Abstract
Recent studies suggest that increased mitochondrial metabolism and the concomitant decrease in NADH levels mediate calorie restriction (CR)-induced life span extension. The mitochondrial inner membrane is impermeable to NAD (nicotinamide adenine dinucleotide, oxidized form) and NADH, and it is unclear how CR relays increased mitochondrial metabolism to multiple cellular pathways that reside in spatially distinct compartments. Here we show that the mitochondrial components of the malate–aspartate NADH shuttle (Mdh1 [malate dehydrogenase] and Aat1 [aspartate amino transferase]) and the glycerol-3-phosphate shuttle (Gut2, glycerol-3-phosphate dehydrogenase) are novel longevity factors in the CR pathway in yeast. Overexpressing Mdh1, Aat1, and Gut2 extend life span and do not synergize with CR. Mdh1 and Aat1 overexpressions require both respiration and the Sir2 family to extend life span. The mdh1Δaat1Δ double mutation blocks CR-mediated life span extension and also prevents the characteristic decrease in the NADH levels in the cytosolic/nuclear pool, suggesting that the malate–aspartate shuttle plays a major role in the activation of the downstream targets of CR such as Sir2. Overexpression of the NADH shuttles may also extend life span by increasing the metabolic fitness of the cells. Together, these data suggest that CR may extend life span and ameliorate age-associated metabolic diseases by activating components of the NADH shuttles.
[Keywords: Calorie restriction (CR), Sir2, aging, NADH shuttles, respiration, metabolism]
Calorie restriction (CR) is the most effective intervention known to extend life span in a variety of species including mammals, suggesting a possible conservation between longevity-regulating pathways in different species (Weindruch and Walford 1998; Roth et al. 2001). CR has also been shown to delay the onset or reduce the incidence of many age-related diseases such as cancer and diabetes (Weindruch and Walford 1998). Recent studies in model systems have identified CR-specific longevity genes, providing insight into the molecular mechanism underlying CR-induced effects (Lin et al. 2000; Bishop and Guarente 2007; Easlon et al. 2007; Panowski et al. 2007). In yeast, CR requires NAD (nicotinamide adenine dinucleotide, oxidized form) and Sir2 (Lin et al. 2000, 2002; Wood et al. 2004; Lamming et al. 2005), a key regulator of life span in both yeast and animals (Kaeberlein et al. 1999; Tissenbaum and Guarente 2001). Sir2 exhibits an NAD-dependent histone deacetylase activity that is conserved among the Sir2-family members (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). By coupling the cleavage of NAD and deacetylation of target proteins, Sir2 serves as a molecular link, relaying the cellular energy state to the machinery of life span regulation. The Sir2 family in higher eukaryotes also plays an important role in CR (Rogina and Helfand 2004; Chen et al. 2005; Nisoli et al. 2005; Wang and Tissenbaum 2006). In addition, the mammalian Sir2 family proteins (SIRT1–7) have been shown to regulate stress response, cell survival, as well as insulin and fat metabolism (Guarente 2006; Dilova et al. 2007), suggesting a role for SIRT proteins in age-related metabolic diseases and perhaps human aging.
The budding yeast Saccharomyces cerevisiae provides an efficient model for studying longevity regulation. Owing to the short life cycle and well-established molecular genetic techniques in this system, yeast mutants and the corresponding genes that alter life span are relatively easy to obtain. Moderate CR can be imposed in the budding yeast S. cerevisiae by reducing the glucose concentration from 2% to 0.5% in rich media (Lin et al. 2000, 2002, 2004; Kaeberlein et al. 2002; Anderson et al. 2003; Lamming et al. 2005; Easlon et al. 2007). Under moderate CR condition, the growth rate remains robust and yeast mother cells show an extended replicative life span (RLS) (division potential) of ∼20%–30%. Variations in CR protocols have also been described in which limitation of amino acids and/or further reduction in carbon source are employed (Jiang et al. 2000, 2002). Many studies have shown that the Sir2 family plays an important role under moderate CR conditions (0.5% glucose) (Lin et al. 2000, 2002; Wood et al. 2004; Lamming et al. 2005; Easlon et al. 2007); however, Sir2 appeared to be dispensable under more severe CR conditions (Jiang et al. 2000, 2002; Kaeberlein et al. 2004; Lamming et al. 2005; Easlon et al. 2007). Although these CR regimens may represent other longevity pathways that function in parallel to moderate CR in rich media, it remains highly possible that moderate CR also induces other proteins in addition to the Sir2 family to extend life span (Easlon et al. 2007).
It has been suggested that moderate CR induces Sir2-dependent life span extension by decreasing the level of nictotinamide (NAM), a byproduct of NAD degradation in the Sir2-mediated deacetylation reaction (Bitterman et al. 2002). Overexpression of the nicotinamidase, Pnc1, increases yeast life span and suppresses the inhibitory effect of NAM on Sir2 deacetylase activity (Anderson et al. 2003; Gallo et al. 2004). Alternatively but not mutually exclusive, this CR condition also induces a shunting of carbon metabolism from fermentation to the mitochondrial TCA cycle (Lin et al. 2002). The concomitant increase in respiration is necessary and sufficient for the activation of Sir2-mediated silencing and extension in life span (Lin et al. 2002). The fact that respiration produces NAD from NADH (Bakker et al. 2001; Cruz et al. 2001), as well as the finding that NADH can function as a competitive inhibitor of Sir2 activity (Lin et al. 2004), reinforce the idea that an increase in the NAD/NADH ratio activates Sir2 during CR. A link between CR and increased mitochondrial metabolism has also been reported in higher eukaryotes including mammals (Nisoli et al. 2005; Bishop and Guarente 2007). It is noteworthy that many metabolic enzymes are regulated by the NAD/NADH ratio and are also likely to mediate CR-induced life span extension. We previously characterized one such metabolic target of CR, PDC, the conserved NAD-dependent pyruvate dehydrogenase complex (Easlon et al. 2007). The PDC acts in the mitochondria to convert pyruvate into acetyl-CoA, (Pronk et al. 1996) an activity regulated by the NAD/NADH ratio (Kresze and Ronfit 1981). We showed that a functional PDC is required for CR-induced life span extension. Overexpressing components of the PDC extended life span, suggesting PDC is a limiting longevity factor.
The pyridine nucleotides NAD and NADH are the primary redox carriers involved in metabolism. NAD also participates in many biological processes such as DNA repair and transcription (Lin and Guarente 2003). The overall cellular levels of NAD is maintained by both de novo and salvage NAD biosynthetic pathways (Lin and Guarente 2003). The oxidation and reduction of NAD and NADH drive both catabolic and anabolic reactions (Bakker et al. 2001). The majority of NADH formation is connected to biomass production: synthesis of proteins, nucleic acids, and highly reduced lipids (Bakker et al. 2001). Under aerobic conditions, the mitochondrial electron transport chain is the major site for reoxidizing NAD from NADH. Since the mitochondrial inner membrane is impermeable to NAD and NADH, the NADH shuttle systems have been suggested to move permeable NAD and NADH redox equivalents across the mitochondrial membrane, thereby balancing the NAD/NADH ratio between the mitochondrial and cytosiolic/nuclear pool (Bakker et al. 2001). As more targets of CR are discovered, it is evident that CR affects multiple cellular pathways that reside in spatially distinct cellular compartments. It also remains unclear whether an increase in the NAD/NADH ratio is the trigger that activates the downstream targets of CR. To determine whether the shuttle components are required for CR-mediated life span extension we further analyzed the role of each component in longevity regulation. Our results demonstrate that the mitochondrial shuttle components play an important role in activating downstream targets of CR.
