Abstract
Pleistocene glacial cycles are thought to have played a major role in the diversification of temperate and boreal species of North American birds. Given that coalescence times between sister taxa typically range from 0.1 to 2.0 Myr, it has been assumed that diversification occurred as populations were isolated in refugia over long periods of time, probably spanning one to several full glacial cycles. In contrast, the rapid postglacial range expansions and recolonization of northern latitudes following glacial maxima have received less attention as potential promoters of speciation. Here we report a case of extremely rapid diversification in the songbird genus Junco as a result of a single continent-wide range expansion within the last 10 000 years. Molecular data from 264 juncos sampled throughout their range reveal that as the yellow-eyed junco (Junco phaeonotus) of Mesoamerica expanded northward following the last glacial maximum, it speciated into the dark-eyed junco (Junco hyemalis), which subsequently diversified itself into at least five markedly distinct and geographically structured morphotypes in the USA and Canada. Patterns of low genetic structure and diversity in mitochondrial DNA and amplified fragment length polymorphism loci found in dark-eyed juncos relative to Mesoamerican yellow-eyed juncos provide support for the hypothesis of an expansion from the south, followed by rapid diversification in the north. These results underscore the role of postglacial expansions in promoting diversification and speciation through a mechanism that represents an alternative to traditional modes of Pleistocene speciation.
Keywords: speciation, postglacial expansion, phylogeography, Holocene, Junco
1. Introduction
Range expansions and colonization of new areas following glacial periods have resulted in some of the most remarkable radiations known in vertebrates (Schluter 2000; Coyne & Orr 2004). In North America, isolation of populations in refugia during Pleistocene glacial cycles is thought to have been largely responsible for the most recent speciation events in birds (Johnson & Cicero 2004; Weir & Schluter 2004), though the overall importance of the Pleistocene relative to older periods remains a topic of considerable debate (Klicka & Zink 1997; Avise & Walker 1998; Zink et al. 2004).
Bird populations underwent rapid continent-wide expansions following glacial maxima as ice sheets receded and climatic amelioration allowed the recolonization of northern latitudes, a process documented for a number of bird species as well as numerous other animal and plant taxa since the last glacial maximum (LGM) 18 000 years ago (Taberlet et al. 1998; Lessa et al. 2003; Hewitt 2004). In birds, intraspecific tests of this ‘postglacial expansion hypothesis’ have documented patterns of low genetic diversity and structure in temperate and boreal populations relative to southern temperate and subtropical populations, demonstrating that the lack of genetic structure is due to shared ancestral polymorphism rather than gene flow (Milá et al. 2006; Ruegg et al. 2006).
Given that genetic distances between sister avian taxa generally correspond to divergence times ranging from 0.1 to 2 Myr (Klicka & Zink 1997; Johnson & Cicero 2004; Weir & Schluter 2007), it has been traditionally assumed that speciation took place in isolated refugia over one to several full glacial cycles (Mengel 1964; Hubbard 1973; Zink & Klicka 2006). The role of postglacial expansions themselves in driving speciation has received much less attention. Since continent-wide range expansions would subject advancing populations to a wide variety of unoccupied habitats with varying selective regimes, relatively short periods of isolation as new areas are colonized could potentially suffice to drive diversification in comparatively short periods of time. Considering that avian postglacial expansions are as short as a few 1000 years (Milá et al. 2006), and that this scenario does not invoke the need for long-term isolation in refugia, the postglacial expansion hypothesis poses an alternative to traditional modes of Pleistocene speciation.
Here we use molecular genetic data to test the postglacial expansion hypothesis in the phenotypically diverse dark-eyed junco (Junco hyemalis), one of the most common and widespread bird species of temperate North America, and its sister species the yellow-eyed junco (Junco phaeonotus) of the Mesoamerican highlands. By assessing patterns of genetic structure and diversity in mitochondrial DNA (mtDNA) and a multilocus genome scan using amplified fragment length polymorphism (AFLP), we document a sudden expansion of the yellow-eyed junco into temperate North America following the LGM and its exceptionally rapid speciation into the polymorphic dark-eyed junco as expanding populations reached temperate latitudes. These results have important implications for our understanding of the tempo of speciation, as well as that of the evolution of phenotypic traits like plumage colouration.
2. Material and methods
(a) Taxonomy of the study group
The genus Junco is currently composed of three species—the dark-eyed junco J. hyemalis, the yellow-eyed junco J. phaeonotus and the volcano junco Junco vulcani—although the actual number of species comprised in the dark-eyed junco complex remains a matter of debate (Nolan et al. 2002). The complex comprises at least five well-differentiated, largely allopatric morphotypes that differ prominently in plumage, beak colour and morphology (figure 1a; Miller 1941; Nolan et al. 2002). Formerly considered species (slate-coloured junco J. hyemalis, white-winged junco Junco aikeni, Oregon junco Junco oreganus, grey-headed junco Junco caniceps and the Guadalupe island junco Junco insularis), these five forms were recently lumped to form the dark-eyed junco (J. hyemalis; American Ornithologists' Union 1973), though their assignment as subspecific ‘groups’ was intended to reflect the marked phenotypic differentiation between them, their allopatric breeding distributions and the stability of hybrid zones that exist between some groups (Nolan et al. 2002). In a recent monograph on the complex, Nolan et al. (2002) assert that with further investigation, the group is likely to be split again into multiple species. Indeed, a recent study on speciation times in birds has already considered two of the dark-eyed junco groups (J. oreganus and J. caniceps) as species based on evidence for lack of interbreeding when found in sympatry at some localities (Johnson & Cicero 2004). The yellow-eyed junco (J. phaeonotus) has been taxonomically stable and is composed of four different subspecies (phaeonotus, palliatus, fulvescens and alticola; Sullivan 1999). Finally, the volcano junco (J. vulcani) of southern Central America is considered a distant relative of other juncos and is included as an out-group for comparison. Given the unresolved status of Junco taxonomy, the number of species comprised by the genus ranges from a minimum of three to as many as seven. For consistency, we follow current nomenclature which considers the genus to be composed of three species (dark-eyed, yellow-eyed and volcano juncos) and examine individuals from six dark-eyed junco morphotypes (slate-coloured Junco hyemalis hyemalis, white-winged Junco hyemalis aikeni, Oregon Junco hyemalis oreganus, pink-sided Junco hyemalis mearnsi, grey-headed Junco hyemalis caniceps and red-backed Junco hyemalis dorsalis), referred to here collectively as the ‘dark-eyed junco complex’, and from three of the four described subspecies of the yellow-eyed junco (phaeonotus, palliatus and alticola).
