Abstract
Silencing of transposable elements occurs during fetal gametogenesis in males via de novo DNA methylation of their regulatory regions. The loss of MILI (miwi-like) and MIWI2 (mouse piwi 2), two mouse homologs of Drosophila Piwi, activates retrotransposon gene expression by impairing DNA methylation in the regulatory regions of the retrotransposons. However, as it is unclear whether the defective DNA methylation in the mutants is due to the impairment of de novo DNA methylation, we analyze DNA methylation and Piwi-interacting small RNA (piRNA) expression in wild-type, MILI-null, and MIWI2-null male fetal germ cells. We reveal that defective DNA methylation of the regulatory regions of the Line-1 (long interspersed nuclear elements) and IAP (intracisternal A particle) retrotransposons in the MILI-null and MIWI2-null male germ cells takes place at the level of de novo methylation. Comprehensive analysis shows that the piRNAs of fetal germ cells are distinct from those previously identified in neonatal and adult germ cells. The expression of piRNAs is reduced under MILI- and MIWI2-null conditions in fetal germ cells, although the extent of the reduction differs significantly between the two mutants. Our data strongly suggest that MILI and MIWI2 play essential roles in establishing de novo DNA methylation of retrotransposons in fetal male germ cells.
[Keywords: Piwi, piRNA, retrotransposon, DNA methylation, spermatogenesis]
Argonaute proteins, also known as PAZ Piwi domain (PPD) proteins, are members of a well-conserved family that is expressed in a variety of organisms, from fission yeasts to humans. The family can be divided into two subfamilies, Piwi and Ago, based on the primary sequence homology and expression pattern of each member. Piwi subfamily members are expressed only in germ lineage cells, whereas members of the Ago subfamily are expressed ubiquitously. The PPD proteins were initially characterized as essential molecules for stem cell self-renewal and maintenance in Drosophila, Caenorhabditis elegans, and certain plant species (Cox et al. 1998; Moussian et al. 1998), and a member of the family is essential for stem cell function during regeneration in planaria (Reddien et al. 2005). There are three Piwi subfamily genes in the mouse genome: Miwi (mouse piwi), Miwi2, and Mili (miwi-like), which are termed Piwil1 (piwi-like homolog 1), Piwil4, and Piwil2, respectively, in the official nomenclature. Although they are expressed only in germ lineage cells, the expression patterns of the three genes are different during germ cell differentiation. However, it is noteworthy that both Mili- and Miwi2-targeted mice are sterile because of impaired spermatogenesis at the pachytene stage (Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007).
Spermatogenesis is one of the most dramatic biological processes, as it involves cellular proliferation, differentiation, and morphogenesis. Primordial germ cells (PGCs), which emerge at embryonic day 7.5 (E7.5), reach the genital ridges up to E11.5. Male germ cells enter mitotic arrest around E13.5 and restart cell proliferation on postnatal day 2 (Sakai et al. 2004). De novo DNA methylation occurs during the quiescent phase in the fetal testes. After birth, the first wave of spermatogenesis proceeds in a synchronous manner. Following mitotic division, the first meiotic division commences on day 10. Several stages of meiosis are distinguished on the basis of differences in the constitution and relationships of the pairs of homologous chromosomes and their degrees of condensation. During differentiation, male germ cells enter the preleptotene/leptotene, zygotene, pachytene, and diplotene stages on days 10, 12, 14, and 17, respectively (Bellve et al. 1977). Thereafter, the second meiotic division takes place, with the round spermatids first appearing around postnatal day 20, and spermatogenesis continues in the adult testis. Therefore, as shown in Figure 1A, the germ cells in male gonads are different in the fetal, neonatal, and adult stages.
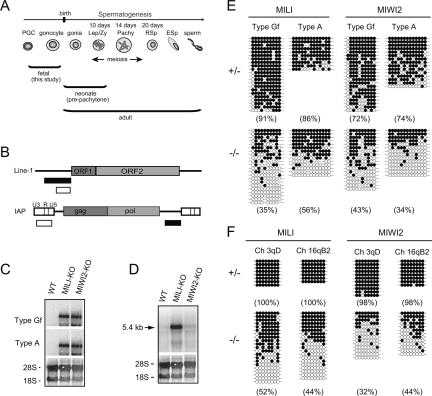
Figure 1.
Expression of the IAP and Line-1 retrotransposons and methylation of their regulatory regions in neonatal prepachytene testes. (A) Scheme for the development of mouse male germ cells. (Gonia) Spermatogonia; (Lep) leptotene; (Zy) zygotene; (Pachy) pachytene; (RSp) round spermatid; (Esp) elongated spermatid. (B) Schematic diagram of the Line-1 and IAP genes. The locations of the probes used for Northern blotting and bisulfite sequencing are indicated by filled and open boxes, respectively. The sequences of the 5′-noncoding regions of the Line-1 genes are different for type Gf and type A. The probe for the 3′-noncoding region of IAP recognizes the full-length and all deletion derivatives of IAP. (C,D) Northern blotting showing transcription of the Line-1 (C) and IAP (D) retrotransposon genes in testes from 2-wk-old MILI- and MIWI2-deficient mice. The 5′-noncoding regions of type Gf and type A Line-1 and the 3′-noncoding region of IAP were used as probes. (E,F) Bisulfite sequencing of Line-1 (E) and IAP (F) in MILI- and MIWI2-deficient germ cells. Spermatogonial cells from 6- to 12-d-old mice were purified from Oct4-EGFP transgenic mouse (Yoshimizu et al. 1999) testes. (E) The 5′-noncoding regions of type Gf and type A Line-1 (nucleotides 874–1156; GenBank accession no. D84391) and nucleotides 1251–1542 of M13002 were analyzed. (F) Two LTR regions from the 5.4-kb IΔ1-type IAP in chromosomes 3qD and 16qB2 were arbitrarily chosen for analysis. Filled and open circles represent methylated and unmethylated CpGs, respectively. The percentage of methylated CpGs is shown in parentheses.
