SUMMARY
Inosine 5’-monophosphate dehydrogenase (IMPDH) catalyzes the rate limiting step in guanine nucleotide biosynthesis. IMPDH has an evolutionary conserved CBS subdomain of unknown function. The subdomain can be deleted without impairing the in vitro IMPDH catalytic activity and is the site for mutations associated with human retinitis pigmentosa. A guanine-prototrophic Escherichia coli strain, MP101, was constructed with the subdomain sequence deleted from the chromosomal gene for IMPDH. The ATP content was substantially elevated in MP101 whereas the GTP content was slighty reduced. The activities of IMPDH, adenylosuccinate synthetase and GMP reductase were 2 – 3 fold lower in MP101 crude extracts compared with the BW25113 wild type strain. Guanine induced a 3-fold reduction in the MP101 ATP pool and a 4-fold increase in the GTP pool within 10 minutes of addition to growing cells; this response does not result from the reduced IMPDH activity or starvation for guanylates. In vivo kinetic analysis using 14-C tracers and 33-P pulse-chasing revealed mutation-associated changes in purine nucleotide fluxes and turnover rates. We conclude that the CBS subdomain of IMPDH may coordinate the activities of the enzymes of purine nucleotide metabolism and is essential for maintaining the normal ATP and GTP pool sizes in E. coli.
Keywords: CBS, IMPDH, Bateman domain, nucleotide pools
INTRODUCTION
Inosine 5’-monophosphate dehydrogenase catalyzes the rate limiting step in guanine nucleotide biosynthesis, the NAD-dependent oxidation of inosine 5’-monophosphate (IMP1) to xanthosine 5’-monophosphate (XMP) (Zalkin and Nygaard, 1996). IMPDH is located at the branch point of the de novo purine biosynthetic pathway, where IMP becomes committed to either GMP or AMP synthesis (Fig. 1); thus, IMPDH has the potential to regulate both guanylate and adenylate nucleotide pools. IMP may originate from de novo purine biosynthesis starting with 1-pyrophosphoribosyl-5-phosphate (PRPP), be regenerated from AMP or GMP, or be salvaged from hypoxanthine (Zalkin and Nygaard, 1996). Depletion of the guanylate nucleotide pool has a strongly detrimental effect on cell proliferation which has led to the establishment of IMPDH as a target for anti-cancer, immunosuppressive, anti-viral and anti-microbial chemotherapies (Pankiewicz and Goldstein, 2003).
Fig. 1. The de novo and salvage pathways for purine nucleotide biosynthesis.
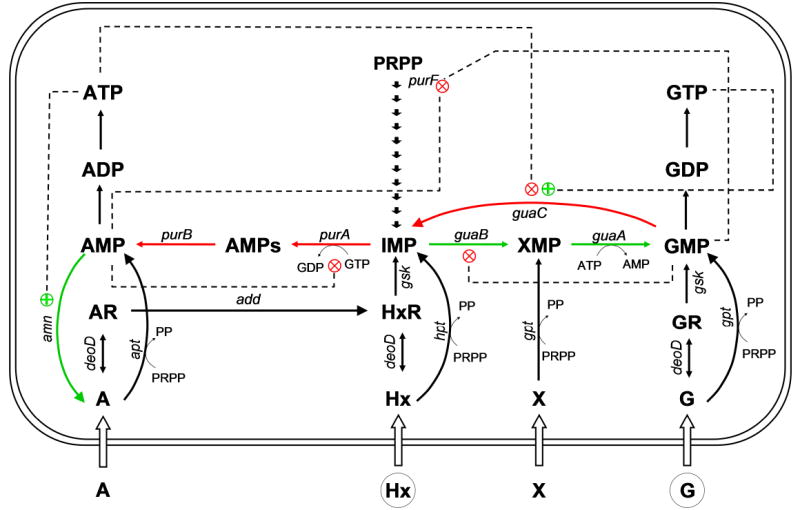
A, adenine; Hx, hypoxanthine; G, guanine; X, xanthine; AR, adenosine; HxR, inosine; GR, guanosine; AMPs, adenylosuccinate. purA, adenylosuccinate synthetase (AMPsS); purB, adenylosuccinate lyase; guaB, IMP dehydrogenase (IMPDH); guaA, GMP synthetase (GMPS); deoD, purine nucleoside phosphorylase; hpt, guanine/hypoxanthine phosphoribosyltransferase; gpt, guanine phosphoribosyltransferase; gsk, guanosine/inosine kinase; apt, adenine phosphoribosyltransferase; add, adenosine deaminase; guaC, GMP reductase (GMPR); purF, amidophosphoribosyl transferase; amn, AMP nucleosidase. Shown in red and green arrows are reactions that may have a lower or a higher flux, respectively, in the conditions of a high GTP pool in the MP101 mutant. Dashed lines reflect feedback regulation.
IMPDH has an unusual structure with a subdomain of ca. 120 residues, composed of two repeats of a sequence known as the cystathionine β-synthase (CBS) domain (Bateman, 1997), inserted within the center of the dehydrogenase sequence (Zhang et al., 1999). CBS domains have been found in many proteins of unrelated function, including ion transporters, protein kinases and other enzymes (see www.sanger.ac.uk/Users/agb/CBS/CBS.html), and generally occur in tandem pairs (Ignoul and Eggermont, 2005). Each CBS pair is also known as a Bateman domain (Bateman, 1997). The physiological importance of Bateman domains is emphasized by the fact that mutations in these domains are associated with a variety of human hereditary diseases, such as homocystinuria, Wolff-Parkinson-White syndrome, myotonia congenital, etc. (for a review see (Ignoul and Eggermont, 2005)). However, the physiological roles of only a few CBS domains have been described and for many proteins the functional significance of their CBS sequences remains a mystery.
AMP-dependent protein kinase (AMPK) and ClC-5 chloride transporter were recently demonstrated to be regulated by binding of adenosine phosphates to their Bateman domains. AMPK is activated by AMP, but this activation is prevented by ATP (Hardie, 2003; Kahn et al., 2005). Hence, AMPK is capable of directly sensing the cellular energy state reflected in the ATP/AMP ratio. Recent crystal structures of AMPK revealed that AMP and ATP alternate in binding to a Bateman domain located in the kinase γ subunit (Townley and Shapiro, 2007). Similarly, a crystal structure of ClC-5 provided evidence for ATP and ADP binding by the CBS pair, but the physiological significance of this interaction remains unclear (Meyer et al., 2007). In contrast, the Bateman domain of cystathionine β-synthase is involved in enzyme activation by S-adenosylmethionine (Jhee and Kruger, 2005) while the Bateman domain of ABC transporter OpuA from Lactococcus lactis has been postulated to act as a sensor for intracellular ionic strength (Biemans-Oldehinkel et al., 2006). Thus, it seems that the physiological functions and binding partners of the Bateman domains may vary considerably between different proteins.