Results
Overexpression of the NADH shuttle components function in the CR pathway to extend life span
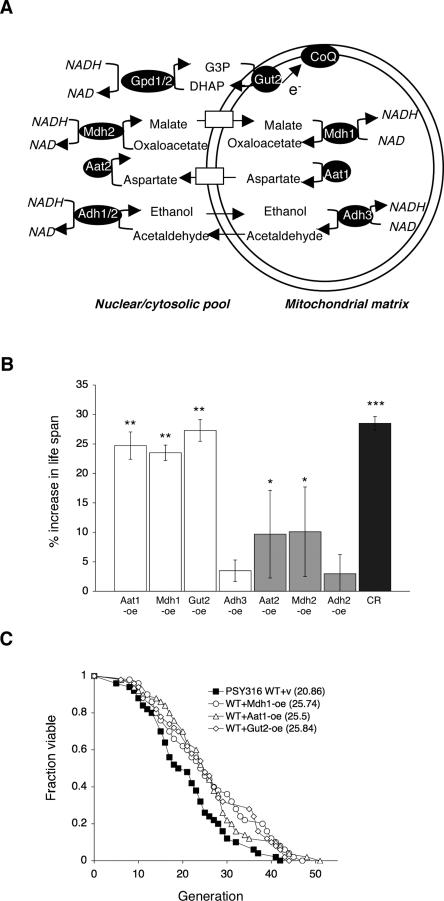
Several mitochondrial NADH shuttles have been identified (Fig. 1A; Bakker et al. 2001). The malate–aspartate shuttle, consisting of aspartate amino transferase (Aat1/Aat2) (Cronin et al. 1991; Morin et al. 1992) and malate dehydrogenase (Mdh1/Mdh2) (Thompson et al. 1988; Minard and McAlister-Henn 1991), acts in concert to exchange cytosolic NADH for mitochondrial NAD. The mitochondiral components, Aat1 and Mdh1, convert mitochondrial malate into aspartate, with the reverse reactions occurring on the cytoplasmic side of the mitochondrial membrane. The ethanol–acetaldehyde shuttle, consisting of mitochondrial alcohol dehydrogenase (Adh3) (Young and Pilgrim 1985; Bakker et al. 2000), and cytosolic alcohol dehydrogenase (Adh1/Adh2) (Thielen and Ciriacy 1991), creates a symmetrical shuttle system for the interconversion of ethanol and acetaldehyde between the mitochondrial and cytoplasmic pools. Because both ethanol and acetaldehyde diffuse freely across biological membranes, it cannot exchange NADH and NAD against a concentration gradient. The glycerol-3-phosphate shuttle is an indirect mechanism that oxidizes cytosolic NADH and transfers the electrons directly into the respiratory chain (Bakker et al. 2001). This shuttle consists of two components—the cytosolic glycerol-3-phosphate dehydrogenase (Gpd1/Gpd2) (Albertyn et al. 1994; Wang et al. 1994; Eriksson et al. 1995), and the mitochondrial glycerol-3-phosphate dehydrogenase (Gut2) (Ronnow and Kielland-Brandt 1993)—located in the inner mitochondrial membrane.
Figure 1.
Overexpressing components of the NADH shuttle systems extend life span. (A) Schematic presentation of the yeast NADH shuttle systems that balance the NAD/NADH ratio between the cytosolic and the mitochondrial pools. For example, in the case of the ethanol–acetaldehyde shuttle, an increase in the mitochondrial NAD/NADH ratio leads to the production of acetaldehyde through mitochondrial alcohol dehydrogenases (Adh3). Mitochondrial acetaldehyde diffuses freely to the cytosol, which is then reduced to ethanol via cytosolic alcohol dehydrogenases (Adh1/2), resulting in an increase in the cytosolic NAD/NADH ratio. In the malate–aspartate shuttle, an increase in the mitochondrial NAD/NADH ratio leads to the production of aspartate from malate through the malate dehydrogenase (Mdh1) and the aspartate aminase (Aat1). Aspartate is transported to the cytosol via the Agc1 carrier, which is then converted to malate by cytosolic malate dehydrogenase (Mdh2) and aspartate aminase (Aat2), resulting in an increase in the cytosolic NAD/NADH ratio. Cytosolic malate enters mitochondria though specific carriers. In the glycerol-3-phosphate shuttle, the mitochondrial glycerol-3-phosphate (G-3-P) dehydrogenase (Gut2) converts G-3-P to DHAP (dihydroxyacetone phosphate), resulting in an increase in the cytosoilc NAD/NADH ratio through the action of the cytosolic G-3-P dehydrogenases (Gpd1/2). Gut2 directly feeds an electron (e−) from G-3-P to coenzyme Q (CoQ), supporting electron transport chain activity. (B) Overexpressing components of the NADH shuttle systems extend life span. Life span analysis of the wild-type cells carrying a control vector and various overexpression DNA constructs. Percent increase in life span relative to wild-type control grown on 2% glucose media is shown. The results show the average of three independent experiments. Error bars denote standard deviations. P values were calculated using the Student’s t test. (*) P < 0.05; (**) P < 0.005; (***) P < 0.001. (C). Mdh1, Aat1, and Gut2 overexpressions also extend life span in a different strain background. Life span analysis of cells overexpressing Mdh1, Aat1, and Gut2 in the PSY316AR strain. One set of representative data is shown. (WT) Wild-type control; (CR) 0.5% glucose; (v) empty vector; (oe) overexpression.
To determine whether and which of the shuttle components are novel longevity regulators we carried out RLS (division potential) analysis in strains overexpressing components of each shuttle system as previously described (Easlon et al. 2007). As shown in Figure 1B, overexpression of Aat1, Mdh1, and Gut2 resulted in a ∼25% increase in life span, similar to life span extension attained by 0.5% CR. However, overexpression of Aat2 and Mdh2 only resulted in a minimal extension (∼15%), and overexpression of Adh2 and Adh3 did not significantly increase life span. It has been reported that Gpd1 overexpression did not extend life span (Kaeberlein et al. 2002). These data suggested that the mitochondrial components of the malate–aspartate (Aat1/Mdh1) and glycerol-3-phosphate (Gut2) shuttles are limiting longevity regulators. In addition, overexpression of Aat1, Mdh1, and Gut2 also extended life span in a different strain background (Fig. 1C). These mitochondrial shuttle components are therefore likely to be conserved longevity factors.
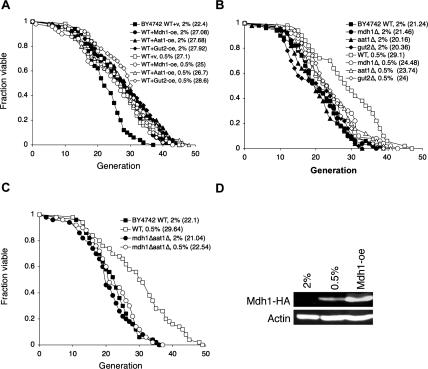
We then examined whether Aat1, Mdh1, and Gut2 overexpressions functioned as CR mimics to extend life span. As shown in Figure 2A, Aat1, Mdh1, and Gut2 overexpressions did not synergize with 0.5% glucose to extend life span, suggesting that these mitochondrial NADH shuttle components function in the same pathway as CR to extend life span. We next determined which shuttle components were required for CR-mediated life span extension. As seen in Figure 2B, the aat1Δ, mdh1Δ, and gut2Δ single mutants partially blocked CR-mediated life span extension. Complete impairment of the malate–aspartate shuttle (the mdh1Δaat1Δ double deletion) blocked CR-mediated life span extension, suggesting the malate–aspartate shuttle played an essential role in 0.5% CR (Fig. 2C). Interestingly, the mdh1Δaat1Δ double mutant (Fig. 2C) and all single mutants (Fig. 2B) showed no significant reduction in life span. It is possible that other functional shuttle components may compensate for the disabled ones under normal growth conditions (2% glucose). Under growth conditions that favor glycolysis, such as growth in the presence of excess glucose, expression of the malate–aspartate shuttle components and tricarbolic acid cycle enzymes are repressed (Minard and McAlister-Henn 1992; Ferea et al. 1999; Grauslund and Ronnow 2000). These glucose-repressed genes resume gene expression when glucose is depleted in the growth media (derepression). Since the glucose concentration used in our CR studies (0.5%) was higher than the glucose concentration utilized in many derepression studies (∼0.01%), we further examined whether the expression of components of the shuttle systems was induced under our CR condition. As shown in Figure 2D, 0.5% glucose was sufficient to induce the expression of Mdh1. Together, these results suggest that CR activates the NADH shuttle components to extend life span.