Recently, it has been reported that PPD proteins play key roles in many gene-silencing phenomena involving small RNAs, including microRNA (miRNA)-directed or siRNA-directed target RNA cleavage, translational repression, and chromatin silencing. Post-transcriptional gene silencing by AGO subfamily-associated miRNAs or siRNAs has been reported (Peters and Meister 2007). MILI and MIWI were recently reported to bind 26-nucleoties (nt) to 31-nt Piwi-interacting small RNAs (piRNAs), respectively (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006) in the adult testis. Recently, MILI-associated piRNAs in the neonatal stage have been described as “prepachytene piRNAs” from day 10 testes (Aravin et al. 2007). However, the mechanisms of biogenesis and the biological functions of piRNAs are poorly understood compared with those of miRNAs and siRNAs.
Nearly half of the mammalian genome is composed of repeated sequences (Lander et al. 2001). Accumulating evidence suggests that PIWI-related mechanisms are involved in repressing the expression of retrotransposons, which are representative of the repeated sequences. Mutations in Piwi family genes have been shown to increase retrotransposon transcription in Drosophila (Sarot et al. 2004; Kalmykova et al. 2005), Trypanosoma (Shi et al. 2004), Neurospora (Nolan et al. 2005), and mice (Aravin et al. 2007; Carmell et al. 2007). Small RNAs against transposon sequences have been cloned in germlines, and the loss of Piwi family gene expression causes down-regulation of these small RNAs in Drosophila, zebrafish, and mice (Saito et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007). Based on these results, it has been suggested that the murine Piwi family genes are involved in retrotransposon gene silencing via small RNAs, probably repeat-associated piRNAs.
Retrotransposons are thought to be maintained in a transcriptionally silent state by DNA methylation (Walsh et al. 1998). During spermatogenesis, the DNA methylation status of the regulatory regions in retrotransposons, such as IAP (intracisternal A particle) and Line-1 (long interspersed nuclear elements), which belong to the long terminal repeat (LTR) and non-LTR retrotransposon families, respectively, changes dynamically (Tanaka et al. 2000; Lane et al. 2003). These regions are demethylated in PGCs around E12.5–E13.5, and the reacquisition of DNA methylation (i.e., de novo methylation) takes place in nondividing prospermatogonia (i.e., gonocytes) in the fetal testis around E16.5–E18.5. This de novo methylation involves either of the two de novo DNA methyltransferases, Dnmt3a and Dnmt3b, as well as their activating factor Dnmt3L, and analyses of Dnmt3L-deficient mice clearly show that this molecule is essential for the process (Bourc’his and Bestor 2004; Hata et al. 2006; Kato et al. 2007). Recently, it was reported that Mili- and Miwi2-targeted mice exhibited enhanced IAP and Line-1 expression, and methylation of the 5′-untranslated region (UTR) region of Line-1 was shown to be reduced in these mice at the neonatal stage (Aravin et al. 2007; Carmell et al. 2007). However, the DNA methylation status and piRNA expression of retrotransposons during de novo DNA methylation have not been examined. Thus, in the present study, we analyzed the piRNA expression and DNA methylation profiles of the IAP and Line-1 retrotransposons in fetal MILI-null and MIWI2-null testes.
Results
Comparison of Line-1 and IAP gene expression in MILI−/− and MIWI2−/− testes
To understand the roles of the mouse PIWI family proteins, we generated and analyzed MILI- (Aravin et al. 2007; Carmell et al. 2007) and MIWI2-deficient mice (Supplemental Fig. S1). In MILI- and MIWI2-null testes, spermatogenesis was arrested at the early prophase of meiosis I (Supplemental Fig. S1E). Affymetrix GeneChip microarray hybridization showed that five genes were up-regulated more than threefold in the MILI−/− testes, and all of these genes belonged to the IAP retrotransposon (Supplemental Fig. S2). The Line-1 and IAP gene expression levels were subsequently examined in the testes of 2-wk-old mice—i.e., testes that contained both premeiotic and meiotic germ cells—by Northern blotting (Fig. 1C,D). The transcripts of two representative Line-1 species, type Gf and type A, which have similar and unique sequences in their coding and 5′-noncoding regions, respectively, were similarly accumulated in both the MILI- and MIWI2-deficient testes. Meanwhile, expression of IAP was strongly and only slightly enhanced in the MILI- and MIWI2-null testes, respectively. Although the mouse genome contains full-length (7.2-kb) IAP elements and variants with deletions of various sizes in the gag-pol area (Ishihara et al. 2004), only the 5.4-kb IΔ1-type IAP transcript was enhanced in both mutants.
Reduced CpG methylation of Line-1 and IΔ1-type IAP in MILI−/− and MIWI2−/− germ cells
We examined the methylation status of the regulatory regions of Line-1 and IAP in MILI- and MIWI2-deficient male germ cells sorted 8–12 d after birth; namely, premeiotic germ cells. The promoter regions of type Gf and type A Line-1 in the sorted male germ cells were examined by bisulfite sequencing. As shown in Figure 1E, a significant reduction in CpG methylation was observed in the MILI- and MIWI2-deficient male germ cells at this stage. The reduction in methylation of type Gf Line-1 was confirmed by methylation-sensitive Southern blotting (Supplemental Fig. S3). Next, we analyzed the methylation pattern of CpG in two arbitrarily chosen LTR regions of the 5.4-kb IΔ1-type IAP on chromosomes 3 and 16. These regions were almost completely methylated in the control germ cells, whereas the levels of methylation were much lower in the MILI- and MIWI2-deficient germ cells (Fig. 1F).