The Bateman domain of IMPDH is highly evolutionarily conserved but has no established function. Crystallographic studies have shown that the subdomain is appended to the (αβ)8-barrel protein core as an independent folding unit (Zhang et al., 1999) and its overall structure is highly similar to the structures of other Bateman domains (Day et al., 2007; Meyer et al., 2007; Miller et al., 2004; Townley and Shapiro, 2007). The subdomain is the site of amino acid substitutions that are associated with the RP10 form of human retinitis pigmentosa (Bowne et al., 2002; Bowne et al., 2006; Kennan et al., 2002). Nevertheless, these mutations, as well as a complete deletion of the subdomain, do not impair the in vitro IMPDH catalytic activity (Bowne et al., 2006; Gan et al., 2002; Mortimer and Hedstrom, 2005; Nimmesgern et al., 1999). Several in vitro properties of the subdomain have been reported, but none have been substantiated in vivo, and many appear unlikely to reflect the in vivo function. For instance, the subdomain has been postulated to be involved in regulation of IMPDH catalytic function via allosteric stimulation by ATP (Scott et al., 2004). However, others have been unable to reproduce this work (Mortimer and Hedstrom, 2005). Non-sequence specific binding of single-stranded nucleic acids of unclear function has also been described in vitro (McLean et al., 2004). A marked decrease in the in vitro nucleic acid binding was observed in human IMPDH containing RP10-linked amino acid substitutions (Mortimer and Hedstrom, 2005). Consistent with the latter, IMPDH has been found to associate with proteins that are involved in RNA processing and splicing regulation in yeast (Butland et al., 2005; Ho et al., 2002). Taken together, these observations suggested that IMPDH has an as yet unappreciated role in RNA metabolism. It has therefore been speculated that the RP10-linked mutations impair the RNA-related function of IMPDH and, perhaps, have no effect on retinal nucleotide pools (McLean et al., 2004; Mortimer and Hedstrom, 2005). Thus, elucidating whether the subdomain is an integral component of IMPDH function in the de novo nucleotide biosynthesis, or serves an unrelated purpose, is central to understanding its biological role and the RP10 pathogenesis.
It is clear that Escherichia coli is the most facile system for in vivo studies of the biological role of the subdomain. Unlike bacteria, most Eucarya, including yeast, have two or more genes for IMPDH which complicates genetic studies. We have excised the CBS-pair coding sequence from the E. coli chromosome leaving the core domain intact and functional. We report results of a comparative analysis of the purine nucleotide pools, fluxes and turnover rates in the mutant guaBΔCBS and wild type guaB+ strains, which indicate for the first time that the CBS subdomain of IMPDH plays an important role in the regulation of purine nucleotide metabolism.
RESULTS
Following the Krogh principle that there is an optimal experimental organism for any physiological question (Krogh, 1920), we used the “recombineering” technology (Court et al., 2002) to precisely delete the CBS subdomain coding sequence of the single chromosomal copy of the E. coli gene for IMPDH (guaB) while retaining the IMPDH activity (Fig. 2A and Fig. S1). The resulting guaBΔCBS strain, MP101, was prototrophic for guanine and grew with an unaltered rate on various minimal and rich media (Fig. S3 and data not shown), indicating that IMPDH catalytic function is sustained.
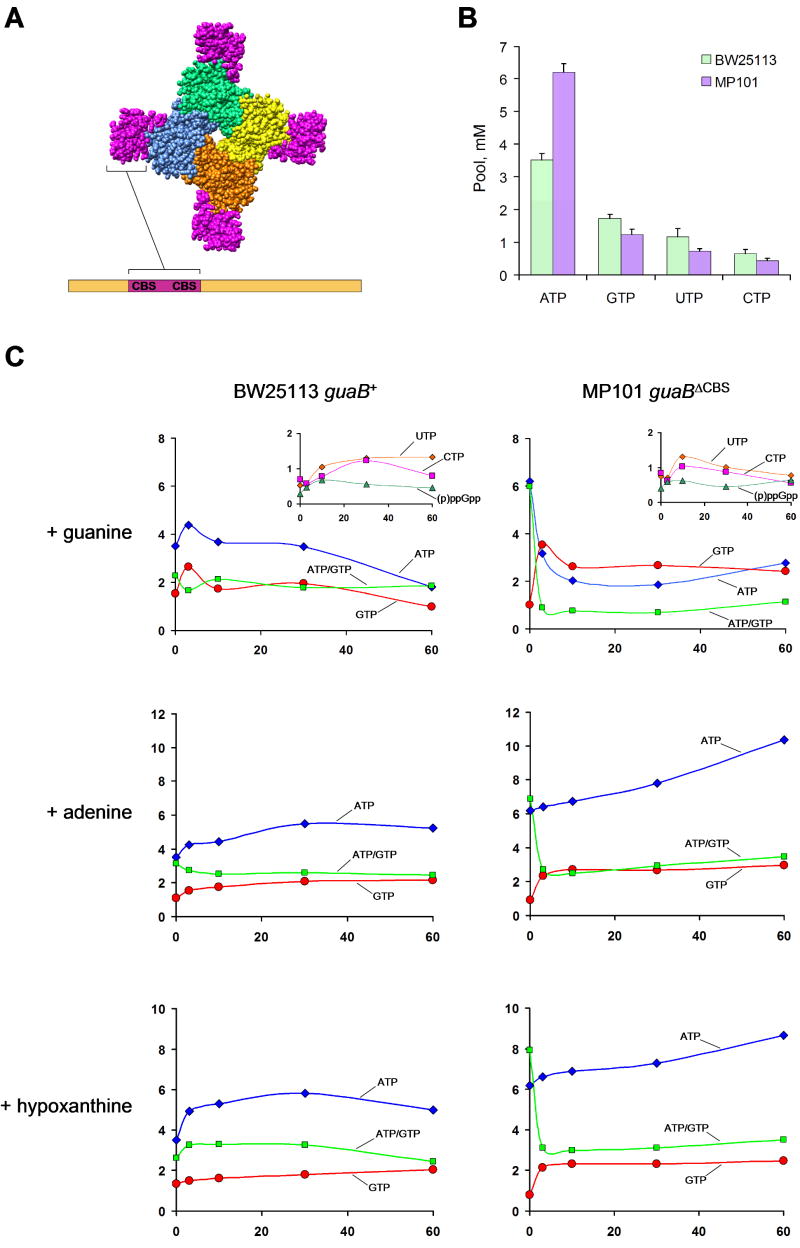
Fig. 2. The effect of guaBΔCBS mutation on nucleotide pools.
A, The structure of Streptococcus pyogenes IMPDH tetramer (PDB code 1ZFJ); the catalytic domains are shown in yellow, blue, orange and green; the deleted CBS domain is shown in each monomer in purple with respect to the sequence homology with E. coli GuaB. The positioning of the CBS pair within the IMPDH sequence is shown as a schematic bar representation (a full ClustalW alignment of E. coli BW25113 guaB+, E. coli MP101 guaBΔCBS and S. pyogenes IMPDH proteins is presented in Fig. S1). B, Nucleotide pools in strains grown on minimal media. The average of 5 independent measurements is given. Error bars are standard deviations. C, Purine-induced nucleotide pool responses in BW25113 guaB+ and MP101 guaBΔCBS. 30 μg/ml of the indicated purine base was added at 0 min. Vertical axis, nucleotide pool (mM); horizontal axis, time (min).