Figure 2.
CR and the NADH shuttle system function in the same pathway. (A) CR does not synergize with the overexpressions to extend life span. Life span analysis of cells overexpressing Mdh1, Aat1, and Gut2 grown under normal and CR (0.5% glucose) conditions. (B) Deletions of single components of the NADH shuttle systems partially inhibit CR-mediated life span extension. (C) Complete inactivation of the malate–aspartate shuttle totally abolishes CR-induced life span extension. (D) CR (0.5% glucose) is sufficient to induce expression of the shuttle component. Western blot analysis of the Mdh1 protein levels in cells grown in 2% and 0.5% glucose. Mdh1 is fused to an HA epitope tag and visualized with the anti-HA antiserum. (WT) Wild-type control; (oe) overexpression. One set of representative data is shown.
Requirement of respiration and the Sir2 family in Aat1, Mdh1, and Gut2 overexpression strains
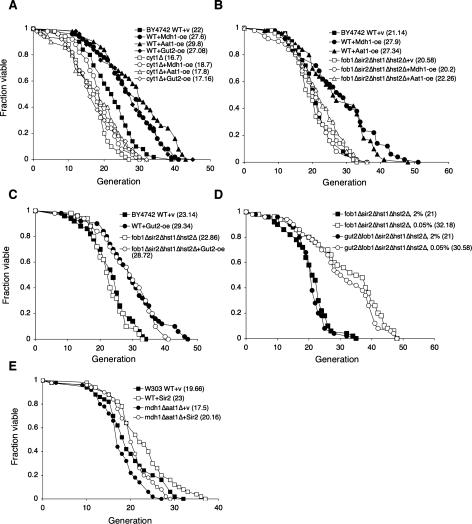
Mitochondrial respiration has been shown to be required for 0.5% CR-mediated life span extension (Lin et al. 2002; Kaeberlein et al. 2005a; Easlon et al. 2007). We therefore determined whether Aat1, Mdh1, and Gut2 overexpressions also required functional mitochondria using the cyt1Δ mutant. Cells devoid of cytochrome c1 (cyt1Δ) are defected in electron transport chain activities (Schneider and Guarente 1991). As expected, Aat1, Mdh1, and Gut2 overexpressions all required a functional electron transport chain to elicit the maximum life span extension (Fig. 3A), reinforcing the importance of respiration in CR. However, it remained possible that respiration was simply required to maintain normal life span (at least in this strain background), since the cyt1Δ deletion significantly shortened life span of wild-type cells. Overall, these results are consistent with the findings that CR increases mitochondrial respiration/activities in multiple organisms (Lin et al. 2002; Nisoli et al. 2005; Bishop and Guarente 2007).
Figure 3.
The requirement of respiration and the Sir2 family in the NADH shuttle overexpression induced life span extension. (A) Mdh1, Aat1, and Gut2 overexpressions require Cyt1 for life span extension. Life span analysis of the wild-type cells, the cyt1Δ mutant, and the cyt1Δ mutant overexpressing Mdh1, Aat1, and Gut2 grown in 2% glucose. (B) Mdh1 and Aat1 overexpression require the Sir2 family to extend life span. Life span analysis of the wild-type, Mdh1-oe, and Aat1-oe cells, the fob1Δsir2Δhst1Δhst2Δ quadruple mutant, and the fob1Δsir2Δhst1Δhst2Δ mutant with Mdh1-oe and Aat1-oe on 2% glucose media. (C) Gut2 overexpression does not require the Sir2 family to extend life span. Life span analysis of the wild-type and Gut2-oe cells, the fob1Δsir2Δhst1Δhst2Δ quadruple mutant, and the fob1Δsir2Δhst1Δhst2Δ mutant with Gut2-oe on 2% glucose media. (D) To extend life span, 0.05% glucose does not require Gut2. Life span analysis of the fob1Δsir2Δhst1Δhst2Δ quadruple mutant and the gut2Δfob1Δsir2Δhst1Δhst2Δ quintuple mutant cells grown in 0.05% and 2% glucose. (E) Expressing one extra copy of Sir2 requires the malate–aspartate shuttle for maximum life span extension in the W303AR strain background. (WT) Wild-type control; (v) empty vector control; (oe) overexpression. One set of representative data is shown.
The Sir2 family members (sirtuins) play important roles in several CR model systems (Lin et al. 2000; Rogina and Helfand 2004; Chen et al. 2005; Nisoli et al. 2005; Wang and Tissenbaum 2006). The use of NAD in the sirtuin-mediated deacetylation reactions provides a link between NAD metabolism and sirtuin-mediated metabolic regulations (Guarente 2006). To determine whether Aat1, Mdh1, and Gut2 overexpressions functioned through the Sir2 family to extend life span, we overexpressed Mdh1, Aat1, and Gut2 in the fob1Δsir2Δhst1Δhst2Δ quadruple mutant, a genetic model for studying the requirement of the Sir2 family for life span (Lamming et al. 2005; Easlon et al. 2007). As seen in Figure 3B, deletions of the Sir2 family abolished the long life span induced by Aat1 and Mdh1 overexpressions. However, the long life span induced by Gut2 overexpression was not affected by deletions of the Sir2 family (Fig. 3C). Interestingly, overexpressing Mdh1, Aat1, and Gut2 all increased Sir2-mediated gene silencing at the ribosomal DNA locus (Supplemental Fig. 1). These studies suggested that although both the malate–aspartate and glycerol-3-phosphate shuttles act to regulate the intracellular NAD/NADH ratio and Sir2 activity, they are likely operating through separate downstream targets to extend life span. We then further examined the effect of the mdh1Δaat1Δ double deletions in Sir2 overexpression-induced life span extension using a previously reported W303 Sir2 overexpression strain (Kaeberlein et al. 1999). As shown in Figure 3E, Sir2 overexpression partially extended life span in the mdh1aat1Δ double mutant, suggesting Sir2 functioned downstream from the malate–aspartate shuttle. However, it remains highly possible that Sir2 is not the only target of the malate–aspartate shuttle.
Another CR pathway induced by 0.05% glucose (Kaeberlein et al. 2004, 2005b; Easlon et al. 2007) has recently been characterized. Interestingly, this severe CR condition does not require respiration or the Sir2 family to extend life span. In fact, it extends life span more robustly in mutants devoid of the Sir2 family or cytochrome c than in wild-type cells (Easlon et al. 2007), suggesting this CR pathway is antagonized by the Sir2 family and mitochondrial respiration. Mitochondrial dysfunction activates retrograde response, which has been linked to life span extension (Jazwinski 2005). It is therefore possible that in the cyt1Δ mutant the retrograde response is activated under 0.05% glucose CR leading to life span extension. However, our preliminary results suggested that retrograde response did not play a role in this pathway (Supplemental Fig. 2). To gain more insight into 0.05% glucose-induced CR, we examined the role of Gut2 in this pathway since, similar to 0.05% glucose, Gut2 overexpression also extended life span in a Sir2-independent manner. Our results showed that 0.05% glucose extended the life span of the fob1Δsir2Δhst1Δhst2Δ quadruple mutant independent of Gut2 (Fig. 3D), suggesting Gut2 did not mediate 0.05% glucose CR. Gut2 is therefore likely to function in a Sir2-independent branch of the 0.5% glucose CR pathway.