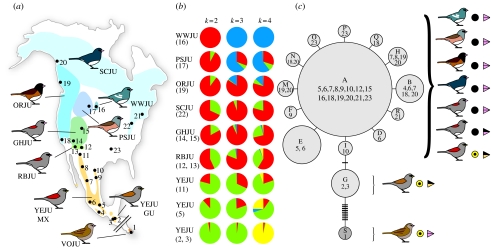
Figure 1.
Phenotypic and genetic variation in the genus Junco. (a) Breeding ranges and phenotypic variation. The eight different taxa included in the genetic analyses are shown. Colours indicate breeding ranges and dots represent sampling localities. From the south, clockwise, with sampling locality number in parentheses: volcano (1), yellow-eyed (2–11), red-backed (12, 13), grey-headed (14, 15), white-winged (16), pink-sided (17), Oregon (18, 19) and slate-coloured juncos (20–23). Site 23 corresponds to a wintering locality of the slate-coloured junco in Alabama. See §2 for specific locality designations. (b) Posterior assignment probabilities of 139 individuals in different Junco populations to K values of 2, 3 and 4 using 75 AFLP loci in the program Structure. Each colour in each pie diagram represents the per cent posterior probability of assignment to a given cluster, averaged across all individuals in that population. Posterior assignment probabilities per individual are provided in the electronic supplementary material, figure S2. Taxon abbreviations are as follows: WWJU, white-winged; PSJU, pink-sided; ORJU, Oregon; SCJU, slate-coloured; GHJU, grey-headed; RBJU, red-backed; YEJU, yellow-eyed. Numbers under taxon codes correspond to sampling localities in (a). (c) Minimum-spanning network of absolute distances between mtDNA control region haplotypes found in 264 individual juncos. Each circle represents a haplotype, with size proportional to the haplotype's overall frequency. Letters designate haplotypes and numbers correspond to the sampling localities where the haplotype was detected. Network branches represent a single nucleotide change and hatch marks along branches represent additional changes. Circular symbols to the right of the bird schematics represent yellow or dark iris colour and triangles depict beak colouration.
(b) Sampling and mtDNA sequencing
Individuals were captured in the field using mist nets, and blood and/or feather samples were collected for genetic analysis. Three digital images of each individual were taken as photographic vouchers. Sampling localities, preceded by locality numbers in figure 1a in brackets, and with sample sizes in parentheses (for mtDNA sequencing and AFLP analysis, respectively), were as follows: volcano junco J. vulcani: [1] La Georgina, Cerro de la Muerte, Costa Rica (obtained from toe-pad tissue of specimens nos. 37851 and 37853 in the Dickey Bird and Mammal Collection at UCLA); yellow-eyed junco Junco phaeonotus alticola: [2] Chichim (11, 11) and [3] Chiantla (8, 9) in Huehuetenango, Guatemala; Junco phaeonotus phaeonotus: [4] San Pedro Mixtepec, Oaxaca (14, 0), [5] La Cima, Distrito Federal (28, 16) and [6] El Rosario, Michoacán (35, 0); J. ph. palliatus: [7] San Diego Tenaenz, Durango (33, 0), [8] El Vergel, Chihuahua (19, 0) and [11] Pinaleno Mountains, Arizona, USA; Junco phaeonotus palliatus/phaeonotus: [9] Galeana, Nuevo León (4, 0) and [10] Sierra del Carmen, Coahuila (8, 0) in Mexico (0, 16); red-backed junco J. h. dorsalis: [12] Greer (6, 11) and [13] Flagstaff, Arizona, USA (0, 5); grey-headed junco J. h. caniceps: [14] Long Valley Junction (0, 8) and [15] Uinta Mountains (7, 7), Utah, USA; white-winged junco J. h. aikeni: [16] Sundance, Black Hills, Wyoming, USA (5, 14); pink-sided junco J. h. mearnsi: [17] Lander, Big Horn Mountains, Wyoming, USA (5, 12); Oregon junco J. h. oreganus: [18] Malibu, California, USA (19, 0) and [19] Banks Island, British Columbia, Canada (15, 14); and slate-coloured junco J. h. hyemalis: [20] Juneau, Alaska (16, 0), [21] Lyme, Connecticut (4, 0), [22] Virginia, USA (0, 19), and [23] Hollins, Alabama (wintering individuals, 24, 0), USA.
Primers for PCR amplification of 349 bp of the hypervariable region I of the mitochondrial control region (CR) were H417 and LGL2 (Tarr 1995). Sequencing of two different coding regions of the mtDNA (610 bp of the cytochrome c oxidase I and 327 bp of cytochrome b genes) yielded no variation across dark-eyed and yellow-eyed individuals (see electronic supplementary material). DNA extraction, amplification and sequencing protocols were as previously described (Milá et al. 2000). We calculated values of haplotype diversity (h) and nucleotide diversity (π) using the program Arlequin v. 3.1. (Schneider et al. 2000). As a precaution against sequencing a nuclear copy of the mtDNA CR, we aligned our most frequent haplotype (‘A’) with a fragment containing partial sections of the CR and the ND6 gene deposited in GenBank by M. D. Sorenson (no. AF407130). The coding ND6 fragment translated into its amino acid sequence with no stop codons or indels, and the CR fragment had identical sequence to our haplotype A. Our haplotype A was also identical to submission no. AY138919 by R. M. Zink, which was amplified from ultrapurified mtDNA (Zink & Weckstein 2003). All sequences in this study have been deposited in GenBank under accession numbers AY995307–AY995315.
