on protein A-agarose beads, and washed three times with an NP-40 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% NaCl, 50 mM NaF) and twice with a ligase assay buffer (25 mM Tris-HCl at pH 7.5, 50 mM sodium chloride, 1 mM EDTA, 0.01% NP-40, 10% glycerol). Immobilized immunocomplexes were eluted by incubating with a molar excess of Myc peptide. For the in vitro TSC2 ubiquitination, the different combinations of CUL4A, CUL1, Fbw5, and Fbl6 immunocomplexes were mixed with GST-TSC2 substrate, and the mixtures were added to an ubiquitin ligation reaction (final volume 30 μL) containing the following: 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM sodium fluoride, 10 nM okadaic acid, 2 mM ATP, 0.6 mM dithiothreitol, 12 μg of bovine ubiquitin, 1 μg of ubiquitin (Sigma), 60 ng of E1, and 300 ng of E2 (hUbc5c). Reactions were incubated for 60 min at 30°C, terminated by boiling for 5 min in an SDS sample buffer containing 0.1 M dithiothreitol, and resolved on an SDS-PAGE gel before immunoblotting with the anti-GST antibody to examine ubiquitin ladder formation.
Drosophila mutants
Cul411L, Cul46AP, and Ddb1PL12c alleles were generated by imprecise excision of the P element in Cul4EP2518 and Ddb1EY01408 chromosomes, respectively, and will be described in detail elsewhere. The Cul4EP2518, Ddb1pic2, Ddb1S026316, and Ddb1EY01408 alleles were obtained from the Bloomington Stock Center. gig192 was a kind gift of D.J. Pan.
Quantification of mean eclosion day and percentage of eclosed flies
DDB1PL12c/TM3 Sb, Ser P[act-GFP] females were crossed to either DDB1pic2/TM3 Sb, Ser P[act-GFP] or DDB1pic2 gigas/TM3 × DDB1pl12c/TM3 males, and Ddb1 mutant and heterozygous sibling progeny were counted. For mean eclosion day calculations, the first day any fly from the culture eclosed established day 1, and the genotype of all flies was scored each day for nine and 12 successive days at 25°C and 18°C, respectively. The experiment was performed in triplicate. A 95% Confidence Interval test was used to obtain the range of the true mean of the combined data at each temperature. A two-tailed t-test assuming unequal variances was used to compare eclosion day data between different genotypes. The percentage of eclosed flies was calculated using the combined data at each temperature, and different genotypes were compared using a Fisher’s exact test.
Acknowledgments
We thank D.J. Pan for providing reagents, Corbin Jones for helping with the statistical analysis of Drosophila genetic data, Bill Marzluff for discussion, Lisa Wolff for technical assistance, and members of the Duronio and Xiong laboratory for discussion and support throughout this work. This study is supported by an American Cancer Society grant RSG-04-179-01-DDC to R.J.D. and an NIH grant (GM067113) to Y.X.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1624008.
References
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J.W., Elledge S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Chong-Kopera H., Inoki K., Li Y., Zhu T., Garcia-Gonzalo F.R., Rosa J.L., Guan K.L. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J. Biol. Chem. 2006;281:8313–8316. doi: 10.1074/jbc.C500451200. [DOI] [PubMed] [Google Scholar]
- Clark S.H., Chovnick A. A selective screen for transposable element mobilization in Drosophila melanogaster. Environ. Mutagen. 1985;7:439–449. doi: 10.1002/em.2860070405. [DOI] [PubMed] [Google Scholar]
- Clark S.H., Chovnick A. Studies of normal and position-affected expression of rosy region genes in Drosophila melanogaster. Genetics. 1986;114:819–840. doi: 10.1093/genetics/114.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino P.B., Nathanson K.L., Henske E.P. The tuberous sclerosis complex. N. Engl. J. Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Engelman J.A., Luo J., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Feldman R.M.R., Correll C.C., Kaplan K.B., Deshaies R.J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Furukawa M., He Y.J., Borchers C., Xiong Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4–ROC1 ubiquitin ligases. Genes & Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. CUL4–DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Hu J., McCall C.M., Ohta T., Xiong Y. Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K., Corradetti M.N., Guan K.L. Dysregulation of the TSC–mTOR pathway in human disease. Nat. Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Jin J., Arias E.E., Chen J., Harper J.W., Walter J.C. A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Liu J., Furukawa M., Matsumoto T., Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1–SKP1 binding and SCF ligases. Mol. Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Michel J.J., Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- Nellist M., Sancak O., Goedbloed M.A., Rohe C., van Netten D., Mayer K., Tucker-Williams A., van den Ouweland A.M., Halley D.J. Distinct effects of single amino-acid changes to tuberin on the function of the tuberin–hamartin complex. Eur. J. Hum. Genet. 2005;13:59–68. doi: 10.1038/sj.ejhg.5201276. [DOI] [PubMed] [Google Scholar]
- Ohta T., Michel J.J., Schottelius A.J., Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Rushlow C.A., Chovnick A. Heterochromatic position effect at the rosy locus of Drosophila melanogaster: Cytological, genetic and biochemical characterization. Genetics. 1984;108:589–602. doi: 10.1093/genetics/108.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway S.D., Li Y., Xiong Y. 14–3–3β binds to and negatively regulates the tuberous sclerosis complex 2 (TSC2) tumor suppressor gene product, tuberin. J. Biol. Chem. 2003;278:2089–2092. doi: 10.1074/jbc.C200499200. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig K., Tyers M., Elledge S.J., Harper J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Zacharek S.J., Xiong Y., Shumway S.D. Negative regulation of TSC1–TSC2 by mammalian D-type cyclins. Cancer Res. 2005;65:11354–11360. doi: 10.1158/0008-5472.CAN-05-2236. [DOI] [PubMed] [Google Scholar]