A functional NADH shuttle is required for multiple genetic models of CR
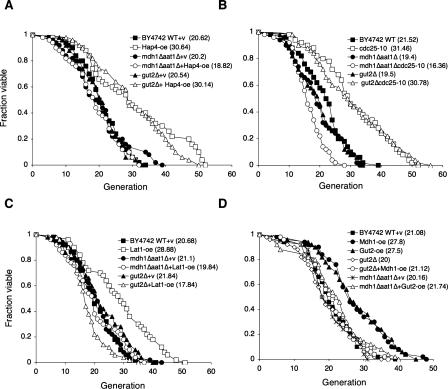
We also analyzed the requirement of the NADH shuttles in several genetic models of CR to further examine the role of NAD metabolism in longevity regulation. Three CR mimics were analyzed: the cdc25-10 (RAS nucleotide exchange factor) mutant, Hap4 overexpression, and Lat1 overexpression strains. In the cdc25-10 mutant, the glucose-responsive cAMP-activated PKA activity is down-regulated, thereby increasing life span (Lin et al. 2000). Hap4 is a component of the Hap complex that binds to the consensus sequences CCAAT and increases the transcription of many genes involved in mitochondrial metabolism (Forsburg and Guarente 1989). Overexpressing Hap4 has been shown to induce a metabolic shift toward respiration and extend life span (Lin et al. 2002). Lat1 (dihydrolipoamide acetyltransferase) is the E2 component of the mitochondrial PDC (Lawson et al. 1991). Lat1 overexpression has been shown to extend life span in the CR pathway (Easlon et al. 2007). As shown in Figure 4, A–C, the mdh1Δaat1Δ double deletion significantly reduced the life span extension by Hap4 overexpression, cdc25-10 mutation, and Lat1 overexpression, suggesting that the malate–aspartate shuttle played an important role in these CR genetic models. The gut2Δ mutation did not affect the long life span in the Hap4-overexpressing (Fig. 4A) and cdc25-10 (Fig. 4B) mutant cells. Interestingly, the long life span induced by Lat1 overexpression was totally abolished by the gut2Δ mutation (Fig. 4C), suggesting this CR model required both functional malate–aspartate and glycerol-3-phosphate shuttles. We then examined whether the life span extension induced by overexpressing one shuttle component required the function of the other. As shown in Figure 4D, the gut2Δ mutation abolished the long life span of Mdh1-overexpressing cells. The mdh1Δaat1Δ mutations also prevented Gut2 overexpression-induced life span extension. Together, these results demonstrated that the NADH shuttle components are required for life span extension in multiple CR genetic models.
Figure 4.
CR genetic models and the NADH shuttle. (A) Overexpression of Hap4 requires the malate–aspartate shuttle for life span extension. Life span analysis of the wild-type and Hap4-oe cells, the mdh1Δaat1Δ double mutant, and gut2Δ single mutant with and without Hap4-oe on 2% glucose media. (B) The cdc25-10 mutation requires the malate–aspartate shuttle for life span extension. Life span analysis of the wild-type and cdc25-10 mutant cells, the mdh1Δaat1Δ double mutant, and gut2Δ single mutant with and without the cdc25-10 mutation on 2% glucose media. (C) Lat1 overexpression requires both the malate–aspartate shuttle and the glycerol-3-phosphate shuttle to extend life span. (D) Mdh1 overexpression requires the glycerol-3-phosphate shuttle to extend life span and Gut2 overexpression requires the malate–aspartate shuttle to extend life span. (WT) Wild-type control; (v) empty vector control; (oe) overexpression. One set of representative data is shown.
Role of the mitochondrial shuttle components in CR-induced changes in the NAD/NADH ratio
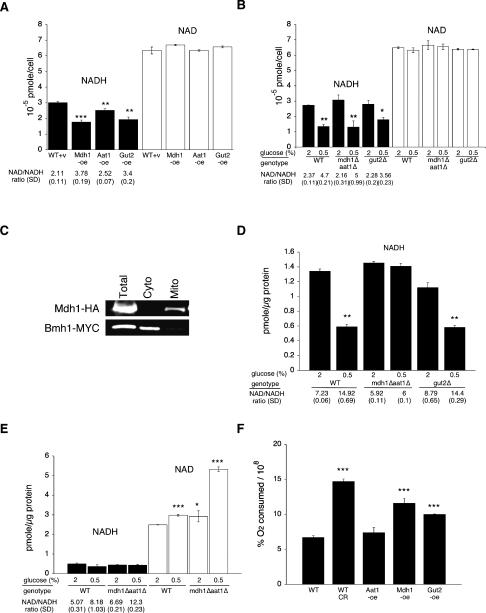
It has been suggested that CR increases the intracellular NAD/NADH ratio, thereby activating NAD-utilizing metabolic targets to extend life span (Lin et al. 2004; Easlon et al. 2007). Although we showed that components of the malate–aspartate and glycerol-3-phosphate shuttles are required for CR-mediated life span extension, it was still unclear whether these shuttle components indeed affected the NAD/NADH ratio under CR. We therefore directly examined whether the malate–aspartate and the glycerol-3-phosphate shuttles mediated CR-induced effects. The levels of total intracellular NAD and NADH were measured in strains overexpressing Mdh1, Aat1, and Gut2. Similar to CR (Fig. 5B; Lin et al. 2004; Easlon et al. 2007), although to a lesser extent, Mdh1, Aat1, and Gut2 overexpressions increased the NAD/NADH ratio by decreasing the levels of NADH in the whole-cell extract (Fig. 5A). We also examined the oxygen consumption rate of the NADH shuttle overexpression strains. As shown in Figure 5F, both Gut2 and Mdh1 overexpressions significantly increased oxygen consumption. Therefore, it was likely that overexpression of these two shuttles extended life span in part by inducing a shift toward respiration. However, Aat1-oe did not increase oxygen consumption, strongly suggesting that overexpression of the NADH shuttles did not simply extend life span by shifting metabolism toward respiration. We then determined whether these shuttle components were required for CR-induced decrease in the NADH levels. The levels of total intracellular NAD and NADH were measured in the gut2Δ and mdh1Δaat1Δ mutant cells grown under normal and CR conditions. Surprisingly, neither the gut2Δ nor the mdh1Δaat1Δ mutants abolished the decrease in total intracellular NADH levels induced by CR (Fig. 5B). It was therefore likely that CR induced the initial decrease of NADH in the mitochondria by activating the respiratory chain, which is then transmitted to the cytosol by the shuttle components. Consistent with this model, the mdh1Δaat1Δ double mutants and the gut2Δ mutant showed increased oxygen consumption under non-CR conditions (data not shown), suggesting these mutants may accumulate higher cytosolic NADH levels, and therefore, mitochondrial respiration was induced as a compensatory effect. To determine whether these shuttle components indeed played important roles in transmitting CR-induced decreases in NADH from the mitochondrial to the cytosolic/nuclear pool, we measured the levels of NAD and NADH in the cytosolic/nuclear fractions of the cell extracts. Consistent with previous reports (Kresze and Ronfit 1981), our fractionation procedure gave rise to satisfactory separation of the cytosolic and mitochondrial fractions (Fig. 5C). As seen in Figure 5D, the mdh1Δaat1Δ mutation totally abolished the decrease in NADH induced by CR. The gut2Δ mutation surprisingly did not affect the CR-induced decrease in the NADH levels, suggesting the malate–aspartate shuttle played a major role under CR. We also examined NAD and NADH levels in mitochondrial fractions. Since the mdh1Δaat1Δ mutations totally abolished the balance of the NAD/NADH ratio between the cytosolic and mitochondrial pools, we expected that in the mdh1Δaat1Δ mutant, CR-induced increases in the NAD/NADH ratio would be trapped inside the mitochondrial fractions. As shown in Figure 5E, the mitochondrial fractions of the mdh1Δaat1Δ mutant showed a higher NAD/NADH ratio under CR compared with wild type under CR. Interestingly, CR increased the NAD/NADH ratio in mitochondria by increasing NAD levels (Fig. 5E). Perhaps the mitochondrial NAD carriers (Todisco et al. 2006) are more active under CR, increasing the inward flux of NAD. It is noteworthy that these values represented relative differences in nucleotides among matched samples rather than precise quantification. Therefore, the increase in the mitochondrial NAD/NADH ratio may not totally account for the increase in the NAD/NADH ratio seen in the total cellular extracts under CR (Fig. 5A). It is also very likely that in the mdh1Δaat1Δ mutant, other factors such as the glycerol-3-phosphate shuttle are activated to affect total cellular pools of NAD and NADH
Figure 5.