(c) AFLP genotyping and analysis
We generated AFLP profiles for 142 individuals (for which blood samples were available) using a protocol modified from Vos et al. (1995). Whole genomic DNA extracted from blood samples using a Qiagen kit was digested with restriction enzymes EcoRI and MseI, and fragments were ligated to oligonucleotide adapters with T4 DNA ligase. A random sample of fragments was obtained through a pre-selective amplification using primers matching the adapters and enzyme restriction sites with an additional selective nucleotide in the 3′ end (E-tag and M-cgt). A final selective amplification was conducted with the same primers modified by the addition of three arbitrarily chosen nucleotides for each enzyme primer (E-tag and M-cgt), and labelled with the fluorescent dye 6FAM. Selectively amplified fragments were run in an ABI 3700 genetic analyzer using a LIZ500 size standard. Peaks were visualized using Genemapper v. 7.0 and scored manually. Only unambiguously scorable loci and individuals were included in the analysis and peaks found in less than 2% of individuals were excluded. The average per locus methodological error rate for the AFLP data (assessed by running a subset of 10 individuals three times from the pre-selective amplification step) was 1.8%, similar to that of other AFLP studies on birds (Mock et al. 2002; Spaulding et al. 2006).
To assess genetic structure among samples, we used the program Structure v. 2.1 (Pritchard et al. 2000) and applied a model of no admixture as recommended for dominant markers (Pritchard & Wen 2004). The optimal number of populations (K) was calculated following the method by Evanno et al. (2005). We calculated Fst using Arlequin v. 3.1., and tested for significance through 1000 random permutations of the dataset (Excoffier et al. 1992). We also obtained Weir & Cockerham's Fst estimator θ (Weir & Cockerham 1984) using Genepop (Raymond & Rousset 1995).
(d) Demographic history
We tested for population expansions using mtDNA sequence data and Fu's test of neutrality (Fu 1997), which is particularly effective at detecting sudden changes in effective population size (Ramos-Onsins & Rozas 2002) by measuring departures from neutrality in situations characterized by an excess of rare alleles and young mutations. Large negative values of Fs reject population stasis. Significance of Fs values was evaluated with 1000 random permutations in Arlequin v. 3.1. In addition, we compared mismatch distributions of pairwise nucleotide differences among CR haplotypes with expectations of a sudden expansion model (Rogers & Harpending 1992) using Arlequin v. 3.1.
(e) Divergence times and diversification rates
Divergence times between taxa were estimated using three different methods. We applied a molecular clock using a range of calibrations to take into account the inter-taxon variability of CR mutation rates (Baker & Marshall 1997; Ruokonen & Kvist 2002). Given the lack of variation in the mtDNA coding regions surveyed (see electronic supplementary material) and the relatively higher levels of CR variation found in this study, we estimated the Junco CR mutation rate to be considerably higher than the 0.02 substitutions per site Myr−1 rate at which coding regions generally evolve (García-Moreno 2004), and we used rates of 0.15, 0.10 and 0.05 substitutions per site Myr−1, which encompass rates generally found in birds (Baker & Marshall 1997). Much faster mutation rates, as those documented for the CR of penguins (0.30 substitutions per site Myr−1; Lambert et al. 2002) or geese (0.21 substitutions per site Myr−1; Quinn 1992), would make our diversification rate estimates even faster.
We also estimated diversification time with a coalescence-based model that generates non-equilibrium estimates of divergence time independent of gene migration (Nielsen & Wakeley 2001). We used Markov chain Monte Carlo simulations with the program Mdiv (Nielsen & Wakeley 2001), which generates maximum likelihood estimates of θ (2Nfeμ); T, the divergence time between two populations scaled by population size; M, the gene migration rate between the two populations and time to the most recent common ancestor (TMRCA). We assumed uniform priors and set maximum values for T and M of 5 and 20, respectively. To allow for the possibility of multiple mutations at the same nucleotide site, we used the HKY model (Hasegawa et al. 1985). We ran four Markov chains of 4 000 000 cycles with different random seeds, each preceded by a ‘burn-in’ period of 500 000 cycles. Estimates of T and TMRCA were converted to years using t=(Tθ/2μk); and tMRCA=TMRCA θ/2μk; where T and θ are generated by the program; μ is the marker-specific mutation rate; and k is the length of the sequence. We assumed a generation time of 1 year (Nolan et al. 2002) and used the three mutation rates mentioned above.
A third estimate of diversification time was obtained from the distribution of pairwise nucleotide differences among individuals (Rogers & Harpending 1992) of pooled dark-eyed junco samples. To estimate the time since the beginning of an expansion, we used τ=2ut; where t is the time elapsed between initial and current population sizes and u=2μk; where μ is the mutation rate and k is the length of the sequence (Rogers & Harpending 1992).
We used estimates of divergence time to calculate the net diversification interval (NDI), defined as the average time elapsing between the origin of a new lineage and the next branching of that lineage (assuming no extinction). NDI is defined as t/ln Nt; where t is the time to a single common ancestor and Nt is the resulting number of species (Coyne & Orr 2004). We calculated NDIs using Nt=2, 3 and 5, and the time divergence values obtained from Mdiv using the three mutation rates described above. An Nt of 5 would correspond to the yellow-eyed junco and a minimum number of dark-eyed forms (grey-headed, Oregon, slate-coloured and white-winged), and an Nt of 2 would represent the most conservative assessment of variation in the group (yellow-eyed and dark-eyed juncos).