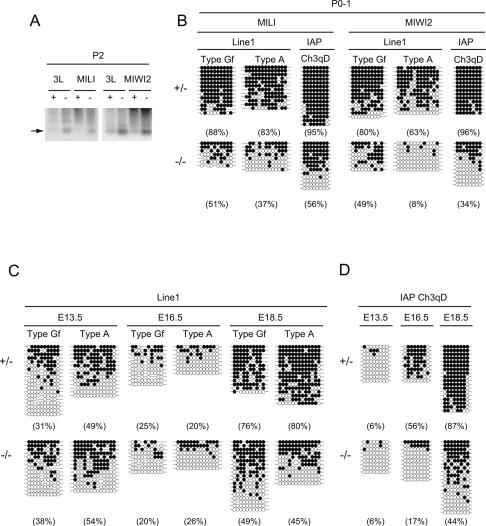
Dnmt3L is essential for the de novo methylation of retrotransposons in fetal testes, and the testicular phenotype of Dnmt3L-null mice is essentially the same as those of MILI- and MIWI2-null mice (Bourc’his and Bestor 2004; Webster et al. 2005; Hata et al. 2006). Thus, we examined the DNA methylation status of the Line-1 regulatory regions in MILI- and MIWI2-deficient neonatal testes, which contain premeiotic germ cells. Methylation-sensitive Southern blot analysis revealed that hypomethylation was already present on day 2 after birth (Fig. 2A), as was seen in Dnmt3L-mutant mice. Bisulfite sequencing confirmed the impaired CpG methylation status of the Line-1 and IΔ1-type IAP regulatory regions in MILI- and MIWI2-deficient germ cells on day 0–1 after birth (Fig. 2B). It seems unlikely that the defective methylation in the MILI-deficient mice was due to decreased expression of Dnmt3L and/or its presumptive cooperating DNA methyltransferase, Dnmt3a2, since the expression of these methylases was unaffected by the mutation in MILI (Supplemental Fig. S4).
Figure 2.
Methylation of the IAP and Line-1 regulatory regions in fetal testes. (A) Methylation-sensitive Southern blot analysis of the Line-1 promoter region. Whole-testis DNA was extracted from 2-d-old heterozygous (+) and homozygous (−) mice, followed by digestion with KpnI and the methylation-sensitive restriction enzyme HpaII. The probe for the type Gf Line-1 5′-noncoding region is the same as that in Figure 1C. (B) Bisulfite analysis of the Line-1 regulatory region in day 0–1 germ cells from MILI- and MIWI2-deficient and control testes. The germ cells were collected as described in Materials and Methods. (C,D) Bisulfite sequencing of the Line-1 (C) and IAP (D) regulatory regions in germ cells from MILI-deficient and control testes between E13.5 and E18.5. The germ cells were collected as described in Materials and Methods.
It is possible that MILI and MIWI2, as well as Dnmt3L, have roles in de novo methylation during male germ cell differentiation. Therefore, we carried out bisulfite sequencing of the regulatory regions of the Line-1 and IAP retrotransposons in MILI-deficient fetal germ cells around de novo methylation. In control germ cells, almost all of the CpGs in the LTRs of IAP were methylated soon after birth (Figs. 1F, 2B), whereas only incomplete methylation was observed in E13.5, E16.5, and E18.5 germ cells (Fig. 2D). De novo methylation of the Line-1 and IAP regulatory regions of control mice occurred after E16.5 and E13.5, respectively (Fig. 2C,D). De novo methylation of Line-1 genes was slightly delayed compared with that of the IAP genes. In the MILI-deficient germ cells, hypomethylation of Line1 and IAP was observed at E18.5 and after E16.5, respectively. Differences in the methylation status of Line-1 in the MILI-deficient mice became apparent somewhat later than that of IAP, which seemingly reflects the delay of de novo methylation in the control germ cells. Our observations indicate that MILI plays an essential role in the timing of de novo methylation of the Line-1 and IAP regulatory regions.
Characterization of small RNAs in fetal testes
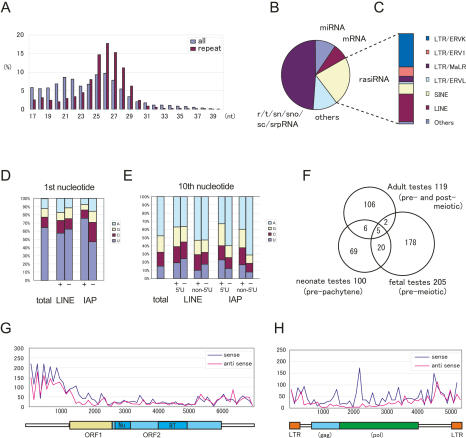
As discussed above, MILI and MIWI2 presumably function in gene silencing through DNA methylation in fetal testes. Based on the role of small RNAs from other organisms in gene silencing (Zaratiegui et al. 2007), it is possible that DNA methylation is mediated by small RNAs. Therefore, we analyzed many small RNA sequences (127,997 clones), 17–40 nt in length, from E12.5–E19.5 fetal male germ cells, to obtain a comprehensive picture of the piRNAs present at this stage, and compared our findings with the results of previous studies on piRNAs in neonatal (prepachytene) testes (Aravin et al. 2007) and adult testes (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). The lengths of the small RNAs from the germ cells showed a bimodal pattern (Fig. 3A). One peak was observed at ∼21 nt, which corresponds to the lengths of miRNAs, and a second was observed at 25–27 nt, which is consistent with the lengths of piRNAs.