Role of the NADH shuttles in the regulation of intracellular NAD/NADH levels. Measurements of the intracellular NAD and NADH levels in wild-type cells, cells overexpressing the shuttle components, and cells carrying deletions in the shuttle components grown in 2% and 0.5% glucose. (A) Mdh1, Aat1, and Gut2 overexpression decreases the total NADH levels. (B) Deletions of the shuttle components do not abolish the decrease in the total NADH levels induced by CR. (C) Western blot analysis of total cellular, cytosolic, and mitochondrial fractions used for NAD/NADH ratio assay analysis. (D) Inactivation of the malate–aspartate shuttle abolishes the decrease in the NADH levels in the cytosolic/nuclear pools induced by CR. (E) Inactivation of the malate–aspartate shuttle further increases the NAD/NADH ratio in mitochondria under CR. (F) Gut2 and Mdh1 overexpressions significantly increase oxygen consumption rates compared with wild-type cells grown in 2% glucose. (WT) BY4742 wild type; (v) empty vector control; (oe) overexpression; (SD) standard deviations. One representative set of three independent experiments, each conducted in quadruplicate (A,B) or triplicate (D–F), is shown. Error bars denote standard deviations. P values were calculated using the Student’s t test. (*) P < 0.05; (**) P < 0.005; (***) P < 0.001.
Components of the malate–aspartate shuttle played an important role in chronological life span (CLS)
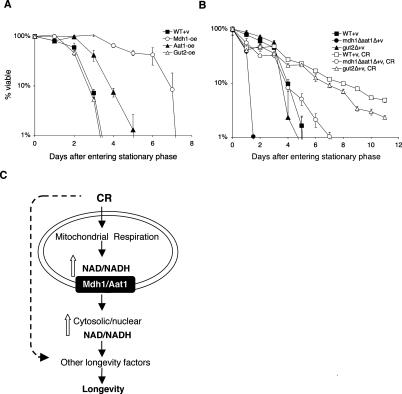
CLS has been suggested to be a model for studying senescence of post-mitotic cells, whereas (RLS resembles the proliferative potential of mitotic cells. CLS measures the length of time a cell survives at a nondividing state. RLS measures the number of cell divisions an individual yeast cell undergoes before senescence. Several longevity factors and pathways have been independently identified through both types of studies. The complex mechanisms underlying CLS and RLS regulation and whether these two pathways share common components are currently unclear. Although it has been shown that RLS and CLS functioned through distinct downstream targets (Fabrizio et al. 2001), certain CR models appeared to extend both RLS and CLS (Fabrizio et al. 2001; Easlon et al. 2007). Therefore, if a longevity factor extends both types of life span regulation, it is more likely to be a conserved longevity regulator. As shown in Figure 6A, both Mdh1 and Aat1 overexpression significantly extended CLS. Interestingly, although Mdh1, Aat1, and Gut2 overexpressions extended RLS to a similar extent, Mdh1 overexpression appeared to extend CLS more robustly than Aat1 overexpression, and Gut2 overexpression did not extend CLS. We also examined the effect of mdh1Δaat1Δ double deletions on CR-induced CLS. As shown in Figure 6B, the mdh1Δaat1Δ double deletions significantly blocked the life span extension by CR. In addition, the double mutant exhibited very short CLS, suggesting the malate–aspartate shuttle was required for normal CLS. Interestingly, CR was able to rescue the short life span of the double mutants back to wild-type level, suggesting the malate–aspartate shuttle was an important but not the only target of CR for CLS extension. It was also possible that in the mdh1Δaat1Δ mutant, CR activated other factors such as the Gut2 shuttle to rescue the growth defects, thereby restoring the short CLS back to wild-type level. Collectively, these results suggested that components of the malate–aspartate shuttle functioned at the interface of both CLS and RLS pathways and were therefore likely to be conserved longevity factors.
Figure 6.
The NADH shuttle systems and life span regulation. (A) Mdh1 and Aat1 overexpressions extend CLS. (B) The malate–aspartate shuttle is required for maximum CLS extension induced by CR. Fractions of viable wild-type, Mdh1-oe, Aat1-oe, Gut2-oe, mdh1Δaat1Δ double mutant, and gut2Δ single mutant cells were determined from cultures grown to stationary phase. One representative set of three independent experiments, each conducted in triplicate, is shown. Error bars denote standard deviations. (WT) BY4742 wild type; (CR) 0.5% glucose; (v) empty vector control; (oe) overexpression. (C) A model for the role of the NADH shuttle in CR. Multiple CR pathways act to affect longevity in yeast. CR activates mitochondrial respiration, which is required for 0.5% CR and CR mimic-induced life span extension. A functional malate–aspartate shuttle is also required for CR-induced life span extension. We propose that under CR, the malate–aspartate shuttle transmits the increase in the NAD/NADH ratio from the mitochondrial pool to the cytosolic/nuclear pool. The increased NAD/NADH ratio in the cytosolic/nuclear pool then activates other longevity factors such as the Sir2 family to extend life span. Overexpression of the shuttle components (such as Gut2) may also extend life span by maintaining mitochondrial activity, thereby increasing metabolic fitness of the cell.
Discussion
The malate–aspartate shuttle regulates NADH compartmentalization and life span
CR increases mitochondrial respiration in multiple model organisms (Lin et al. 2002; Nisoli et al. 2005; Bishop and Guarente 2007). In yeast, increased respiration leads to a higher rate of electron transport, which in turn, increases the reoxidation of NADH to NAD in the mitochondria (Lin et al. 2004). This CR-induced increase in the NAD/NADH ratio is suggested to activate downstream metabolic targets such as the Sir2 family to extend life span (Fig. 6C). It has been suggested that Sir2 extends RLS by increasing genomic stability at the rDNA loci (Sinclair and Guarente 1997). Sir2 may also extend life span by increasing the fitness of newborn cells (Aguilaniu et al. 2003; Erjavec and Nystrom 2007; Erjavec et al. 2007). It has been shown that oxidatively damaged proteins are retained in mother cells and not inherited by daughter cells during cytokinesis. Sir2 acts in concert with multiple chaperone proteins and the actin cytoskeleton to prevent the inheritance of oxidatively damaged proteins to daughter cells upon separation from the mother. As more downstream targets of CR are identified, it is becoming more apparent that these downstream targets create a web of interconnected pathways to alter stress resistance, genomic stability, and life span. In this report, we showed that the malate–aspartate and glycerol-3-phosphate NADH shuttles were novel longevity regulators required for CR-mediated life span extension. Overexpression of the mitochondrial components of these shuttles (Mdh1, Aat1, and Gut2) extended life span and did not synergize with CR, whereas deletion of these components abolished life span extension induced by CR. Consistent with our model, Aat1, Mdh1, and Gut2 overexpressions all required mitochondrial respiration for life span extension (Fig. 3A). Our results also showed that the malate–aspartate shuttle played a major role in shuttling NAD and NADH redox equivalents across the mitochondrial membrane under CR (Fig. 6C).
In mammalian mitochondria, the malate–aspartate shuttle plays an important role in the oxidation of cytosolic NADH (Meijer and Van Dam 1974; Bremer and Davis 1975). In yeast, the existence and importance of the malate–aspartate shuttle has been debatable until the transporters required for the malate–aspartate shuttle to operate were identified (Palmieri et al. 1996, 1999, 2001; Cavero et al. 2003). Here our data showed that the malate–aspartate shuttle played a major role in mediating life span extension under CR as well as in several genetic mimics of CR. Although the mitochondrial shuttle components (Mdh1, Aat1) appeared to be the major limiting longevity regulator, the cytosolic components (Mdh2, Aat2) were expected to also play an important role, since a functional shuttle require both. However, overexpression of the cytosolic components only resulted in minimal life span extension. It is possible that the cytosolic components are subject to additional regulation, which may block the effects of overexpressing the proteins. For instance, Mdh2 is rapidly degraded in the presence of excess glucose (Minard and McAlister-Henn1992).