3. Results
(a) mtDNA sequence and population expansion analysis
We examined mtDNA CR sequence from 102 individuals of the dark-eyed junco complex, 160 yellow-eyed and 2 volcano juncos. Remarkably, most individuals from all dark-eyed junco morphotypes and most Mexican yellow-eyed juncos share a single mtDNA CR haplotype (A in figure 1c), suggesting a very recent burst of phenotypic diversification. The high frequency of haplotype A and its close relationship to multiple low-frequency haplotypes suggests a sudden and recent population expansion (Avise 2000). Specifically, populations making up the dark-eyed junco complex show low levels of haplotype and nucleotide diversity (h=0.2722±0.0584, π=0.0008±0.0009), a high frequency of ancestral haplotype A, and a large negative Fs value (Fs=−12.219, p<0.0001; table 1), which are also evidence of a sudden expansion in effective population size. In contrast, yellow-eyed juncos in Mexico and Guatemala show a higher genetic diversity (h=0.6312±0.0330, π=0.0028±0.0021), a lower frequency of haplotype A and a non-significant Fs value (Fs=−0.085, p=0.490).
Table 1.
Genetic diversity in the mtDNA CR and population expansion in Junco. (***p<0.0001; **p<0.01; n.s.: p>0.05; n.a.: not applicable as only one haplotype was found.)
| taxon | n | haplotypes (number) | ha | πb | Fsc |
|---|---|---|---|---|---|
| dark-eyed junco (Junco hyemalis) | 102 | 0.2722±0.0584 | 0.0008±0.0009 | −12.219*** | |
| Oregon junco (J. h. oreganus) | 50 | A(39), B(3), H(2), M(3), N(2), Q(1) | 0.3886±0.1003 | 0.0012±0.0013 | −3.832** |
| pink-sided junco (J. h. mearnsi) | 5 | A(5) | 0.0000±0.0000 | 0.0000±0.0000 | n.a. |
| slate-coloured junco (J. h. hyemalis) | 28 | A(25), O(1), P(1), R(1) | 0.2063±0.1005 | 0.0006±0.0008 | −3.266*** |
| white-winged junco (J. h. aikeni) | 6 | A(6) | 0.0000±0.0000 | 0.0000±0.0000 | n.a. |
| grey-headed junco (J. h. caniceps) | 7 | A(6), I(1) | 0.2857±0.1964 | 0.0008±0.0011 | −0.095 n.s. |
| red-backed junco (J. h. dorsalis) | 6 | A(6) | 0.0000±0.0000 | 0.0000±0.0000 | n.a. |
| yellow-eyed junco (J. phaeonotus) | 160 | 0.6312±0.0330 | 0.0028±0.0021 | −0.850 n.s. | |
| northern Mexico yellow-eyed junco (J. ph. palliatus) | 64 | A(60), B(1), H(2), F(1) | 0.1215±0.0554 | 0.0004±0.0006 | −4.045*** |
| southern Mexico/Guatemala yellow-eyed junco (J. ph. phaeonotus and alticola) | 96 | A(29), B(15), D(1), E(32), G(19) | 0.7417±0.0163 | 0.0040±0.0027 | 1.497 n.s. |
| volcano junco (J. vulcani) | 2 | S(2) | 0.0000±0.0000 | 0.0000±0.0000 | n.a. |
Haplotype diversity.
Nucleotide diversity.
Fu's (14) test of neutrality.
A closer examination of patterns of genetic diversity within the yellow-eyed junco reveals an expansion in northern Mexico populations as evidenced by a significant Fs value and low genetic diversity values (due to a high frequency of haplotype A; table 1). Mismatch distributions further suggest a history of expansion in the dark-eyed junco complex and northern Mexican (sites 7–10 in figure 1a) yellow-eyed juncos, in contrast to southern Mexican (sites 4–6) yellow-eyed juncos (see figure S1 in the electronic supplementary material). Indeed, the Fst value between the dark-eyed junco complex and northern Mexico yellow-eyed juncos (Fst=0.003, p=0.418) was much lower than that between the dark-eyed junco complex and southern Mexico yellow-eyed juncos (Fst=0.272, p<0.001, table 2). These results suggest that yellow-eyed juncos from southern Mexico expanded through northern Mexico, rapidly diversifying into the various dark-eyed morphotypes after they arrived in temperate North America.
Table 2.
Differentiation between dark-eyed and yellow-eyed juncos based on mtDNA CR sequence data (Fst) and AFLP loci (Fst and Weir and Cockerham's θ). Pairwise comparisons of all dark-eyed junco populations combined (DEJU) and yellow-eyed junco populations (northern sites, NYEJU; and southern sites, SYEJU). (***p<0.001; n.s.: p>0.05.)
| mtDNA | AFLP | ||
|---|---|---|---|
| Fst | Fst | θ | |
| DEJU versus NYEJU | 0.003 n.s. | 0.129*** | 0.129 |
| DEJU versus SYEJU (Mex only) | 0.272*** | 0.200*** | 0.199 |
| DEJU versus SYEJU (Mex+Gua) | 0.193*** | 0.202*** | 0.202 |
(b) AFLP analysis
A Bayesian analysis of population structure among all individuals using 75 AFLP markers revealed marked genetic similarity and an absence of structure among most dark-eyed junco morphotypes. The optimal number of populations in the assignment probability analysis using Structure was K=4 (figure 1b), with the four clusters corresponding broadly to Guatemala (yellow), Mexico (green), the white-winged junco population in the Black Hills and some pink-sided juncos from the nearby Bighorn Mountains (blue) and all remaining dark-eyed junco forms (red).
Also evident in the Structure results is the gradual pattern of differentiation between yellow-eyed and dark-eyed populations. When individuals are assigned to one of two clusters (K=2, figure 1b), each one corresponds roughly to yellow-eyed juncos (green) and dark-eyed juncos (red), with average assignment probabilities changing gradually with latitude. Interestingly, red-backed junco individuals (sites 12 and 13 in figure 1a) show similar assignment probabilities to the two clusters as predicted by their intermediate phenotype. For the optimal K=4 (figure 1b), yellow-eyed individuals from Arizona (site 11) show a higher average assignment probability to the dark-eyed cluster (red colour) than those in southern Mexico (site 5), which show a higher assignment probability to the Guatemalan cluster (yellow colour). This pattern is reflected in the Fst analysis, with values between dark-eyed and northern yellow-eyed juncos being lower than between dark-eyed and southern yellow-eyed juncos (table 2).