Figure 3.
piRNAs in fetal premeiotic germ cells. (A) In total, 127,997 small RNAs were sequenced from E12.5–E19.5 fetal germ cells. The size distributions (in nucleotides) of the total small RNAs and rasiRNAs are shown by blue and purple bars, respectively. (B,C) Genomic annotation of the small RNAs (B) and the ratio of piRNA sequences of fetal premeiotic germ cells (C). Detailed results are listed in Supplemental Table S2. (D,E) Comparison of the first (D) and tenth (E) nucleotides of the total, sense (+), and antisense (−) piRNAs. Nucleotide biases were calculated for the Gf type Line-1 and IAP piRNAs analyzed in Supplemental Table S2. (E) The piRNA classes that contain or lack a 5′-end U are shown separately. (F) Venn diagram of the piRNAs in adult pre- and post-meiotic (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006), neonatal prepachytene (Aravin et al. 2007), and fetal (this study) testes. (G,H) Distribution of piRNAs corresponding to type Gf Line-1 (G) and IAPIΔ1 (H).
Annotation of the small RNAs revealed that the library was enriched with retrotransposon sequences, with the exception of the breakdown products of rRNAs and other noncoding RNAs (tRNA/snRNA/snoRNA/scRNA/srpRNA) (Fig. 3B; Supplemental Table S1). The repeat-associated small RNAs (rasiRNAs) in the library, which reportedly bind MILI, showed a single peak at 25–27 nt (Fig. 3A). The relative abundance of repeat-associated RNAs (23%) was similar to that of the MILI-associated prepachytene piRNAs identified in neonatal testis (26%) (Fig. 3B; Supplemental Table S1; Aravin et al. 2007). However, detailed characterization of the repeat-derived small RNAs revealed that there were some differences between the fetal germ cell piRNAs and the reported prepachytene piRNAs (Aravin et al. 2007). As shown in Supplemental Table S2 and Figure 3C, the majority of the rasiRNAs in the E16.5 male germ cells were LTR retrotransposons (55%); namely, ERVK (37%), ERV1 (10%), MaLR (6%), and ERVL (2%). The others were LINEs (30%) and SINEs (11%). In contrast, among the prepachytene piRNAs, the SINE frequency (49%) was higher than the LTR (33.8%) or LINE (15.8%) (Aravin et al. 2007) frequency. Unique pachytene piRNAs were scarcely detected in the fetal male germ cells (0.1%) (Supplemental Table S1). Next, as the small RNAs that correspond to Line-1 and IAP were abundant, we examined the nucleotide composition of the piRNAs corresponding to type Gf Line-1 and IAP retrotransposons. The first and tenth nucleotides of the piRNAs are shown in Figure 3, D and E. Most of the piRNAs started with uridine and had an adenine at the tenth position, which is similar to the characteristics of prepachytene piRNAs (Aravin et al. 2007).
Adult and neonatal piRNAs are clustered within the genome (Aravin et al. 2006, 2007; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). Therefore, we performed a cluster analysis of the male fetal gonadal small RNAs and detected 205 clusters (Supplemental Table S3), only seven and 25 of which were identified in adult and prepachytene testes, respectively (Fig. 3F). About 75% of the clustered small RNAs were 24–28 nt in length, and the percentage of small RNAs with U as the first nucleotide was high (71.5%). Therefore, we conclude that most of the clusters are piRNA clusters, and that the set of piRNAs expressed at the stage of de novo methylation is quite different from the piRNA sets expressed in the neonate and adult.
The distribution and frequency plots of the piRNAs that correspond to the type Gf Line-1 and IAPIΔ1 genes are shown in Figure 3, G and H, respectively. For Line-1, the number of piRNAs that corresponded to the regulatory region was higher than that corresponding to the coding region, and the distributions of sense and antisense piRNAs were indistinguishable. Meanwhile, for the IAPIΔ1 genes, the number of sense piRNAs was slightly higher than the number of antisense piRNAs, and there were many piRNAs that corresponded to the coding region as well as to the regulatory region.
Reduced levels of piRNAs in MILI- and MIWI2-deficient fetal testes
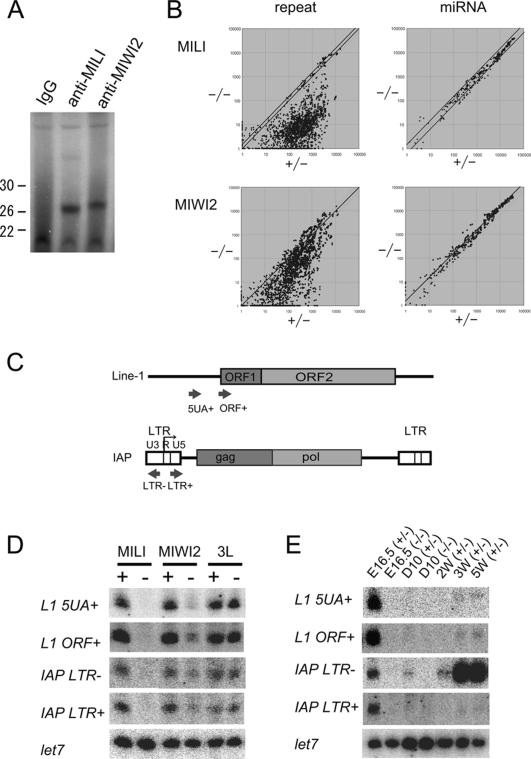
Although both Mili and Miwi2 were expressed in fetal testes, the expression period of Miwi2 was more restricted (Supplemental Fig. S5) than that of Mili (Kuramochi-Miyagawa et al. 2001). Miwi2 expression was reduced after birth, whereas Mili expression continued at essentially the same level throughout life. To examine the functions of MILI and MIWI2 in fetal testes, we determined whether the proteins bound to small RNAs. Ribonucleoprotein complexes from E16.5 testicular lysates were immunoprecipitated, the associated RNAs were isolated, and 32P-labeling was carried out. As shown in Figure 4A, several RNAs of length 26–27 nt were coimmunoprecipitated with MILI and MIWI2.