Nucleotide pools and enzyme activities in crude extracts
When intracellular nucleotide pools of MP101 and the parent strain BW25113 were compared by 33Pi labeling followed by 2-D TLC (Bochner and Ames, 1982), a ca. 1.8 fold higher ATP content was found in the guaBΔCBS mutant (Fig. 2B and Table 1). The pools of GTP, UTP and CTP were only slightly lower in the mutant strain which may result from a mild deficit of PRPP imposed by the increased ATP synthesis (alternatively, the pyrimidine nucleotide pool may be affected due to the described regulatory effect of changing ATP and GTP levels on the expression of enzymes of pyrimidine nucleotide synthesis (Jensen, 1979; Jensen, 1989). The intracellular nucleotide concentrations in the wild type strain were similar to the previously reported values (Bochner and Ames, 1982; Neidhardt et al., 1987). The IMPDH and adenylosuccinate synthetase (AMPsS) activities were 2 times lower in the guaBΔCBS mutant (Table 2) whereas the activity of GMP reductase (GMPR) was lower in the mutant strain by a factor of 3. The reduced AMPsS activity contrasts with the elevated ATP pool in the guaBΔCBS mutant and suggests that other mechanisms, such as allosteric regulation, may be involved.
Table 1. Nucleotide pool sizes in BW25113 and MP101 grown on MOPS minimal media.
The pool values are averaged from 5 independent measurements.
| Strain | |||
|---|---|---|---|
| Pool (mM) ± σ | BW25113 | MP101 | p |
| ATP | 3.50 ± 0.20 | 6.19 ± 0.27 | 2.4E-07 |
| ADP | 0.220 ± 0.022 | 0.264 ± 0.057 | 0.193 |
| AMP | 0.215 ± 0.078 | 0.255 ± 0.090 | 0.523 |
| dATP | 0.193 ± 0.025 | 0.278 ± 0.019 | 0.001 |
| GTP | 1.726 ± 0.132 | 1.228 ± 0.179 | 0.002 |
| GDP | 0.069 ± 0.011 | 0.060 ± 0.014 | 0.691 |
| dGTP | 0.109 ± 0.019 | 0.086 ± 0.031 | 0.226 |
| IMP | 0.386 ± 0.064 | 0.513 ± 0.153 | 0.164 |
| NAD | 0.610 ± 0.094 | 0.861 ± 0.154 | 0.024 |
| UTP | 1.163 ± 0.253 | 0.732 ± 0.150 | 0.019 |
| CTP | 0.662 ± 0.119 | 0.443 ± 0.068 | 0.013 |
| dCTP | 0.157 ± 0.049 | 0.105 ± 0.049 | 0.168 |
| NADPH | 0.071 ± 0.016 | 0.086 ± 0.003 | 0.273 |
| (p)ppGpp | 0.366 ± 0.045 | 0.285 ± 0.045 | 0.149 |
| XMP | 0.147 ± 0.015 | 0.203 ± 0.053 | 0.129 |
Table 2. Enzyme activities in the crude extracts of BW25113 guaB+ and MP101 guaBΔCBS grown on MOPS minimal media.
An average of two measurements conducted on extracts obtained from separate cultures is given along with the arithmetic +/- values.
| Enzyme activity (nmol/h/mg protein)
| |||
|---|---|---|---|
| Strain | IMPDH | AMPsS | GMPR |
| BW25113 | 25.6 ± 3.4 | 295.6 ± 12.4 | 43.9 ± 11.1 |
| MP101 | 11.8 ± 1.4 | 124.4 ± 15.2 | 14.6 ± 2.9 |
While the previous studies indicate that deletion of the CBS subdomain is unlikely to render IMPDH less catalytically active (Gan et al., 2002; Nimmesgern et al., 1999), the mutation may alter protein folding or predispose the enzyme to proteolysis, leading to the observed 2 fold reduction in the IMPDH activity in MP101. Low IMPDH activity (10% of normal) in a leaky guaB mutant has been shown to cause a marked decrease in the GTP pool and an increase in ATP and, especially, IMP pools (Jensen, 1979). We note that GMP synthetase (GMPS) activity is increased in the guaBΔCBS mutant, which indicates an increased expression of the guaB-guaA operon (Fig. S6). This response may result from an insufficient IMPDH activity in the guaBΔCBS mutant and some level of starvation for guanylate nucleotides. In line with this interpretation, incubation of MP101 with 10 μg/ml xanthine decreases the GMPS activity in this strain, although it still remains ca. 2 fold higher compared with the isogenic guaB+ strain. To test whether the purine nucleotide pool distortion observed in the guaBΔCBS mutant could be a (non-specific) effect of the 2 fold decrease in IMPDH activity, rather than relate specifically to the CBS subdomain deletion, we overexpressed the mutant enzyme in MP101 from the pGUA22 plasmid containing the IMPDHΔCBS gene under the control of a constitutive lacUV5 promoter (Fig. S2). The IMPDH activity in the MP101/pGUA22 strain was 2 fold higher compared with the isogenic guaB+ strain and 4 fold higher than in MP101 cells transformed with the “empty” pCR2.1-TOPO-E plasmid (see. Fig. 3A). The overexpression of IMPDHΔCBS completely normalized the GTP pool in the guaBΔCBS mutant and decreased the ATP pool to 4.6 mM, which is still 1.1 mM higher than the wild type ATP level (Fig. 3B). Further, leaky guaB mutants have been shown to accumulate very high IMP levels (at least 10 fold higher than the wild type), in addition to the ATP pool expansion and GTP pool reduction (Jensen, 1979), whereas only slight, if any, IMP accumulation was observed in MP101 (Table 1). These results suggest that the reduced IMPDH activity may lead to a mild starvation for guanylate nucleotides but additional mechanisms are implicated in the altered regulation of the ATP pool in the guaBΔCBS mutant.
Fig. 3. Effect of IMPDHΔCBS overexpression on MP101 guaBΔCBS purine nucleotide pools.
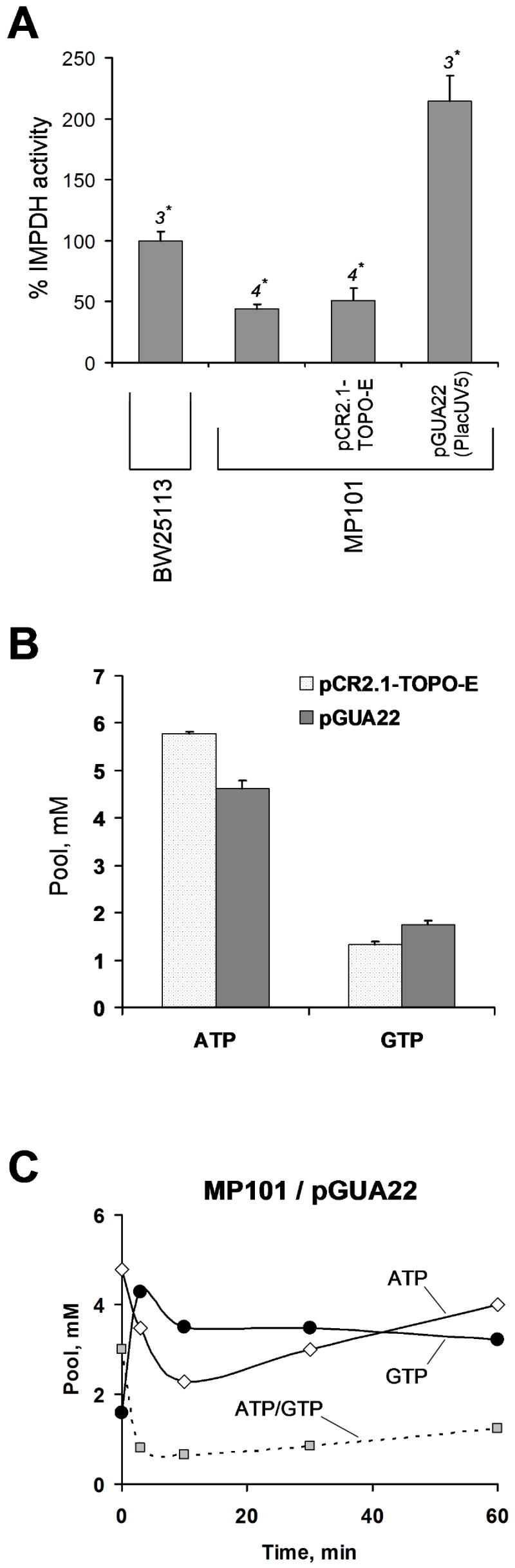
A. Constitutive overexpression of IMPDHΔCBS in MP101 guaBΔCBS. MP101 cells containing the pGUA22 plasmid were grown to a mid-exponential phase in MOPS media supplemented with 100 μg/ml carbenicillin and harvested by centrifugation. BugBuster reagent (Novagen) containing 30 μg/ml PMSF was used for protein extraction (1 ml per 100 OU600). The extract was cleared by centrifugation and IMPDH activity per mg of protein was measured using the 14C-IMP-based assay as described in the Experimental Procedures section. The asterisk indicates the number of independent measurements. B. ATP and GTP pools of minimal-media grown MP101 cells containing the pGUA22 plasmid for IMPDHΔCBS overexpression versus the pCR2.1-TOPO-E control vector. C. Guanine-induced purine nucleotide pool response in minimal media-grown MP101 guaBΔCBS cells overexpressing IMPDHΔCBS. Vertical axis, nucleotide pool (mM); horizontal axis, time from addition of 30 μg/ml guanine (min).