(c) Divergence times and diversification rates
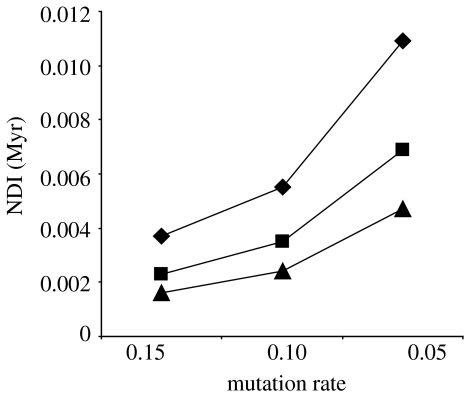
The average genetic distance in mtDNA CR between the dark-eyed junco complex and yellow-eyed juncos was 0.019% when corrected for intraspecific polymorphism (Nei 1987). Applying a molecular clock, this value corresponds to divergence times ranging between 1300 and 3800 years BP, using mutation rates of 15% Myr−1 and 5% Myr−1, respectively. The non-equilibrium estimates of time divergence between populations obtained with Mdiv were 2548–7620 years BP, and TMRCA, or the time when genes last shared a common ancestor ranged between 26 764 and 80 028 years. Diversification time obtained from the distribution of pairwise differences among individuals provided comparable time estimates of 4046–11 976 years BP to the time of the population expansion, depending on mutation rate. Using a generation time of 2 years instead of 1 year, which might apply to small passerines with relatively high mortality rates, would double the time divergence estimates above. Finally, NDIs ranged from 0.0109 to 0.0016 Myr, depending on the mutation rate and the number of new taxa (Nt) considered to have been produced (figure 2).
Figure 2.
Estimates of NDIs in the yellow-eyed and dark-eyed juncos based on mtDNA CR sequence, using three mutation rates (0.15, 0.10 and 0.05 substitutions per site Myr−1) and three values for the number of diversified taxa (Nt), 2 (filled diamonds), 3 (filled squares) and 5 (filled triangles).
4. Discussion
Results from the mtDNA analyses provide strong support for a rapid postglacial expansion of the yellow-eyed junco from Mexico into temperate North America, and for a subsequent rapid burst of diversification that gave rise to at least five distinct morphotypes of the dark-eyed junco as expanding populations reached temperate latitudes. An alternative explanation for the genetic pattern found among dark-eyed junco morphotypes is the occurrence of a selective sweep by an adaptive mtDNA haplotype, which could have spread across previously differentiated morphotypes by crossing hybrid zones. The ubiquity of haplotype A in yellow-eyed junco populations (with whom dark-eyed juncos are not known to hybridize), the limited amount of gene flow evident in southern populations (e.g. haplotypes D and E are restricted to central Mexico, and G is restricted to Guatemala, see table 1) and the gradual latitudinal increase in the frequency of ancestral haplotype A, all strongly suggest that the predominance of haplotype A in northern populations reflects shared ancestral polymorphism resulting from a rapid population expansion, and not gene flow (Milá et al. 2006).
However, in order to confidently reject the selective sweep hypothesis, a congruent pattern of low genetic structure and diversity in dark-eyed juncos must be confirmed with unlinked nuclear markers. This is because factors such as bottlenecks and population expansions should affect the entire genome equally, whereas selective sweeps should only be detected in the locus under selection (and tightly linked loci; Galtier et al. 2000). The lack of clear structure in AFLP loci within the dark-eyed junco complex despite marked phenotypic differentiation and considerable geographical distances separating them is congruent with the mtDNA results and supports the hypothesis of rapid diversification following a recent expansion. The differentiation of the white-winged junco population was driven largely by the presence of three nearly diagnostic loci and could be due to founder events and the subsequent fixation of rare alleles in a population of small effective size, although the influence of natural selection cannot be ruled out. The fact that some pink-sided juncos from the nearby Bighorn Mountains of Wyoming and even some Oregon juncos cluster with the white-winged juncos suggests that the alleles involved appeared (or became fixed) in the small Black Hills population and spread west through gene flow across hybrid zones.
Several phenotypic characteristics of juncos are consistent with a history of diversification through a northward expansion from southern forms. The red-backed and grey-headed juncos of Arizona and the southern Rocky Mountains, in particular, are phenotypically intermediate between yellow-eyed and dark-eyed juncos. For example, the loss of the yellow iris in the red-backed junco (otherwise remarkably similar to its yellow-eyed neighbour), and the loss of the bicoloured beak in the grey-headed junco just to the north (figure 1c), appear to represent successive steps in the gradual evolution of dark-eyed forms, which subsequently and rapidly acquired their diverse plumage patterns (often with a prominent increase of eumelanins and phaeomelanins). Recent divergence with fast plumage evolution (since the LGM) has been recently documented in Icterus orioles (Baker et al. 2003; Kondo et al. 2004), redpolls (Seutin et al. 1995), bluethroats (Questiau et al. 1998; Zink et al. 2003), yellow wagtails (Ödeen & Björklund 2003) and yellow-rumped warblers (Milá et al. 2007). However, given the higher number and marked distinctiveness of plumage morphs and the short amount of time involved, the dark-eyed junco complex represents an exceptional case of fast plumage diversification.