Figure 4.
piRNA expression in MILI- and MIWI2-deficient fetal testes. (A) MILI- and MIWI2-bound piRNAs. The immunoprecipitated RNAs from E16.5 testicular lysates were 32P-end-labeled and separated in a 15% denaturing urea–polyacrylamide gel. (B) Microarray analysis of the repeat-associated piRNAs and miRNAs. A microarray that contained 672 types of repeat-associated piRNAs and 150 types of miRNAs, both of which are expressed in fetal testes, was produced. RNA samples isolated from E16.5 control, MILI-deficient, and MIWI2-deficient mice were used. The expression of each small RNA was examined three times, and the mean values are plotted to compare the control and MILI- or MIWI2-deficient samples. Diagonal lines indicate a 1.5-fold difference in expression. (C–E) Expression of four arbitrarily selected piRNAs. piRNAs corresponding to the type A Line-1 promoter sense strand (5UA+), Line-1 ORF sense strand (ORF+), IAP LTR sense strand (LTR+), and IAP LTR antisense strand (LTR−) were arbitrarily selected. (C) The site and orientation of each piRNA is indicated by red arrows. (D) Northern blot analysis of the piRNAs in E16.5 control, MILI-deficient, MIWI2-deficient, and Dnmt3L-deficient testis. (E) Time-course analysis of the expression of the four piRNAs in control and MILI-null testes.
Next, we used microarray analysis to test the roles of MILI and MIWI2 in the production of small RNAs in fetal testes. Using this method, we examined the expression patterns of 670 types of repeat-associated piRNAs and 150 types of miRNAs. As shown in Figure 4B, the vast majority of the piRNAs were significantly down-regulated in MILI-null fetal testes. Although this down-regulation was also observed in MIWI2-null testes, the mean fold reduction was smaller by one order of magnitude. In MILI-null fetal germ cells, ∼88% of the piRNAs were present at less than one-eighth of the level in the control cells. In contrast, only 29% of the piRNAs in the MIWI2-null fetal germ cells were reduced to this level.
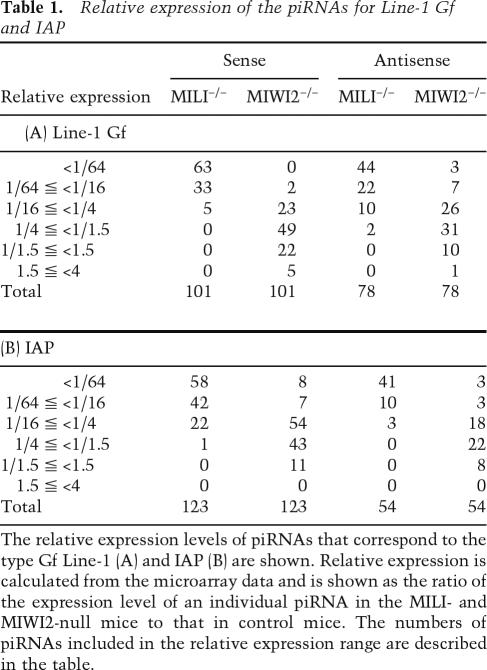
The relative levels of expression of the piRNAs that correspond to type Gf Line-1 and IAP are summarized in Table 1. In MILI-null testes, the expression levels of all of the type Gf Line-1 sense piRNAs and the vast majority of the type Gf Line-1 antisense piRNAs were less than one-quarter of the control. In contrast, the level of reduction was significantly lower in MIWI2-null testes. About 25% (27 of 101) and 14% (11 of 78) of the sense and antisense piRNAs, respectively, did not show significantly reduced expression in MIWI2-null testes. Notably, all of the piRNAs whose expression was unaffected by the MIWI2-null mutation were located in ORF1 and the endonuclease region of ORF2, whereas none of these piRNAs were located in the reverse transcriptase region of ORF2. As with the type Gf Line-1 piRNAs, the expression levels of almost all of the piRNAs for IAP in the MILI-null testes were less than one-quarter of the control (Table 1), and the reduction was significantly less pronounced in the MIWI2-null testes. Unlike the case of Line-1, these piRNAs were not located in any special region.
Table 1.
Relative expression of the piRNAs for Line-1 Gf and IAP
The relative expression levels of piRNAs that correspond to the type Gf Line-1 (A) and IAP (B) are shown. Relative expression is calculated from the microarray data and is shown as the ratio of the expression level of an individual piRNA in the MILI- and MIWI2-null mice to that in control mice. The numbers of piRNAs included in the relative expression range are described in the table.
We examined in greater detail the expression levels of four arbitrarily chosen piRNAs by Northern blot analysis using antisense oligonucleotides against small RNAs of 25–28 nt in length: the sense strand 5′-UTRs of type A (5UA+) and ORF (ORF+) for Line-1, and the sense and antisense strands of LTR (LTR+ and LTR−, respectively) for IAP (Fig. 3C). The sequences of LTR+ and LTR− were identical to those of the U3 region and R plus U5 region of LTR, respectively, and the expression levels of the four small RNAs were found to be quite similar in the E16.5 testes. As shown in Figure 4D, the four small RNAs were not detected in MILI-null testes, although they were weakly detected in MIWI2-null testes and expressed normally in Dnmt3L-null testes. The expression of the piRNAs was further examined in control and MILI-null testes during development, and 5UA+, ORF+, and LTR+ showed similar expression patterns (Fig. 4E). All three were expressed in fetal germ cells but were almost undetectable on day 10 and 2 wk after birth. Weak expression was detected 3 and 5 wk after birth. In contrast, the expression level of LTR− was higher 3 wk after birth than at E16.5.