IMPDH is not activated by ATP
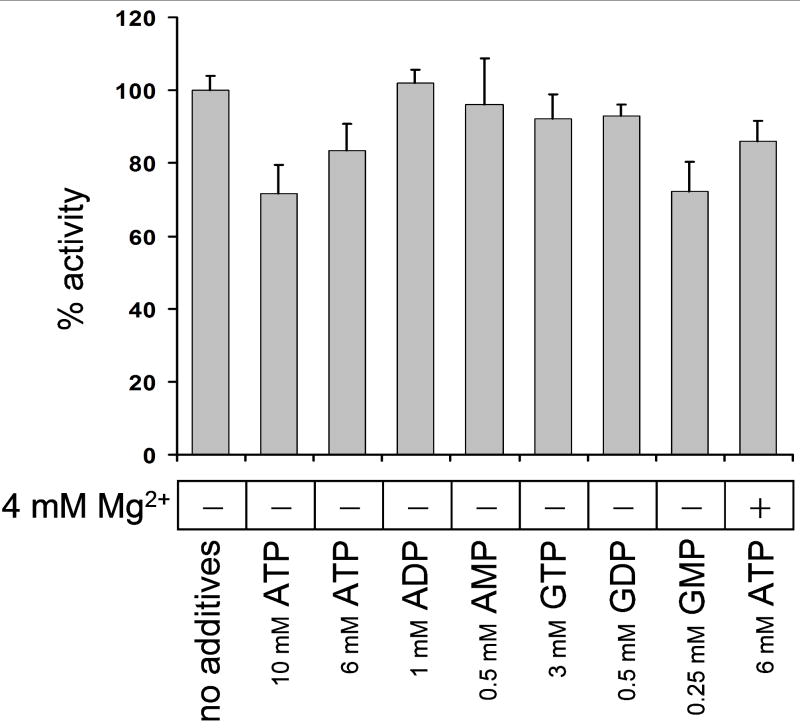
Despite the fact that IMPDH has been postulated to be akin to AMPK and ClC proteins in their activation by ATP (Scott et al., 2004), others could not reproduce this work (Mortimer and Hedstrom, 2005). However, the recently published crystal structures of AMPK and ClC-5 complexed with nucleotides revealed that the Bateman domains bind a Mg2+-free form of ATP (Day et al., 2007; Meyer et al., 2007; Townley and Shapiro, 2007). We speculated that the large Mg2+ concentrations previously used in ATP activation experiments on IMPDH (Mortimer and Hedstrom, 2005) prevented researchers from observing an effect of ATP on IMPDH catalysis. Therefore, we tested whether IMPDH enzymatic activity is influenced by ATP in the absence of Mg ions. The results of this experiment are shown in Fig. 4. Addition to the IMPDH reaction mix of 6 – 10 mM ATP, both in the presence and in the absence of 4 mM magnesium acetate, did not activate IMPDH. In fact, 10 mM ATP had a slightly inhibitory effect; we did not investigate whether this effect was specific. Addition of 0.25 – 3 mM of other adenosine and guanosine nucleotides did not significantly influence the IMPDH activity in the E. coli BW25113 extract. Our use of a crude dialyzed extract, as opposed to a purified enzyme preparation, eliminates the possibility that ATP-mediated regulation requires a third binding partner that is removed during purification. Our results offer support to the study by Mortimer and Hedstrom in which the regulation of human IMPDH by ATP was not found (Mortimer and Hedstrom, 2005).
Fig. 4. The effect of adenylate and guanylate nucleotides on IMPDH activity in E. coli BW25113 guaB+ extract.
The concentrations of nucleotide additives were chosen to be 2 – 5 times greater than physiological with the exception of GMP which was used at a concentration at least 10 times greater than physiological (Neidhardt et al., 1987). The presence in the reaction of 4 mM Mg(CH3COO)2 is indicated with a plus sign. The experiment was conducted in 4 replicates. Error bars are standard deviations.
Purine-induced changes in nucleotide pools
Next, we asked whether the guaBΔCBS mutation affected the response of the intracellular nucleotide pools to the presence of salvageable purine bases in the culture medium. In the first series of experiments, shown in Figs. 2C and S4, the cells were grown in MOPS minimal media and the nucleotide pools were measured before and immediately after addition to the growth culture of a purine base. A remarkable result was obtained with guanine (Fig. 2C). Within the first 3 – 10 min after addition of 30 μg/ml guanine to the guaBΔCBS mutant culture, we observed a 3-fold decrease in the ATP pool whereas the GTP pool increased by a factor of 4, reaching 4 mM, thus resulting in a marked decrease in the ATP/GTP ratio from 6 to 0.7. No such changes were observed in the isogenic guaB+ strain; apart from a slight and transient increase in the GTP and ATP pools, the wild type strain displayed the expected ability to maintain the ATP/GTP ratio within a narrow physiological range (Petersen, 1999). Both strains demonstrated similar changes in the concentrations of UTP, CTP and (p)ppGpp in response to guanine. The initial slight decrease in the UTP and CTP content probably reflects a transient deficit of PRPP which normally happens in the first minutes after addition of a base or nucleoside as a result of an increased demand for PRPP by the purine phosphorybosyl transferase reaction (Zalkin and Nygaard, 1996). It further suggests that the depletion of the PRPP pool and the efficiency of guanine salvage are not substantially different in the two strains. Importantly, no accumulation of AMP or ADP was observed either in MP101 or the wild type strain (data not shown). Therefore, the decrease in ATP content is not simply a result of a decreased adenylate energy charge due to increased ATP consumption in the reactions of guanylate de novo biosynthesis and salvage (Chapman and Atkinson, 1977). Further, simultaneous addition of 30 μg/ml adenine and 30 μg/ml guanine lessened the [ATP] decrease but not the [GTP] increase (Fig. S4); this substantiates the absence of severe changes in energy metabolism in the guaBΔCBS mutant, which has apparently retained the ability to salvage adenine into AMP and phosphorylate it to ATP. Responses of BW25113 and MP101 to 60 μg/ml guanosine were identical to those seen with guanine (Fig. S4). In contrast, addition to the media of similar molar concentrations of adenine or hypoxanthine had almost no effect on BW25113 or MP101 adenylate nucleotide pools. A reproducible trend of the guaBΔCBS mutant to quickly but moderately increase its GTP pool in response to adenine or hypoxanthine was noted, which tended to normalize the ATP/GTP ratios (Fig. 2C). The ability of hypoxanthine and adenine to increase the GTP pool in the guaBΔCBS mutant to a value even slightly higher than the normal substantiates the absence of a catalytic defect in the IMPDHΔCBS enzyme since an active IMP dehydrogenase is essential for the conversion of adenylate nucleotides and IMP into guanylates (see Fig. 1). Further, a 4 fold overexpression of IMPDHΔCBS from the pGUA22 plasmid did not normalize the MP101 guanine response, despite its ability to relieve the mild GTP starvation and partially decrease the ATP pool (Fig. 3C). Together, these results indicate that the decreased IMPDH activity per se has a limited, if any, role in the purine nucleotide pool distortion observed in the guaBΔCBS mutant.