Large-scale range expansions into formerly glaciated areas following glacial maxima have been reported for numerous animal and plant taxa during the Pleistocene (Hewitt 2000) and speciation during this period is thought to have been driven in part by isolation in glacial refugia (Weir & Schluter 2004). Previous studies on avian speciation rates have shown that coalescence times to the closest common ancestor of extant species pairs are typically in the range of 0.1–2.0 Myr (Avise & Walker 1998; Lovette & Bermingham 1999; Johnson & Cicero 2004; Weir & Schluter 2007) and often pre-date the Pleistocene (Klicka & Zink 1999). In a recent molecular study of 39 sister-species pairs of birds (Johnson & Cicero 2004), only two pairs showed splits younger than 50 000 years (Spizella breweri/Spizella taverneri, two weakly differentiated taxa, and Junco oreganus/J. caniceps, two of the taxa in the dark-eyed junco complex examined here). In contrast, our molecular data indicate that diversification of dark-eyed juncos took place within the time span of a single postglacial expansion within the last 10 000 years, as ancestral yellow-eyed junco populations expanded their range through northern Mexico and into temperate North America following the LGM.
The occurrence of hybridization along contact zones between some forms in the dark-eyed junco complex (except for, at least, caniceps/hyemalis, caniceps/aikeni, and insularis), suggests that reproductive isolation between some morphotypes is not yet complete. However, the marked phenotypic differentiation between forms, their largely allopatric distribution, and the fact that most hybrid zones are of limited width and apparently stationary over at least a century (Miller 1941; Nolan et al. 2002), all suggest that dark-eyed junco forms are on distinct evolutionary trajectories and may represent a case of speciation in action. The rate of diversification presented here, even if only two species are considered, is among the fastest observed in vertebrates (Coyne & Orr 2004) and is similar to that reported for the cichlid fish species flock of Lake Nabugabo in East Africa, where five species evolved in less than 4000 years, which is thought to represent the fastest radiation within cichlids (NDI=0.004 Myr; Coyne & Orr 2004). Because the Junco diversification is so recent, the probability of extinction of new taxa is low, which could account for the unusually low NDI values obtained. Potentially, similarly fast diversification bouts could have taken place in older lineages, yet because only a fraction of the original number of species has survived, speciation rates might be underestimated. However, given that none of the previously studied avian groups show similar recent bouts of diversification, and the fact that several bird taxa show similar signatures of postglacial population expansion over North America yet lack a comparable amount of phenotypic diversification (Ball et al. 1988; Zink & Dittman 1993; Milot et al. 2000; Kimura et al. 2002; Ruegg & Smith 2002; Veit et al. 2005; Milá et al. 2006), the rapid diversification in Junco appears to be exceptional.
Selection-driven evolutionary change can be especially fast in peripheral isolates such as those found at the leading edge of an expansion (Garcia-Ramos 1997; Gavrilets et al. 2000), and given the marked differences in plumage characters, sexual selection may have acted as the mechanism of divergence (Fisher 1930; Lande 1981; Barraclough et al. 1995). Indeed, a recently established Oregon junco population in southern California has evolved marked differences in a sexually selected trait (amount of white on outer tail feathers) after only several generations (Yeh 2004). The relative roles of female choice and male–male competition in the phenotypic divergence of juncos is unclear, although recent research on the slate-coloured junco suggests that both might be involved (McGlothlin et al. 2005). Our results underscore the role of postglacial expansions in driving diversification and speciation, and suggest that the colonization of new areas following glacial periods can lead to exceptionally rapid rates of evolution.
Acknowledgments
E. Berg, O. Espinosa, S. Larios, A. Lee, A. Gutiérrez, F. Hertel, A. Oliveras, M. Ramírez, V. Rodríguez and R. Van Buskirk provided excellent help in the field, and The Institute for Bird Populations' MAPS Program, G. Baluss, V. Bladen, W. Sakai and M. Wilson generously contributed feather samples. E. Ketterson kindly provided blood samples from slate-coloured juncos. A. Navarro-Sigüenza at the Museo de Zoología A. Herrera at UNAM, the Museo de las Aves in Saltillo, and K. Molina at UCLA's Dickey Bird and Mammal Collection facilitated blood and tissue samples. S. Lopez and N. Timber helped with laboratory work. Financial support was provided by a UC MEXUS Program Dissertation grant, UCLA Latin American Center, an American Ornithologists' Union's Wetmore Award, the Turner Foundation, the Environmental Protection Agency and the National Science Foundation. R. Calsbeek and two anonymous reviewers provided helpful comments on the manuscript.