Overexpressing components of the malate–aspartate shuttle may also affect the glutamine levels. Glutamine activates the TOR signaling pathways that regulate a variety of cellular processes such as cell growth, metabolism, and cell cycle progression, with negative consequences on life span (Crespo et al. 2002; Martin et al. 2004; Kaeberlein et al. 2005b). Glutamine is produced by glutamine synthase from glutamate. In addition to converting oxaloacetate to aspartate, Aat1 also converts glutamate to α-ketoglutarate. It is therefore possible that overexpressing Aat1 may mimic the tor1Δ mutation by depleting glutamate and, hence, the glutamine levels. However, Aat1 overexpression also generates aspartate, which is transported to the cytosol by the Agc1 carrier in exchange for glutamate (Cavero et al. 2003). Therefore, it is more likely that Aat1 and Mdh1 overexpression increases the flux of glutamate in and out of the mitochondria rather than depleting it. The activity of aspartate amino transferase has been shown to be essential for glutamate (neurotransmitter glutamate) production in glutamatergic neurons (Palaiologos et al. 1988). In addition, we were not been able to observe RLS extension by the tor1Δ mutation in our strain background, although the tor1Δ mutation does extend CLS (E. Easlon and S.-J. Lin, unpubl.). In contrast, overexpressing Mdh1 and Aat1 extend both RLS and CLS. It is therefore unlikely that the life span extension seen by overexpressing components of the malate–aspartate shuttle resulted from decreased TOR activity.
Several lines of evidence suggest that Gut2 overexpression extends life span through an alternate pathway to that of Aat1 and Mdh1 overexpressions. Gut2 is the mitochondrial component of the glycerol-3-phosphate shuttle, which is characterized as a Type I shuttle that does not result in a translocation of NAD and NADH redox equivalents across the mitochondrial membrane but does reoxidize cytosolic NADH independently of NADH dehydrogenases (Bakker et al. 2001). Gut2 overexpression extended life span independently of the Sir2 family, whereas the Aat1 and Mdh1 overexpressions required the Sir2 family (Fig. 3A,B). Gut2 was not required for either Hap4 overexpression or cdc25-10 CR mimics to extend life span even though a functional malate–aspartate shuttle was required for both CR mimics (Fig. 4A,B). Although the gut2Δ mutation largely abolished CR-induced life span extension, it did not prevent a CR-induced decrease in the levels of cytosolic NADH. It is therefore likely that Gut2 supports CR by enhancing the mitochondrial activity through delivering electrons from glycerol-3-phosphate directly to the mitochondrial ubiquinone pool. Consistent with this model, the yeast gut2Δ mutant grows slowly on glycerol (a nonfermentable carbon source) containing media (E. Easlon and S.-J. Lin, unpubl.). The gut2Δ mutants in plants also showed reduced mitochondrial respiration rates (Shen et al. 2006). Finally, Gut2 overexpression required a functional mitochondrial respiratory chain to extend life span (Fig. 3A), and overexpression of Gut2 increases mitochondrial oxygen consumption (Fig. 5F).
The NADH shuttles, metabolic fitness, and age-associated diseases
One underlying theme for the life span extension by CR or overexpression of Gut2, Mdh1, and Aat1 is the decrease in total cellular NADH levels. Perhaps the decreased NADH levels reflect increased metabolic activity. It is possible that CR operates through these NADH shuttles not only to regulate the compartmentalization of NAD/NADH but also to allow for heightened mitochondrial activity. The mdh1Δ, aat1Δ, and gut2Δ mutations block 0.5% CR-mediated life span extension, and hence may also be attributed to decreased TCA cycle activity. It is noteworthy that the redox equivalent produced by these shuttles are also metabolic intermediates in the TCA cycle. A 50% reduction in the activity of the TCA cycle is observed in mice deficient of the glycerol-3-phosphate and malate–aspartate shuttle systems (Eto et al. 1999). Our previous studies on Lat1 are also in line with this model. Lat1 is the core component of the PDC. Overexpressing Lat1 allows efficient replacement of the selectively damaged Lat1 subunits in the PDC to maintain the metabolic fitness of the cell (Easlon et al. 2007). Interestingly, Lat1 overexpression required both shuttles to extend life span, suggesting a role for the NADH shuttles in metabolic fitness-mediated life span extension (Fig. 4C). Impairment of mitochondrial metabolism and NADH shuttles has also been implicated in age-associated diseases such as diabetes. The mitochondiral protein SIRT4 (a mammalian Sir2 homolog) represses insulin secretion by ribosylation and inhibition of glutamate dehydrogenase (GDH) enzyme activity. Similar to Aat1, GDH converts glutamate into α-ketoglutarate. Upon CR, Sirt4 repression is alleviated and GDH activity is restored to boost the amino acid-stimulated insulin secretion (Haigis et al. 2006). Eto et al. (1999) have also shown that the NADH shuttles are essential for glucose-induced insulin secretion, linking the NADH shuttles to diabetes. In conclusion, the NADH shuttles play an important role in mitochondrial activity, metabolic fitness, and life span regulation. CR may prevent or delay the onset of age-associated metabolic diseases by activating the NADH shuttle components.
Materials and methods
Strains and media
Yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 was acquired from Open Biosystems (Brachmann et al. 1998). W303AR MATa ura3-1,leu2-3, 112 his3-11, 15 trp1-1 ade2-1 RDN1∷ADE2 can1-100, and PSY316AR MATα ura3-52 leu2-3,112 his3-200 ade2-101 lys2-801 RDN1∷ADE2 have been described previously (Kaeberlein et al. 1999; Lin et al. 2000). Rich-media YPD and synthetic media were made as described (Sherman et al. 1978). Medium used for life span analysis was YEP (2% bacto peptone, 1% yeast extract, 1.5% agar) supplemented with filter-sterilized glucose at a final concentration of 2%, 0.5%, or 0.05%. All gene deletions in this study were generated in our laboratory by replacing the wild-type genes with the reusable Kanr marker as described in Easlon et al. (2007) and verified by PCR using oligonucleotides flanking the genes of interest. The Mdh1 overexpression construct pADH1-Mdh1 was made as follows: A pair of oligonucleotides were designed to amplify the Mdh1 coding region (from start codon to stop codon) via PCR. This pair of oligonucleotides also added a Not1 site to the 5′ end and a Nhe1 site to the 3′ end of the Mdh1 gene. After PCR amplification, DNA was digested with Not1 and Nhe1, then ligated to ppp81 digested with the same enzymes, resulting in pADH1-Mdh1. All other overexpression constructs were made the same way. All constructs were verified by DNA sequencing. We chose the stable integrative ADH1 promoter construct ppp81 for gene overexpression for the following reasons: We showed previously that under our CR condition (0.5% glucose), the ADH1 promoter was not significantly down-regulated (Easlon et al. 2007). Other commonly used overexpression promoters—for example, the GAL promoter (induced by galactose) and the CUP1 promoter (induced by copper)—require using another type of carbon source or adding nutrients into the growth media that will interfere with our CR conditions. Using the multicopy 2μ plasmids allows gene expression under native promoter; however, it has been shown that the 2μ plasmids shorten yeast life span (Falcón and Aris 2003). The cdc25-10 mutants were made by two-step allele replacement as described previously (Lin et al. 2000). The Hap4-overexpressing construct pADH1-HAP4 has been described previously (Lin et al. 2002). The pRS305-Sir2 plasmid carrying a single copy of the SIR2 gene and the promoter region has been described previously (Kaeberlein et al. 1999). Strains overexpressing the shuttle components, Hap4, Sir2, and the control vector were made as described previously (Kaeberlein et al. 1999; Easlon et al. 2007). Mdh1 was tagged by the HA epitope tag in the genome using the pFA6a-3HA-KanMX6 plasmid as described previously (Longtine et al. 1998). Yeast strains expressing the Myc-tagged Bmh1 proteins were made by introducing the pBmh1-13MycXX plasmid. The pBmh1-13MycXX plasmid was made as follows: A pair of oligonucleotides were designed to amplify the coding and promoter regions of Bmh1 via PCR. The pair of oligonucleotides added a Not1 site to the 5′ end and a Xba1 site to the 3′ end of the Bmh1 gene. After PCR amplification, DNA was digested with Not1 and Xba1, then ligated to pRS316-13MycXX plasmid digested with the same enzymes, resulting in pBmh1-13MycXX. pRS316-13MycXX was constructed by amplifying the 13Myc epitope tag in the pFA6a-13Myc-kanMX6 plasmid. The pair of oligonucleotides created an Xba1 site in the 5′ end and an Xho1 site in the 3′ end of the 13Myc epitope tag. After PCR amplification, DNA was digested with Xho1 and Xba1 and ligated to pRS316 digested with the same enzymes, resulting in pRS316-13MycXX.