The differential guanine-induced nucleotide pool responses occur rapidly after purine addition (within 3 – 10 min), which suggests mutation-associated changes at the level of enzyme activity rather than in a transcriptional response of the guaBΔCBS mutant. The decline in the ATP pool did not impose starvation of MP101 for adenylates, resulting in no detectable effect on cell growth with glucose as a carbon source (Fig. S3). Apparently, even the minimal ATP concentration of 2 mM reached after addition of guanine in the guaBΔCBS mutant is sufficient for normal growth. In addition, we reproducibly observed a trend for normalization of the ATP content by 60 min after addition of guanine (Figs. 2C, 3C and 5B). This is likely achieved by the well-described purine-induced transcriptional response mediated primarily by the interaction of intracellular purine bases with the PurR transcriptional repressor (Zalkin and Nygaard, 1996). In this case, the transcriptional response will compensate for the changes in the enzyme activity and the system will achieve a new steady state. Consistent with this interpretation, simultaneous addition of 10 μg/ml chloramphenicol, an inhibitor of translation and, hence, the transcriptional response, with 30 μg/ml guanine completely blocked the delayed increase in ATP content; hence, the ATP/GTP ratio decreased to 0.3 by 60 min (Fig. S4).
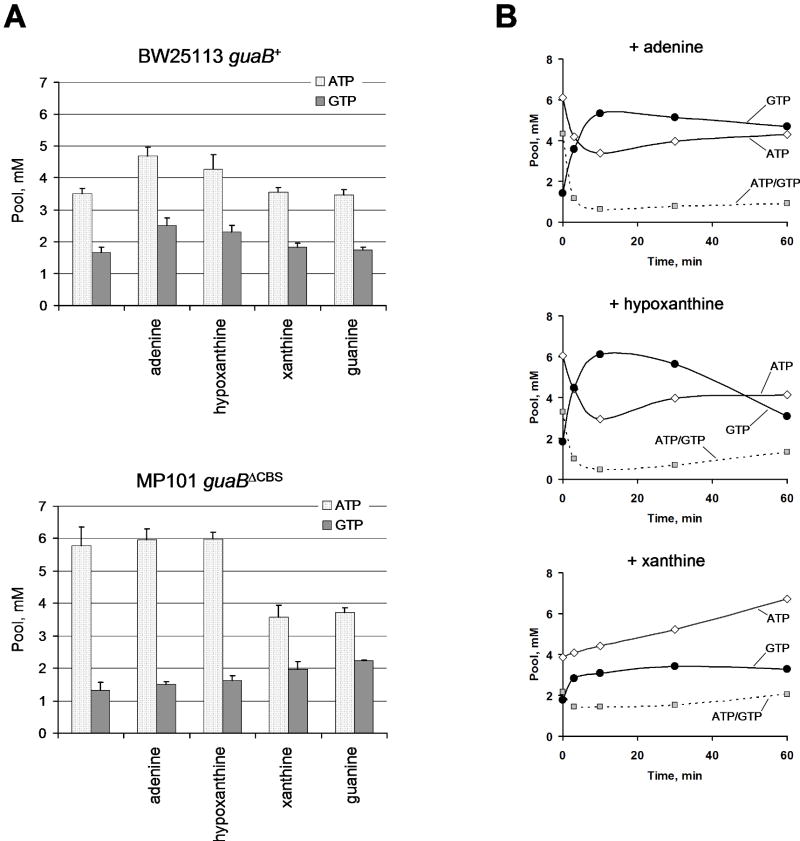
Fig. 5. Purine nucleotide pools in BW25113 guaB+ and MP101 guaBΔCBS during steady-state growth on minimal media supplemented with indicated purine bases.
The cells were cultured in the presence of a purine base and 33Pi for at least 2 hrs before nucleotide pool measurement. A. Nucleotide pools during purine-supplemented steady state growth. B. Guanine-induced purine nucleotide pool responses in MP101 guaBΔCBS cells grown in the presence of adenine, hypoxanthine or xanthine. 30 μg/ml guanine was added at 0 min to MP101 cells growing on MOPS minimal media supplemented with 10 μg/ml of the indicated compounds.
Addition of decoyinine, an inhibitor of GMP synthetase (Vasantha and Freese, 1980), simultaneously with guanine limited the guanine-induced increase in GTP pool in the mutant and completely prevented such an increase in the wild type strain (Fig. S4). Furthermore, a higher ATP content was observed in both strains upon guanine addition in the presence of decoyinine than in its absence. Apparently, the inhibition of GMP biosynthesis by decoyinine allowed more IMP to be converted into adenine nucleotides, resulting in an increase in the ATP pool in the wild type and partially preventing the ATP concentration decrease in MP101. This result shows that IMPDH enzymatic activity is not quickly reduced upon guanine addition, and that IMPDH plays an important role in the guanine-induced swelling of the guanylate nucleotide pool both in the wild type and in the mutant strain.
Two antagonistic enzymes are thought to be involved in the regulation of the guanylate nucleotide pool in E. coli: IMPDH and GMP reductase (GMPR), which converts GMP to IMP (Zalkin and Nygaard, 1996). Together with GMPS the three constitute a complete cycle (see Fig. 1). To our knowledge, individual fluxes through the steps in this cycle have never been reported; rather, the net flux from IMP to GMP has been inferred based on the rates of nucleic acid synthesis (Chapman and Atkinson, 1977). Much of how this cycle operates in vivo has been inferred from the in vitro behavior of individual enzymes. IMPDH is inhibited by GMP; this inhibition is competitive with respect to IMP (Gilbert et al., 1979). GMPR is inhibited by physiological concentrations of ATP; this inhibition is completely reversed by GTP (Mager and Magasnik, 1960). Thus, conditions leading to a decreased ATP/GTP ratio should normally inhibit GMP synthesis and increase the conversion of GMP to IMP, restoring the balance. Apparently, the guaBΔCBS mutant is unable to convert excess guanylate nucleotides into adenylate nucleotides which probably results from the reduced GMPR activity, as shown in Table 2. In turn, the reduction of the ATP pool can result from either decreased de novo synthesis or increased degradation. The total adenylate pool turns over in less than a minute (Chapman and Atkinson, 1977) and even a small change in ATP production or utilization can quickly result in large pool size perturbations. One possible cause of the observed decrease in the ATP pool is an inhibition of amidophosphoribosyltransferase (PurF), the first committed enzyme in the de novo purine biosynthesis pathway, by an increased GMP concentration (Messenger and Zalkin, 1979; Yamaoka et al., 2001; Zalkin and Nygaard, 1996). However, this appears unlikely because in our experiments GMP never accumulated to a detectable level (ca. 50 μM) whereas GMP concentrations above 0.5 mM are needed to significantly inhibit PurF in vitro (Messenger and Zalkin, 1979).