Supplementary Material
References
- American Ornithologists' Union. Thirty-second supplement to the American Ornithologists' Union check list of North American Birds. Auk. 1973;90:411–419. [Google Scholar]
- Avise J.C. Harvard University Press; Cambridge, MA: 2000. Phylogeography: the history and formation of species. [Google Scholar]
- Avise J.C, Walker D. Pleistocene phylogeographic effects on avian populations and the speciation process. Proc. R. Soc. B. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. doi:10.1098/rspb.1998.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Marshall H. Mitochondrial control region sequences as tools for understanding evolution. In: Mindell D.P, editor. Avian molecular evolution and systematics. Academic Press; San Diego, CA: 1997. pp. 49–80. [Google Scholar]
- Baker J.M, López-Medrano E, Navarro-Sigüenza A.G, Rojas-Soto O.R, Omland K.E. Recent speciation in the orchard oriole group: divergence of Icterus spurius spurius and Icterus spurius fuertesi. Auk. 2003;120:848–859. doi:10.1642/0004-8038(2003)120[0848:RSITOO]2.0.CO;2 [Google Scholar]
- Ball R.M, Freeman S, James F.C, Bermingham E, Avise J.C. Phylogeographic population structure of red-winged blackbirds assessed by mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1988;85:1558–1562. doi: 10.1073/pnas.85.5.1558. doi:10.1073/pnas.85.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough T.G, Harvey P.H, Nee S. Sexual selection and taxonomic diversity in passerine birds. Proc. R. Soc. B. 1995;259:211–215. doi:10.1098/rspb.1995.0031 [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates, Inc; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. doi:10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P.E, Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. Oxford University Press; Oxford, UK: 1930. The genetical theory of natural selection. [Google Scholar]
- Fu Y.X. Statistical neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Depaulis F, Barton N.H. Detecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics. 2000;155:981–987. doi: 10.1093/genetics/155.2.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno J. Is there a universal mtDNA clock for birds? J. Avian Biol. 2004;35:465–468. doi:10.1111/j.0908-8857.2004.03316.x [Google Scholar]
- Garcia-Ramos G. Genetic models of adaptation and gene flow in peripheral populations. Evolution. 1997;51:21–28. doi: 10.1111/j.1558-5646.1997.tb02384.x. doi:10.2307/2410956 [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Li H, Vose M.D. Patterns of parapatric speciation. Evolution. 2000;54:1126–1134. doi: 10.1111/j.0014-3820.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. doi:10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. The genetic legacy of the Quaternary ice ages. Nature (London) 2000;405:907–913. doi: 10.1038/35016000. doi:10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- Hewitt G.M. Genetic consequences of climatic oscillations in the Quaternary. Phil. Trans. R. Soc. B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. doi:10.1098/rstb.2003.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J.P. Avian evolution in the aridlands of North America. Living Bird. 1973;12:155–196. [Google Scholar]
- Johnson N.K, Cicero C. New mitochondrial DNA data affirm the importance of Pleistocene speciation in North American birds. Evolution. 2004;58:1122–1130. doi: 10.1111/j.0014-3820.2004.tb00445.x. [DOI] [PubMed] [Google Scholar]
- Kimura M, Clegg S.M, Lovette I.J, Holder K.R, Girman D.J, Milá B, Wade P, Smith T.B. Phylogeographical approaches to assessing demographic connectivity between breeding and overwintering regions in a Nearctic–Neotropical warbler (Wilsonia pusilla) Mol. Ecol. 2002;11:1605–1616. doi: 10.1046/j.1365-294x.2002.01551.x. doi:10.1046/j.1365-294X.2002.01551.x [DOI] [PubMed] [Google Scholar]
- Klicka J, Zink R.M. The importance of recent ice ages in speciation: a failed paradigm. Science. 1997;277:1666–1669. doi:10.1126/science.277.5332.1666 [Google Scholar]
- Klicka J, Zink R.M. Pleistocene effects on North American songbird evolution. Proc. R. Soc. B. 1999;266:695–700. doi:10.1098/rspb.1999.0691 [Google Scholar]
- Kondo B, Baker J.M, Omland K.E. Recent speciation between the Baltimore oriole and the black-backed oriole. Condor. 2004;106:674–680. doi:10.1650/7496 [Google Scholar]
- Lambert D.M, Ritchie P.A, Millar C.D, Holland B, Drummond A.J, Baroni C. Rates of evolution in ancient DNA from Adélie penguins. Science. 2002;295:2270–2273. doi: 10.1126/science.1068105. doi:10.1126/science.1068105 [DOI] [PubMed] [Google Scholar]
- Lande R. Models of speciation by sexual selection on polygenic traits. Proc. Natl Acad. Sci. USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. doi:10.1073/pnas.78.6.3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa E.P, Cook J.A, Patton J.L. Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proc. Natl Acad. Sci. USA. 2003;100:10 331–10 334. doi: 10.1073/pnas.1730921100. doi:10.1073/pnas.1730921100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovette I.J, Bermingham E. Explosive speciation in the New World Dendroica warblers. Proc. R. Soc. B. 1999;266:1629–1636. doi:10.1098/rspb.1999.0825 [Google Scholar]
- McGlothlin J.W, Parker P.G, Nolan V.J, Ketterson E.D. Correlational selection leads to genetic integration of body size and an attractive plumage trait in dark-eyed juncos. Evolution. 2005;59:658–671. [PubMed] [Google Scholar]
- Mengel R. The probable history of species formation in some northern wood warblers (Parulidae) Living Bird. 1964;3:9–43. [Google Scholar]
- Milá B, Girman D.J, Kimura M, Smith T.B. Genetic evidence for the effect of a postglacial population expansion on the phylogeography of a North American songbird. Proc. R. Soc. B. 2000;267:1033–1040. doi: 10.1098/rspb.2000.1107. doi:10.1098/rspb.2000.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milá B, Smith T.B, Wayne R.K. Postglacial population expansion drives the evolution of long-distance migration in a songbird. Evolution. 2006;60:2403–2409. [PubMed] [Google Scholar]
- Milá B, Smith T.B, Wayne R.K. Speciation and rapid phenotypic differentiation in the yellow-rumped warbler Dendroica coronata complex. Mol. Ecol. 2007;16:159–173. doi: 10.1111/j.1365-294X.2006.03119.x. doi:10.1111/j.1365-294X.2006.03119.x [DOI] [PubMed] [Google Scholar]
- Miller A. Speciation in the avian genus Junco. Univ. Calif. Publ. Zool. 1941;44:173–434. [Google Scholar]
- Milot E, Gibbs H.L, Hobson K.A. Phylogeography and genetic structure of northern populations of the yellow warbler (Dendroica petechia) Mol. Ecol. 2000;9:667–681. doi: 10.1046/j.1365-294x.2000.00897.x. doi:10.1046/j.1365-294x.2000.00897.x [DOI] [PubMed] [Google Scholar]
- Mock K.E, Theimer T.C, Rhodes O.E.J, Jr, Greenberg D.L, Keim P. Genetic variation across the historical range of the wild turkey (Meleagris gallopavo) Mol. Ecol. 2002;11:643–657. doi: 10.1046/j.1365-294x.2002.01467.x. doi:10.1046/j.1365-294X.2002.01467.x [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, V. J., Ketterson, E. D., Cristol, D. A., Rogers, C. M., Clotfelter, E. D., Titus, R. C., Schoech, S. J. & Snajdr, E. 2002 Dark-eyed junco (Junco hyemalis). In The birds of North America, vol. 716 (eds A. Poole & F. Gill), pp. 1–44. Philadelphia, PA: The Birds of North America, Inc.