Discussion
In the present study, we analyzed the mechanism of impaired retrotransposon silencing in MILI- and MIWI2-null male germ cells. DNA methylation of the regulatory regions of two retrotransposon species, Line-1 and IAP, was impaired from the stage of de novo methylation. Given that piRNAs are involved in gene silencing, we performed large-scale sequencing of the small RNAs in fetal male germ cells. Our data clearly indicate that the small RNA composition of fetal male germ cells is quite different from those of adult and neonatal prepachytene male germ cells. Finally, we compared the effects of MILI- and MIWI2-null mutations on the expression of repeat-associated piRNAs and found that these two mouse PIWI family proteins play similar but distinct roles in piRNA expression.
De novo DNA methylation and the murine PIWI proteins MILI and MIWI2
RT–PCR analysis showed that Miwi2 expression commenced at E15.5 in male gonadal tissue (Supplemental Fig. S5). Meanwhile, Mili RNA was detected in male gonadal tissues beginning at E12.5. Therefore, both MILI and MIWI2 are expressed during de novo DNA methylation. Male germ cells from both MILI- and MIWI2-null mutant mice exhibited impaired DNA methylation in the regulatory regions of Line-1 and IAP soon after birth (Fig. 2A,B), which suggests that these two proteins play crucial roles in DNA methylation at an early stage of germ cell development. In addition, impaired de novo methylation was observed in MILI-deficient fetal germ cells. The significant reduction in piRNAs against the repeat-associated piRNAs in MILI- and MIWI2-deficient fetal testes implies that piRNAs are active in de novo DNA methylation.
Changes in piRNA content during male germ cell development
The compositions of the piRNAs and piRNA clusters were quite different during the process of male germ cell development; i.e., at the embryonic, neonatal prepachytene (Aravin et al. 2007), and adult stages (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). Although the fetal and neonatal piRNAs were similar in that each included a significant number of repeat-associated RNAs, they shared only 10% of the species in their piRNA clusters. Recently, it has been proposed that piRNA production involves a ping-pong amplification cycle for retrotransposon-related piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). The high proportion of piRNAs with U at the first nucleotide position and A at the tenth nucleotide position in our analysis of fetal repeats corresponding to piRNAs fits well with this hypothesis. However, the different compositions of the embryonic, neonatal, and adult piRNAs suggest that the cycle does not continue throughout male germ cell development, since the piRNAs in fetal germ cells are not maintained in adult germ cells (Fig. 4E). Therefore, some other mechanism(s) of piRNA biogenesis must operate during the pachytene phase to establish piRNA expression in adult germ cells.
We arbitrarily chose four fetal piRNAs and examined their expression in detail (Fig. 4E). All four piRNAs were either undetectable or weakly expressed in neonatal germ cells, although expression was somewhat increased after a couple of weeks. This also suggests that the molecular mechanism of piRNA production changes during development. At the same time, it implies that the function of piRNAs in embryos is different from that in neonates and adults.
MILI and MIWI2 have different functions
Impaired de novo methylation was detected in both MILI- and MIWI2-null male fetal germ cells. Although a general reduction in piRNA expression was observed in the mutants, the extent of the reduction was significantly different. As shown in Figure 4B, the reduction in numbers of piRNAs against repetitive sequences was much greater in the fetal germ cells of MILI-null testis than in those of MIWI2-null testis. This may reflect a difference in the molecular mechanisms of piRNA biogenesis between MILI and MIWI2.
The expression of several piRNAs was not reduced at all in MIWI2-null germ cells. In fact, 27 of 101 sense and 11 of 78 antisense Line-1 piRNAs were found to belong to this group (i.e., relative expression ≥1/1.5 in Table 1A). In contrast, the expression of these same piRNAs was significantly reduced (i.e., relative expression <1/1.5 in Table 1A) in MILI-null germ cells. It is noteworthy that the vast majority of the piRNAs were located in ORF1 and the endonuclease region of ORF2 (Fig. 3G, ORF1 and Nu, respectively), although the significance of this result remains unclear. These data suggest that MILI and MIWI2 play different roles in the production, stabilization, and/or amplification cycle of piRNA.
Line-1 expression was similarly enhanced in MILI- and MIWI2-null testes (Fig. 1B), and bisulfite sequencing revealed that a significant reduction in DNA methylation occurred in both MILI- and MIWI2-null male germ cells (Fig. 2C). Considering that DNA methylation greatly influences retrotransposon transcription, the enhanced expression of Line-1 was correlated with DNA hypomethylation of the mutant male germ cells. The situation was a little different for IAPIΔ1 (Fig. 1B,D). Although defective DNA methylation was detected in both MILI- and MIWI2-null germ cells, IAPIΔ1expression was strongly increased in MILI-null testis, while it was either not increased or only slightly increased in MIWI2-null testis. These results imply different functions for MILI and MIWI2. Although it is difficult to explain the discrepancy, an as-yet-unknown post-transcriptional silencing mechanism may reduce IAP expression in MIWI2-null germ cells.
Molecular mechanisms of piRNA regulation
As described above, several aspects of piRNA regulation change significantly during development. For example, the piRNA composition differed among fetal, neonatal, and adult male germ cells. In addition, the first and tenth nucleotides and the distributions of piRNAs against the Line-1 or IAP sequence were different. Furthermore, although both MILI-null and MIWI2-null mice exhibited sterility owing to arrested spermatogenesis, the reductions in the percentages of piRNAs in their germ cells were significantly different. In addition, the regulatory mechanisms controlling IAPIΔ1 and Line-1 expression were different. It remains to be determined how the expression of IAPIΔ1, but not that of full-length IAP, is influenced by a null mutation of MILI or MIWI2. Finally, the mechanism of de novo methylation, presumably through piRNAs, remains an open question. Currently, it is difficult to propose a unified molecular mechanism to explain our results. Additional analyses will clarify the mechanisms underlying piRNA production and gene silencing.