It can be inferred from the guanine-induced alterations in purine nucleotide pool kinetics in the guaBΔCBS mutant that the effects of the subdomain deletion from IMPDH probably extend to other enzymes in the pathway. To further test if the subdomain function is intimately tied to IMPDH enzymatic activity, we examined if the effects of the guaBΔCBS mutation could be complemented by a wild type IMPDH gene supplied on a single copy plasmid. Indeed, the pGUAB plasmid (containing the wild type guaB gene under control of the native promoter), but not the pGUA4 plasmid (containing guaBΔCBS), restored the normal ATP pool and the normal response to guanine (Fig. S5). It should be noted that although pGUA4 plasmid was unable to restore the wild type response to guanine, it reduced the mutant’s ATP pool. This result is likely explained by an increased IMPDH activity due to the doubled guaBΔCBS gene dosage in the MP101/pGUA4 strain. The trans-complementation of the guaBΔCBS mutation by guaB+ suggests that the function of the subdomain may not be directly associated with the IMPDH enzymatic activity and that IMPDH may act allosterically to regulate functions of other enzymes in the purine metabolic pathway. Alternatively, wild type IMPDH could rescue the guaBΔCBS mutation-associated phenotype by formation of “mixed” enzyme tetramers in which one or more wild type IMPDH subunits affect the function of the entire tetramer.
Next, we measured the nucleotide pools in the conditions of steady state growth in the minimal media supplemented with various purine bases. In the experiment shown in Fig. 5A, the cells were incubated with 10 μg/ml of the indicated purine for at least 2 hours prior to pool measurement. In the wild type strain, the ATP and GTP pools were increased in the presence of hypoxanthine and, especially, adenine. Xanthine and guanine had no effect on the purine nucleotide pools of the guaB+ isogenic strain. In the guaBΔCBS mutant, adenine and hypoxanthine slightly increased the guanylate nucleotide pool in the guaBΔCBS mutant but had no effect on its ATP pool. In contrast, guanine and xanthine completely normalized the ATP pool in the mutant strain and increased its GTP pool slightly above the wild type level.
An interesting result was obtained when the purine nucleotide pools were measured immediately after 30 μg/ml guanine was added to MP101 guaBΔCBS cultures growing in minimal media supplemented with indicated purine bases (Fig. 5B). Pre-incubation of the guaBΔCBS strain with xanthine, but not adenine or hypoxanthine, almost completely rescued the abnormal response to guanine (the rapid decrease in [ATP] and marked increase in [GTP]) although a reproducible trend for a slight swelling of the GTP pool was still evident. The ability of a prolonged growth with xanthine to rescue the steady state nucleotide pool distortion and to normalize the guanine-induced [ATP] kinetics is puzzling in light of our previous observations which suggested that the decreased IMPDH activity and slightly reduced GTP pool in the guaBΔCBS strain do not play a direct role in the [ATP] dynamics. Importantly, the guaBΔCBS mutant growth with hypoxanthine has a GTP pool (1.62 ± 0.15 mM) that is identical to the GTP pool of the guaB+ isogenic strain grown on minimal media (1.67 ± 0.15 mM; see Fig. 5A), yet the hypoxanthine-cultivated mutant strain still displays abnormalities in the ATP content and guanine-induced purine nucleotide kinetics. Thus, it is evident that the starting GTP pool per se does not dictate the nucleotide pool dynamics observed upon guanine addition. Similarly, since the GMPS activity in the guaBΔCBS mutant was still increased in the presence of xanthine in the growth media (Fig. S6), it appears to be a poor predictor of the nucleotide pool response to guanine.
Rates of 14C-purine incorporation
Measurement of the metabolite pools with the use of 33Pi provides a series of “snapshots” of a complex interplay of reactions in which the metabolite participates as a reactant or a product. Alternatively, selected interconversions can be monitored with specific radioactive precursors. These experiments were intended to reveal which enzymatic reactions are involved in the guanylate-mediated changes in nucleotide pools in the guaBΔCBS mutant.
Two radiolabeled purines were used as tracers. Incorporation of [8-14C]-guanine reflected the kinetics of guanine salvage, whereas [14C(U)]-hypoxanthine is an immediate precursor of IMP and should allow measurement of IMP partitioning into the adenylate and guanylate nucleotide pools. To monitor guanine-induced changes in IMP partitioning, we used a small amount of 14C-hypoxanthine together with the addition of a larger quantity of unlabeled guanine to the growth media.
With the purine bases as tracers, the acid-soluble intracellular fraction of the label was almost entirely observed in the pathway end products, adenine and guanine nucleotides (Fig. 6). These labeling kinetics are consistent with the accepted model where precursor assimilation is rate limiting in purine salvage (Roy-Burman and Visser, 1975) and intermediates such as IMP are high-turnover compounds characterized by low pools and high fluxes (Yuan et al., 2006). Lowering the temperature to 15°C allowed observation of the early events in the labeling dynamics and revealed the near-linear time-course of label incorporation for up to 20 min, before the system reached an apparent steady state (Fig. 6). These initial rates reflect mostly the forward flux of tracer compound utilization whereas the steady state reflects the sum of forward and reverse fluxes. It should be noted that absolute fluxes can not be calculated from these experiments since no information is available on the fraction of the total pool that is labeled; rather, they provide an estimate of relative changes in fluxes for the purpose of comparison of strains and conditions.
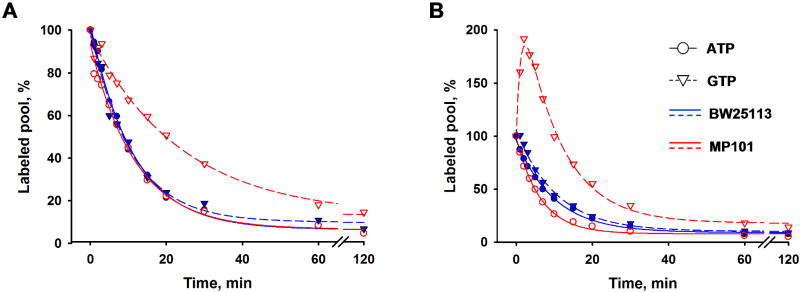
Fig. 6. The kinetics of 14C-purine labeling.