- Ödeen A, Björklund M. Dynamics in the evolution of sexual traits: losses and gains, radiation and convergence in yellow wagtails (Motacilla flava) Mol. Ecol. 2003;12:2113–2130. doi: 10.1046/j.1365-294x.2003.01883.x. doi:10.1046/j.1365-294X.2003.01883.x [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. & Wen, W. 2004 Documentation for Structure software: version 2. See http://pritch.bsd.uchicago.edu
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Questiau S, Eybert M.-C, Gaginskaya A.R, Gielly L, Taberlet P. Recent divergence between two morphologically differentiated subspecies of bluethroat (Aves: Muscicapidae: Luscinia svecica) inferred from mitochondrial sequence variation. Mol. Ecol. 1998;1998:239–245. doi: 10.1046/j.1365-294x.1998.00345.x. doi:10.1046/j.1365-294x.1998.00345.x [DOI] [PubMed] [Google Scholar]
- Quinn T.W. The genetic legacy of mother goose—phylogeographic patterns of lesser snow goose Chen caerulescens caerulescens maternal lineages. Mol. Ecol. 1992;1:105–117. doi: 10.1111/j.1365-294x.1992.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S.E, Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (v. 1.2): a population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rogers A.R, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ruegg K.C, Smith T.B. Not as the crow flies: a historical explanation for circuitous migration in Swainson's thrush (Catharus ustulatus) Proc. R. Soc. B. 2002;269:1375–1381. doi: 10.1098/rspb.2002.2032. doi:10.1098/rspb.2002.2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg K.C, Hijmans R.J, Moritz C. Climate change and the origin of migratory pathways in the Swainson's thrush, Catharus ustulatus. J. Biogeogr. 2006;33:1172–1182. doi:10.1111/j.1365-2699.2006.01517.x [Google Scholar]
- Ruokonen M, Kvist L. Structure and evolution of the avian mitochondrial control region. Mol. Phylogenet. Evol. 2002;23:422–432. doi: 10.1016/s1055-7903(02)00021-0. doi:10.1016/S1055-7903(02)00021-0 [DOI] [PubMed] [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. Arlequin 2.0: a software for population genetics data analysis. [Google Scholar]
- Seutin G, Ratcliffe L.M, Boag P.T. Mitochondrial DNA homogeneity in the phenotypically diverse redpoll finch complex (Aves: Carduelinae: Carduelis flammea hornemanni) Evolution. 1995;49:962–973. doi: 10.1111/j.1558-5646.1995.tb02331.x. doi:10.2307/2410418 [DOI] [PubMed] [Google Scholar]
- Spaulding A.W, Mock K.E, Schroeder A, Warheit K.I. Recency, range expansion, and unsorted lineages: implications for interpreting neutral genetic variation in the sharp-tailed grouse (Tympanuchus phasianellus) Mol. Ecol. 2006;15:2317–2332. doi: 10.1111/j.1365-294X.2006.02935.x. doi:10.1111/j.1365-294X.2006.02935.x [DOI] [PubMed] [Google Scholar]
- Sullivan, K. A. 1999 Yellow-eyed junco (Junco phaeonotus). In The birds of North America, vol. 464 (eds A. Poole & F. Gill). Philadelphia, PA: The Birds of North America, Inc.
- Taberlet P, Fumagalli L, Wust-Saucy A.-G, Cosson J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. doi:10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- Tarr C.L. Primers for amplification and determination of mitochondrial control-region sequences in oscine passerines. Mol. Ecol. 1995;4:527–529. doi: 10.1111/j.1365-294x.1995.tb00251.x. [DOI] [PubMed] [Google Scholar]
- Veit M.L, Robertson R.J, Hamel P.B, Friesen V.L. Population genetic structure and dispersal across a fragmented landscape in cerulean warblers (Dendroica cerulea) Conserv. Genet. 2005;6:159–174. doi:10.1007/s10592-004-7831-9 [Google Scholar]
- Vos P, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. doi:10.1093/nar/23.21.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Weir J.T, Schluter D. Ice sheets promote speciation in boreal birds. Proc. R. Soc. B. 2004;271:1881–1887. doi: 10.1098/rspb.2004.2803. doi:10.1098/rspb.2004.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J.T, Schluter D. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science. 2007;315:1574–1576. doi: 10.1126/science.1135590. doi:10.1126/science.1135590 [DOI] [PubMed] [Google Scholar]
- Yeh P.J. Rapid evolution of a sexually selected trait following population establishment in a novel habitat. Evolution. 2004;58:166–174. doi: 10.1111/j.0014-3820.2004.tb01583.x. [DOI] [PubMed] [Google Scholar]
- Zink R.M, Dittman D.L. Population structure and gene flow in the chipping sparrow and a hypothesis for evolution in the genus Spizella. Wilson Bull. 1993;105:399–413. [Google Scholar]
- Zink R.M, Klicka J. The tempo of avian diversification: a comment on Johnson and Cicero. Evolution. 2006;60:411–412. [PubMed] [Google Scholar]
- Zink R.M, Weckstein J.D. Recent evolutionary history of the fox sparrows (genus: Passerella) Auk. 2003;120:522–527. doi:10.1642/0004-8038(2003)120[0522:REHOTF]2.0.CO;2 [Google Scholar]
- Zink R.M, Drovetski S.V, Questiau S, Fadeev I.V, Nesterov E.V, Westberg M.C, Rohwer S. Recent evolutionary history of the bluethroat (Luscinia svecica) across Eurasia. Mol. Ecol. 2003;12:3069–3075. doi: 10.1046/j.1365-294x.2003.01981.x. doi:10.1046/j.1365-294X.2003.01981.x [DOI] [PubMed] [Google Scholar]
- Zink R.M, Klicka J, Barber B.R. The tempo of avian diversification during the Quaternary. Phil. Trans. R. Soc. B. 2004;359:215–220. doi: 10.1098/rstb.2003.1392. doi:10.1098/rstb.2003.1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.