RLS analysis
All life span analyses in this study were carried out on YEP plates supplemented with different concentrations of glucose with 50 cells per strain per experiment as described previously (Park et al. 2002) with a few modifications. Fresh growing cells were patched daily onto a fresh YEP plate with 2% glucose at least 3 d prior to life span analysis. Cells from frozen stocks were grown on normal media (repatched daily) for at least 2 wk prior to life span analysis. On the day of life span analysis, cells were repatched onto the side of the life span plates and then allowed to grow. After 2 h, 10 groups of cells were moved from the initial patch and arrayed onto the center of the life span plate (one strain per plate) using a micromanipulator and allowed to grow for 1.5 h (one to two divisions). Virgin cells were then selected and subjected to life span analysis. To reduce the chance of human errors and biases, life span assays were measured at least three times by two to three researchers. Statistical analysis was carried out using the JMP statistics software (SAS), and the Wilcoxon rank-sum test P values were calculated for each pair of life spans as shown in Supplemental Table 1.
NAD and NADH measurements
Total intracellular levels of the NAD and NADH nucleotides were determined as described previously (Lin et al. 2001; Easlon et al. 2007) with a few modifications. In brief, cells were grown to OD600 of ∼0.8 in synthetic media, then 107 cells were harvested in duplicate by centrifugation in two 1.5-mL tubes. Acid extraction was performed in one tube to obtain NAD, and alkali extraction was performed in the other to obtain NADH. Three to five microliters of neutralized cell extract (∼105 cells) were used for enzymatic cycling reaction as described previously (Lin et al. 2001). The concentration of nucleotides was measured fluorometrically with excitation at 365 nm and emission monitored at 460 nm. The cytosolic/nuclear factions and the mitochondrial fractions of the cell extract were obtained as described in Kresze and Ronfit (1981) with a few modifications. Harvested cells (∼1 × 109 cells) were incubated in a lysis solution of 20 mM BME, 1.2 M sorbitol, and 0.5 mg of yeast lytic enzyme for 20 min at 37°C. The cytosolic and crude mitochondrial fractions were separated by centrifugation as described previously (Kresze and Ronfit 1981). The resultant supernatant was used as the cytosolic fractions for NAD and NADH quantification. The mitochondrial pellet was further washed with buffer containing 250 mM sucrose, 1 mM EDTA, and 10 mM MOPS adjusted to a pH of 7.2. After washing, the mitochondrial pellet was lysed in buffer containing 1 mM MgCl2 and 20 mM HEPES at a pH of 7.4.
CLS analysis
Three single colonies from each strain were analyzed in each experiment as described previously (Fabrizio and Longo 2003) with a few modifications. Cells were grown in synthetic media supplemented with 2% glucose and 4× auxotrophic amino acids (uracil, histidine, lysine, and leucine) at a starting OD600 of 0.1. Cell viability was monitored daily or every other day by plating a fraction of the culture onto fresh media to determine the colony-forming units (CFU). The rate of cell survival was calculated by normalizing the CFU to the cell number of day 2 (the stationary phase) for each strain.
Protein extraction and Western blot analysis
For total protein extraction, 50 mL of cells were grown in YPD containing 2% or 0.5% glucose as indicated to mid-logarithmic phase (OD600 of 1–2), concentrated by centrifugation. Total protein extract was obtained using an NaOH extraction method (Sekito et al. 2000). In brief, cells were incubated in 0.255 M NaOH and 1% BME on ice with the addition of 50% TCA. The resulting pellet was washed with 1 M Tris-HCl (pH 6.8). The pellet was heated to 65°C in sample buffer containing 20% of the protease inhibitor cocktail tablet from Roche. Approximately 15 μg of total protein were loaded in each lane. After electrophoresis, the protein was transferred to nitrocellulose membrane from Whatman. The membranes were then washed, blotted with either anti-HA antiserum (Covance), anti-actin antiserum (Abcam), or anti-MYC antiserum (Covance). Protein was visualized using the Amersham anti-mouse or rabbit antiserum conjugate to the horseradish peroxidase and the ECL-reagents (Pierce). The chemiluminescent image was analyzed using the Alpha Innotech imaging system and software provided by the manufacturer. For determining the purity of the fractionation samples, 50 mL of cells were grown in SD complete to mid-logarithmic phase (OD600 of ∼1), and were prepared as described above for cytosolic/mitochondrial fractionation for NAD and NADH quantification. Ten micrograms of proteins from each fraction were loaded in each lane followed by electrophoresis and Western blot analysis as described above.
Oxygen consumption assays
To determine oxygen consumption rates, 25 mL of cells grown in YPD containing either 2% or 0.5% glucose were grown to mid-logarithmic phase (OD600 of ∼1) with vigorous shaking. Cells (2 × 108 to 4 × 108) were harvested and washed twice in YEP then resuspended in 700 μL of YEP. Glucose was spiked into each sample to a final concentration of 2% right before the analysis. Oxygen consumption was monitored using a Clark-type electrode, and data were recorded every second.
Acknowledgments
We thank members of the Lin laboratory for discussions and suggestions and Dr. T. Power for providing the pFA6a-3HA-KanMX6 and pFA6a-13Myc-kanMX6 plasmids. We also thank members of the Parales laboratory for their assistance with oxygen consumption assays. This study was supported by the National Institute on Aging, the Ellison Medical Foundation, and the American Cancer Society IRG Award. S.-J.L. is a new research scholar of the Ellison Medical Foundation. We declare that no conflicts of interest exist.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1648308.