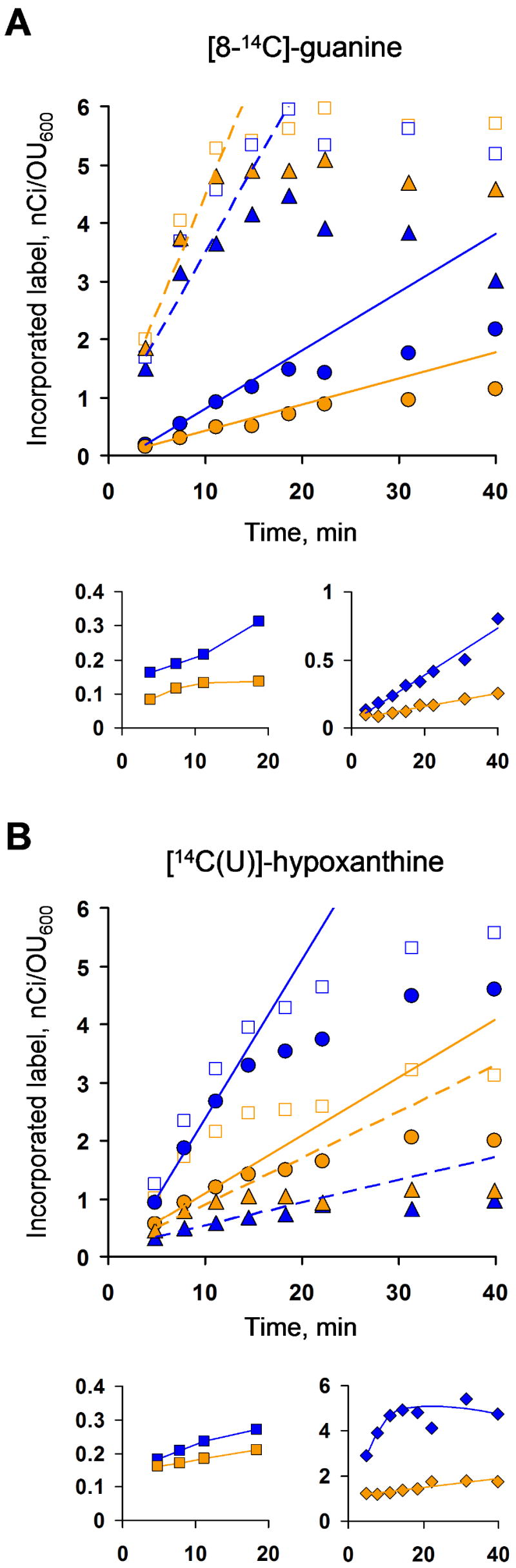
Approximate initial rates were obtained from linear fits to the first three data points, as shown A, Kinetics of guanine salvage. [8-14C]-guanine (57 mCi/mmol) was added to the cell culture media to a final concentration of 32 μM. B, Salvage of hypoxanthine and IMP partitioning. 1.36 μM [14C(U)]-hypoxanthine (291 mCi/mmol) was added to the media simultaneously with 159 μM (24 μg/ml) unlabeled guanine. Blue symbols and lines, BW25113 guaB+; orange symbols and lines, MP101 guaBΔCBS. “●”, solid lines, adenylate nucleotides (ATP + ADP + AMP); “▲”, dashed lines, guanylate nucleotides (GTP + GDP + GMP); “□”, the sum of adenylate nucleotides and guanylate nucleotides; “■”, IMP; “♦”, adenylate/guanylate ratio.
The initial rate of [8-14C]-guanine incorporation into the adenylate pool (the sum of ATP, ADP and AMP) was reduced ca. 2 fold in the guaBΔCBS mutant, whereas its guanylate pool accumulated only slightly more label (Fig. 6A). The total accumulated label (in adenylates plus guanylates) was identical in the two strains. Thus, it appears that MP101 and BW25113 salvaged similar amounts of guanine from the media, but the mutant strain converted slightly less of the salvaged material into the adenylate nucleotides; the unconverted label quantitatively remained in the guanylate pool. The mutant strain also accumulated 2 – 3 fold less label in IMP. The observation that guanine salvage was no more efficient in the guaBΔCBS mutant than in the wild type strain confirms that the guanine-induced swelling of the GTP pool in MP101 (as shown in Fig. 2C) results from a shift of the equilibrium in the IMPDH-GMPS-GMPR cycle toward synthesis of guanine nucleotides.
Unexpectedly, the rate of [14C(U)]-hypoxanthine incorporation into the adenylate pool of the guaBΔCBS mutant was up to 4 times lower than in the wild type strain, whereas the rate of the guanylate pool labeling increased by only a factor of 1.5 (Fig. 6B). The total amount of [14C(U)]-hypoxanthine accumulated in nucleotides was lower in the mutant by a factor of 2. The increased labeling rate of the mutant’s guanylate pool by 14C-hypoxanthine is indicative of a larger flux through the IMPDH reaction which, together with the reduced GMPR flux, could contribute to the guanine-induced increase of the GTP pool in the mutant. The lower labeling of adenylates by [14C(U)]-hypoxanthine in the mutant strain (Fig. 6B) is the most striking finding which indicates a lower flux through the AMPsS reaction and, therefore, a decreased synthesis of adenylate nucleotides from IMP. Our observation of a reduced AMPsS activity in the guaBΔCBS mutant (Table 2) supports this conclusion. However, it should be noted that, despite the lower AMPsS activity, the guaBΔCBS mutant demonstrated an elevated ATP pool in the absence of guanine, which suggests that other mechanisms of specific subdomain-mediated regulation of the adenylate homeostasis are present. The lower labeling of the adenylate pool by [14C(U)]-hypoxanthine also demonstrates that PurF inhibition by increased GMP concentration was not the primary reason for the guanine-induced [ATP] decrease in the guaBΔCBS mutant. Significant inhibition of AMPsS by increased [GMP] in the mutant strain is unlikely since GMP is not a strong inhibitor of AMPsS at physiological concentrations (Stayton et al., 1983).
Another significant observation is the difference in the ratios of the label incorporation into adenylate versus guanylate pools (Fig. 6). It is evident that the guaB+ isogenic strain modulates A/G ratios as the time-course of purine salvage proceeds and more guanylate nucleotides accumulate; in other words, the relative incorporation of the label into the adenylate pool versus the guanylate pool gradually changes in response to the availability of guanine in the media. The guaBΔCBS mutant shows considerably less modulation.
33Pi pulse-chase labeling
The kinetic flux profiling method (Yuan et al., 2006) can be used to analyze the turnover rates of the phosphate groups of nucleotides by the means of a pulse-chase 33Pi labeling experiment based on two assumptions: 1) that the rate of Pi assimilation (transport) is faster than the rate of turnover of nucleotide phosphates and 2) that the nucleotide pool size remains constant after addition of a large quantity of unlabeled Pi in the chase part of the experiment. The gross flux of Pi through a nucleotide triphosphate is a function of the rates of all reactions consuming and regenerating its three phosphates and can be calculated based on its pool size and the rate of disappearance of the labeled nucleotide after excess unlabeled Pi is added (Yuan et al., 2006). The measured turnover rates of the phosphate groups of ATP were similar in the isogenic guaB+ strain and guaBΔCBS mutant (Fig. 7A), with a 2 fold difference in calculated fluxes (Table 3). Quite curiously, GTP turnover was 2 fold slower in the mutant, corresponding to a 3 fold lower flux (Fig. 7A) compared with the wild type. Such a substantial variation of the GTP turnover rate was surprising given that no major changes in energy metabolism and growth characteristics were seen in the guaBΔCBS mutant.
Fig. 7. Turnover rates of ATP and GTP phosphates.