Materials and methods
RNA extraction, RT–PCR, and Northern blot analysis
Total RNA samples were prepared from testes using Sepasol-RNA I Super (Nacalai Tesque), treated with DNase I, and subjected to RT–PCR with the ThermoScript RT–PCR System (Invitrogen) and random hexamers. Each reaction was performed using HotMaster Taq DNA polymerase and specific primers (Supplemental Table S6).
Northern blot analysis was performed at 65°C in 0.2 M NaHPO4 (pH 7.2), 1 mM EDTA, 1% BSA, and 7% SDS. The membranes were washed with a 0.2× SSC/0.1% SDS solution at 65°C. The subcloned PCR products were labeled with [α-32P]-dCTP and used as probes. The sequences used for PCR primers were as follows: the 3′-noncoding region of IAP (GenBank accession no. X04120), nucleotides 4489–4793; and the 5′-noncoding region of type Gf (D84391), nucleotides 874–1156, and A Line-1 (M13002), nucleotides 531–1642.
Methylation-sensitive Southern blot analysis
Whole-testis DNA was extracted from 2-d-old mice, and 3 μg of DNA were digested with KpnI and the methylation-sensitive restriction enzyme HpaII or the methylation-insensitive enzyme MspI. The type Gf Line-1 5′-noncoding region shown in Figure 1A was used as the probe.
Germ cell isolation
MILI mutant mice (Kuramochi-Miyagawa et al. 2004) and MIWI2 mutant mice were crossed with Oct-4/GFP transgenic mice (Yoshimizu et al. 1999) to obtain GFP-positive spermatogonia. Testis cells from embryos or pups were collected by two-step enzymatic digestion (Meistrich 1993), and the GFP-positive cells were sorted by FACS Aria (Becton Dickinson). Genomic DNA was extracted from the sorted germ cells.
Bisulfite methylation analysis
Bisulfite treatment of the genomic DNA isolated from fetal germ cells was performed using the EpiTect bisulfite kit (Qiagen). Two LTR regions of IAP (5.4-kb, IΔΙ-type) on chromosomes 3qD and 16qB2 were arbitrarily selected for analysis by nested PCR, and the products were sequenced. The first and second rounds of PCR were carried out using the primers F1 and R1, and F2 and R2, respectively. PCR amplification of the 5′-region of Line-1 (types Gf and A) was carried out using specific primers. The sequence of each primer is listed in Supplemental Table S6.
Anti-MILI and anti-MIWI2 antibodies
Affinity-purified anti-MILI-N2 (anti-mouse PIWIL2 [MILI], PM044; MBL Co., Ltd.) and anti-MIWI2-N1 polyclonal antibodies against MILI and MIWI2 were generated by immunization with peptides derived from MILI (amino acids 107–122: VRKDREEPRSSLPDPS) and MIWI2 (amino acids 31–45: TSASPGDSEAGGGTSC), respectively (MBL).
Small RNA cloning and sequencing
To isolate small RNAs, 20 μg of total RNA from E12.5–E19.5 male germ cells were gel-fractionated, and species 17–40 nt in length were gel-purified. The small RNAs were sequentially ligated to 3′- and 5′-adapters and then amplified by RT–PCR using a small RNA cloning kit (RR065; Takara Bio). Sequencing of the small RNA library was achieved using the 454 Life Science sequencer.
Annotation of the small RNAs
To identify the small RNAs that corresponded to various repeats (e.g., rRNA, tRNA, retrotransposon, and DNA transposon), the genomic positions of the repeats were retrieved from the University of California at Santa Cruz (UCSC) Web site (http://hgdownload.cse.ucsc.edu/downloads.html) and compared with the genomic positions of the small RNAs. When the genomic position of a particular small RNA overlapped with that of any repeat by up to 15 nt, the small RNA was considered to be repeat-derived. Repeat names were retrieved from all the positions to which the small RNA mapped, and when multiple repeat names were retrieved, the class (e.g., LTR/MaLR and rRNA), and subclass (e.g., IAP), where applicable, were determined by the repeat with the greatest number of positions (Supplemental Table S2). When the top two repeats had the same number of positions, we did not determine the class or subclass. To identify small RNAs that corresponded to tRNAs, rRNAs, snRNAs, snoRNAs, scRNAs, miRNAs, piRNAs (known species from adult testes), and mRNAs based on sequence similarity, we extracted the sequences of the RNAs from GenBank (flat files, ftp://ftp.ncbi.nlm.nih.gov/genbank) and downloaded the remaining sequences from the following databases: for tRNAs, Genomic tRNA database (http://lowelab.ucsc.edu/GtRNAdb/Mmusc); for rRNAs, European ribosomal RNA database (http://www.psb.ugent.be/rRNA/index.html); for snoRNAs, snoRNA database (http://www-snorna.biotoul.fr) and RNA database (http://jsm-research.imb.uq.edu.au/rnadb); for piRNAs, RNA database (http://jsm-research.imb.uq.edu.au/rnadb); for miRNAs, miRBase (http://microrna.sanger.ac.uk/sequences/index.shtml); and for mRNAs, Refseq Genes (http://ftp.ncbi.nih.gov/refseq) and Ensemble Genes (http://www.ensembl.org/index.html). A BLAST search (http://ftp.ncbi.nih.gov/blast) was subsequently performed using our small RNA sequences as queries and our downloaded sequences as a database. Since sequence alignments using the BLAST program are local, a gap can hamper the alignment of short homologous sequences flanking the gap. Therefore, we extracted the sequences of the aligned regions from the data set with an extra 5 nt at both ends and realigned them with the small RNA sequences, using a global alignment program developed in-house that uses plural gap penalty parameter sets. We then classified the small RNAs using a 90% sequence identity threshold. Finally, the repeat annotations based on genomic position and the annotations based on sequence similarity were combined. For small RNAs with more than one annotation, we used the following order of priority: rRNA, tRNA, snoRNA, sc/srpRNA, miRNA, rasiRNA, piRNA, and mRNA. The unannotated sequences were classified as unknown.