References
- Aguilaniu H., Gustafsson L., Rigoulet M., Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Albertyn J., Hohmann S., Thevelein J.M., Prior B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., Bitterman K.J., Wood J.G., Medvedik O., Sinclair D.A. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B.M., Bro C., Kotter P., Luttik M.A., van Dijken J.P., Pronk J.T. The mitochondrial alcohol dehydrogenase Adh3p is involved in a redox shuttle in Saccharomyces cerevisiae. J. Bacteriol. 2000;182:4730–4737. doi: 10.1128/jb.182.17.4730-4737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B.M., Overkamp K.M., van Maris A.J., Kotter P., Luttik M.A., van Dijken J.P., Pronk J.T. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- Bishop N.A., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bitterman K.J., Anderson R.M., Cohen H.Y., Latorre-Esteves M., Sinclair D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bremer J., Davis E.J. Studies on the active transfer of reducing equivalents into mitochondria via the malate–aspartate shuttle. Biochim. Biophys. Acta. 1975;376:387–397. doi: 10.1016/0005-2728(75)90161-9. [DOI] [PubMed] [Google Scholar]
- Cavero S., Vozza A., del Arco A., Palmieri L., Villa A., Blanco E., Runswick M.J., Walker J.E., Cerdan S., Palmieri F., et al. Identification and metabolic role of the mitochondrial aspartate–glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 2003;50:1257–1269. doi: 10.1046/j.1365-2958.2003.03742.x. [DOI] [PubMed] [Google Scholar]
- Chen D., Steele A.D., Lindquist S., Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Crespo J.L., Powers T., Fowler B., Hall M.N. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin V.B., Maras B., Barra D., Doonan S. The amino acid sequence of the aspartate aminotransferase from baker’s yeast (Saccharomyces cerevisiae) Biochem. J. 1991;277:335–340. doi: 10.1042/bj2770335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F., Villalba M., Garcia-Espinosa M.A., Ballesteros P., Bogonez E., Satrustegui J., Cerdan S. Intracellular compartmentation of pyruvate in primary cultures of cortical neurons as detected by (13)C NMR spectroscopy with multiple (13)C labels. J. Neurosci. Res. 2001;66:771–781. doi: 10.1002/jnr.10048. [DOI] [PubMed] [Google Scholar]
- Dilova I., Easlon E., Lin S.J. Calorie restriction and the nutrient sensing signaling pathways. Cell. Mol. Life Sci. 2007;64:752–767. doi: 10.1007/s00018-007-6381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon E., Tsang F., Dilova I., Wang C., Lu S.P., Skinner C., Lin S.J. The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J. Biol. Chem. 2007;282:6161–6171. doi: 10.1074/jbc.M607661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P., Andre L., Ansell R., Blomberg A., Adler L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995;17:95–107. doi: 10.1111/j.1365-2958.1995.mmi_17010095.x. [DOI] [PubMed] [Google Scholar]
- Erjavec N., Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes & Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K., Tsubamoto Y., Terauchi Y., Sugiyama T., Kishimoto T., Takahashi N., Yamauchi N., Kubota N., Murayama S., Aizawa T., et al. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Longo V.D. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pozza F., Pletcher S.D., Gendron C.M., Longo V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Falcón A.A., Aris J.P. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:41607–41617. doi: 10.1074/jbc.M307025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferea T.L., Botstein D., Brown P.O., Rosenzweig R.F. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S.L., Guarente L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes & Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gallo C.M., Smith D.L., Smith J.S. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauslund M., Ronnow B. Carbon source-dependent transcriptional regulation of the mitochondrial glycerol-3-phosphate dehydrogenase gene, GUT2, from Saccharomyces cerevisiae. Can. J. Microbiol. 2000;46:1096–1100. doi: 10.1139/w00-105. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- Haigis M.C., Mostoslavsky R., Haigis K.M., Fahie K., Christodoulou D.C., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Karow M., Blander G., et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jazwinski S.M. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene. 2005;354:22–27. doi: 10.1016/j.gene.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Jiang J.C., Jaruga E., Repnevskaya M.V., Jazwinski S.M. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Jiang J.C., Wawryn J., Shantha Kumara H.M., Jazwinski S.M. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp. Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Andalis A.A., Fink G.R., Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Kirkland K.T., Fields S., Kennedy B.K. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:1381–1387. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Hu D., Kerr E.O., Tsuchiya M., Westman E.A., Dang N., Fields S., Kennedy B.K. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005a;1:614–621. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R.W., Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kresze G.B., Ronft H. Pyruvate dehydrogenase complex from baker’s yeast. 1. Purification and some kinetic and regulatory properties. Eur. J. Biochem. 1981;119:573–579. doi: 10.1111/j.1432-1033.1981.tb05646.x. [DOI] [PubMed] [Google Scholar]
- Lamming D.W., Latorre-Esteves M., Medvedik O., Wong S.N., Tsang F.A., Wang C., Lin S.J., Sinclair D.A. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson J.E., Niu X.D., Reed L.J. Functional analysis of the domains of dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Biochemistry (Mosc.) 1991;30:11249–11254. doi: 10.1021/bi00111a009. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr. Opin. Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin S.S., Manchester J.K., Gordon J.I. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- Lin S.-J., Kaeberlein M., Andalis A.A., Sturtz L.A., Defossez P.-A., Culotta V.C., Fink G.R., Guarente L. Calorie restriction extends life span by shifting carbon toward respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Ford E., Haigis M., Liszt G., Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes & Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martin D.E., Soulard A., Hall M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Meijer A.J., Van Dam K. The metabolic significance of anion transport in mitochondria. Biochim. Biophys. Acta. 1974;346:213–244. doi: 10.1016/0304-4173(74)90001-9. [DOI] [PubMed] [Google Scholar]
- Minard K.I., McAlister-Henn L. Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: Evidence for three isozymes of yeast malate dehydrogenase. Mol. Cell. Biol. 1991;11:370–380. doi: 10.1128/mcb.11.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard K.I., McAlister-Henn L. Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J. Biol. Chem. 1992;267:17458–17464. [PubMed] [Google Scholar]
- Morin P.J., Subramanian G.S., Gilmore T.D. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1992;1171:211–214. doi: 10.1016/0167-4781(92)90124-i. [DOI] [PubMed] [Google Scholar]
- Nisoli E., Tonello C., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Palaiologos G., Hertz L., Schousboe A. Evidence that aspartate aminotransferase activity and ketodicarboxylate carrier function are essential for biosynthesis of transmitter glutamate. J. Neurochem. 1988;51:317–320. doi: 10.1111/j.1471-4159.1988.tb04872.x. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Palmieri F., Runswick M.J., Walker J.E. Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett. 1996;399:299–302. doi: 10.1016/s0014-5793(96)01350-6. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Vozza A., Agrimi G., De Marco V., Runswick M.J., Palmieri F., Walker J.E. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J. Biol. Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Agrimi G., Runswick M.J., Fearnley I.M., Palmieri F., Walker J.E. Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 2001;276:1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- Panowski S.H., Wolff S., Aguilaniu H., Durieux J., Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Park P.U., McVey M., Guarente L. Separation of mother and daughter cells. Methods Enzymol. 2002;351:468–477. doi: 10.1016/s0076-6879(02)51865-6. [DOI] [PubMed] [Google Scholar]
- Pronk J.T., Yde Steensma H., Van Dijken J.P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnow B., Kielland-Brandt M.C. GUT2, a gene for mitochondrial glycerol 3-phosphate dehydrogenase of Saccharomyces cerevisiae. Yeast. 1993;9:1121–1130. doi: 10.1002/yea.320091013. [DOI] [PubMed] [Google Scholar]
- Roth G.S., Ingram D.K., Lane M.A. Caloric restriction in primates and relevance to humans. Ann. N. Y. Acad. Sci. 2001;928:305–315. doi: 10.1111/j.1749-6632.2001.tb05660.x. [DOI] [PubMed] [Google Scholar]
- Schneider J.C., Guarente L. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 1991;11:4934–4942. doi: 10.1128/mcb.11.10.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T., Thornton J., Butow R.A. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Wei Y., Dauk M., Tan Y., Taylor D.C., Selvaraj G., Zou J. Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell. 2006;18:422–441. doi: 10.1105/tpc.105.039750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Lawrence C.W. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1978. [Google Scholar]
- Sinclair D.A., Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Brachmann C.B., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen J., Ciriacy M. Biochemical basis of mitochondrial acetaldehyde dismutation in Saccharomyces cerevisiae. J. Bacteriol. 1991;173:7012–7017. doi: 10.1128/jb.173.21.7012-7017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.M., Sutherland P., Steffan J.S., McAlister- Henn L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry (Mosc.) 1988;27:8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Todisco S., Agrimi G., Castegna A., Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tissenbaum H.A. Overlapping and distinct functions for a Caenorhabditis elegansSIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang H.T., Rahaim P., Robbins P., Yocum R.R. Cloning, sequence, and disruption of the Saccharomyces diastaticus DAR1 gene encoding a glycerol-3-phosphate dehydrogenase. J. Bacteriol. 1994;176:7091–7095. doi: 10.1128/jb.176.22.7091-7095.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch W., Walford R.L. The retardation of aging and diseases by dietary restriction. Charles C. Thomas; Springfield, IL: 1998. [Google Scholar]
- Wood J.G., Rogina B., Lavu S., Howitz K., Helfand S.L., Tatar M., Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Young E.T., Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:3024–3034. doi: 10.1128/mcb.5.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]