Distribution and frequency of piRNAs corresponding to IAP and Line-1
The sequences of IAP1 (M17551) and L1_MdA (nucleotides 588–7713 of M13002) were retrieved from the flat files of GenBank. All of the small RNA sequences cloned were BLAST-searched against the IAP1 and L1_MdA sequences, using a cutoff E-value of 0.0001. The number of hits was determined every 100 nt.
Cluster analysis of small RNAs
We performed a cluster analysis of the male fetal gonadal small RNAs, with the following conditions: (1) The cluster should contain at least 11 types of small RNAs, and (2) the relative abundance of small RNAs between 24 and 28 nt in length should be greater than that of small RNAs between 19 and 23 nt in length.
Immunoprecipitation of the MILI ribonucleoprotein complex, and the isolation, labeling, and detection of small RNAs
E16.5 testes were homogenized in lysis buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.1% Nonidet P-40, 1 mM DTT) that contained protease inhibitors. Protein–RNA complexes were immunoprecipitated using anti-MILI-N2 and anti-MIWI2-N1 antibodies or normal rabbit serum as a control with Protein G Sepharose beads, and the RNAs were purified using ISOGEN-LS (Nippon Gene). The RNAs were labeled with [γ-32P]-ATP using T4 polynucleotide kinase for 15 min at 37°C and then separated by 15% denaturing PAGE.
Microarray analysis
Total RNA from E16.5–E17.5 testes was purified using Sepasol-RNA I Super. For the rasiRNA probes, those sequences that were cloned more than six times and were >22 nt in length were selected from among the small RNAs that were annotated as repeats. In those cases where the small RNAs overlapped by 23 nt at their 5′-ends, a representative sequence was selected according to the number of clones. For the miRNA probes, we chose sequences that were cloned more than five times and were <23 nt in length from among the small RNAs that were annotated as miRNAs. In those cases where the small RNAs overlapped by 17 nt at their 5′-ends, a representative sequence was selected according to the number of clones. The target sequences for the microarray are listed in Supplemental Table S5. Microarray analysis was performed by a service provider (LC Sciences).
Small RNA Northern blotting
Northern blots were used to detect small RNAs, as described previously (Aravin et al. 2006), with 10 μg of total RNA loaded per well. The sequences of the probes used to detect the small RNAs are described in Supplemental Table S6. Hybridization was performed at 42°C in the same buffer used for regular Northern blotting. The membrane was washed twice with a 2× SSC/0.1% SDS solution at 42°C.
Acknowledgments
We thank Dr. Y. Matsui for providing the Oct-4/GFP mice, Dr. S. Tajima for providing the anti-Dnmt antibodies, and Ms. A. Kawai and Mr. K. Yoshinaga for assistance. We also thank Dr. K. Horie and Dr. Y. Kato for helpful discussions, and Ms. A. Mizokami for secretarial work. This work was supported in part by grants from the Japan Society for the Promotion of Science, the Ministry of Education, Culture, Sports, Science and Technology, the 21st Century COE Program “CICET,” and the Uehara Memorial Foundation, Japan.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1640708.
References
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Bellve A.R., Cavicchia J.C., Millette C.F., O’Brien D.A., Bhatnagar Y.M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J., Bourc’his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Baker J., Chang L., Qiao D., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hata K., Kusumi M., Yokomine T., Li E., Sasaki H. Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Mol. Reprod. Dev. 2006;73:116–122. doi: 10.1002/mrd.20387. [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Ishihara H., Tanaka I., Wan H., Nojima K., Yoshida K. Retrotransposition of limited deletion type of intracisternal A-particle elements in the myeloid leukemia Clls of C3H/He mice. J. Radiat. Res. (Tokyo) 2004;45:25–32. doi: 10.1269/jrr.45.25. [DOI] [PubMed] [Google Scholar]
- Kalmykova A.I., Klenov M.S., Gvozdev V.A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T. Two mouse piwi-related genes: miwi and mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lane N., Dean W., Erhardt S., Hajkova P., Surani A., Walter J., Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Meistrich M.L. Cell and molecular biology of the testis. Oxford University Press; New York: 1993. [Google Scholar]
- Moussian B., Schoof H., Haecker A., Jurgens G., Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Braccini L., Azzalin G., De Toni A., Macino G., Cogoni C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 2005;33:1564–1573. doi: 10.1093/nar/gki300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L., Meister G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Reddien P.W., Oviedo N.J., Jennings J.R., Jenkin J.C., Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Suetake I., Shinozaki F., Yamashina S., Tajima S. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Brain Res. Gene Expr. Patterns. 2004;5:231–237. doi: 10.1016/j.modgep.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Sarot E., Payen-Groschene G., Bucheton A., Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ullu E., Tschudi C. Function of the Trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J. Biol. Chem. 2004;279:49889–49893. doi: 10.1074/jbc.M409280200. [DOI] [PubMed] [Google Scholar]
- Tanaka S.S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes & Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster K.E., O’Bryan M.K., Fletcher S., Crewther P.E., Aapola U., Craig J., Harrison D.K., Aung H., Phutikanit N., Lyle R., et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Scholer H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]