A, no purine supplementation; B, 30 μg/ml guanine added simultaneously with excess unlabeled phosphate (see Materials and Methods). With the exception of the curve for MP101 GTP in the presence of guanine, the curves are single-exponential fits with offset.
Table 3. Turnover rates of ATP and GTP phosphates.
The apparent first-order rate constants (k) were derived from the single-exponential fits to the data presented in Fig. 7. The absolute flux values were calculated as described (Yuan et al., 2006) by multiplying the nucleotide pool and the apparent first-rate constant values; the resulting absolute flux values are given for wild type/mutant comparison only and are calculated based on the nucleoside triphosphate pools and not the concentrations of ATP and GTP phosphate groups which equal ([NTP] × 3). Where indicated, 30 μg/ml guanine was added simultaneously with the unlabeled Pi as described in the Experimental Procedures section; absolute fluxes cannot be calculated from this experiment since nucleotide pools are not constant after guanine addition (Yuan et al., 2006).
| Strain/Metabolite | No purine supplementation
|
30 μg/ml guanine
|
||
|---|---|---|---|---|
| k (min-1) | Pool (mM) | Flux (mM/min) | k (min-1) | |
| BW25113 | ||||
| ATP | 0.088 | 3.5 | 0.31 | 0.101 |
| GTP | 0.094 | 1.7 | 0.16 | 0.096 |
| MP101 | ||||
| ATP | 0.088 | 6.2 | 0.54 | 0.155 |
| GTP | 0.042 | 1.2 | 0.05 | 0.086 |
To evaluate how nucleotide turnover rates changed when the cells were challenged with guanine, we added 30 μg/ml guanine simultaneously with the excess unlabeled Pi (Fig. 7B and Table 3). No major changes in the nucleotide turnover rates were seen in the wild type strain, compared to the experiment where no guanine was used. This is consistent with the observation that the wild type strain is able to maintain its nucleotide pools within a fairly narrow range (see Fig. 2C). A distinctly different GTP time course was observed in the guaBΔCBS mutant. In the first 3 minutes the rate of GTP synthesis (for which ATP is the phosphate donor) apparently prevailed over GTP utilization resulting in the ascending character of the curve in the first three minutes of the time-course. This apparently reflects the rapid increase in GTP pool observed upon addition of guanine to the guaBΔCBS mutant cells. The rate of ATP turnover in the mutant increased by a factor of 1.7 compared with the guaB+ strain.
DISCUSSION
Adenine and guanine nucleotides are involved in almost every metabolic pathway. This reinforces the importance of our findings of the changes in purine nucleotide fluxes upon deletion of the CBS subdomain of IMPDH, since a very substantial change in the flux through a particular reaction must occur to perceptibly impact the total rate of nucleotide turnover. The large change in ATP and GTP fluxes upon deletion of the subdomain suggests that the domain has a widespread role in regulating purine utilization. The observed flexibility of the GTP turnover rates in providing identical growth characteristics is striking.
The γ and β phosphates of ATP and GTP are removed and restored mostly in the reactions where these nucleotides participate as energy donors. These processes do not affect the total adenylate and guanylate concentrations and, at least for ATP, the turnover rate of the terminal phosphate is two orders of magnitude greater than that of the adenosine monophosphate moiety itself, i.e. the processes that interconvert purine nucleotides with respect to the identity of the base and are partially reflected by the dynamics of the α phosphate-labeled species (Chapman and Atkinson, 1977). Based on identical growth characteristics of BW25113 and MP101, the flux of purine triphosphates into macromolecules is likely to be the same. The consumption of GTP by AMPsS and ATP utilization by GMPS are unlikely to be major factors in the observed flux differences, since these reactions constitute a minute fraction of the total γ phosphate turnover. It is, therefore, plausible to assign the observed differences in ATP and GTP turnover rates between BW25113 and MP101 to variation in the total adenylate and guanylate nucleotide pool turnover rate, which is reflected by the disappearance of α phosphate-labeled species. The relatively large time-scale of the observed decay supports the assignment of the differences to the relatively slow turnover of the total nucleotide pool. The decreased rate of GTP turnover may be the result of a lower flux through the IMPDH-GMPS-GMPR cycle. This interpretation is in agreement with our observation of reduced activities of IMPDH and GMPR in the mutant strain. We propose that the flux in the IMPDH-GMPS-GMPR cycle is reduced by the guaBΔCBS mutation, although the GTP pool remains similar to that of the guaB+ isogenic strain.
The guanine-induced 2-fold increase in the ATP turnover rate in the guaBΔCBS mutant is an effect of the decreased ATP synthesis, since the shrinking intracellular ATP content increases turnover of the remaining pool. It may also result from an increased ATP decay through the AMP nucleosidase reaction, the primary pathway for purine nucleotide degradation in E. coli (Leung and Schramm, 1980); this would serve as an alternative mechanism for the guanine-induced decrease of the ATP pool in the MP101 mutant. An increase in ATP turnover because of a higher demand for ATP in reactions of guanine salvage appears unlikely since the measured efficiency of guanine salvage is similar in the two strains (Fig. 6A).
We demonstrate that replacement of the subdomain of E. coli IMPDH with a 24 amino acid “scar” sequence results in a major dysregulation of the purine nucleotide pools, enzyme activities and the balance between ATP and GTP. The mutation affects both the ATP pool of the cells grown on minimal media and the way the cell responds to purine bases and nucleosides. Compared with the BW25113 wild type strain, minimal media-grown MP101 guaBΔCBS has a 1.8-fold higher ATP pool and slightly (but significantly; see Table 1) decreased GTP, UTP and CTP pools. When guanine is added to the guaBΔCBS mutant grown on minimal media, a reduction of the cellular ATP pool and an increase of the GTP pool rapidly ensue, inverting the ATP/GTP ratio. A 4 fold overexpression of the mutant IMPDHΔCBS enzyme in the MP101 strain or addition to the growth media of adenine or hypoxanthine increase the GTP pool back to normal but are unable to fully rescue either the increased ATP pool or the guanine-induced inversion of the ATP/GTP ratio. Thus, it seems that the swollen adenylate nucleotide pool and characteristic guanine-induced response of the guaBΔCBS mutant do not result from an insufficient IMPDH activity or the reduced guanylate nucleotide pool size and the physiological mechanism of the nucleotide pool dysregulation following deletion of the IMPDH subdomain is to be sought elsewhere.
To gain further insights into the mechanism of this phenomenon, we conducted a series of in vivo kinetic measurements. The cumulative results of the radioactive tracing and Pi turnover experiments may be summarized as following: i) the guanine-induced swelling of the GTP pool in the guaBΔCBS mutant results from the IMPDH-GMPS-GMPR cycle equilibrium being shifted toward synthesis of GMP; ii) the rapid shrinkage of the ATP pool observed in the guaBΔCBS mutant in response to guanine represents a direct effect of the guaBΔCBS mutation on the adenylate branch of IMP metabolism and primarily results from a decreased adenylate nucleotide synthesis from IMP (but an increased ATP degradation could also play a role); iii) the efficiency of guanine salvage is not affected by the guaBΔCBS mutation.
Interestingly, steady state growth with xanthine, but not adenine or hypoxanthine, fully normalizes both adenylate and guanylate nucleotide pools and prevents the inversion of the ATP/GTP ratio following guanine addition. The mechanism of this observation is not entirely clear in light of the previous experiments which ruled out guanylate nucleotide starvation as a key factor in the nucleotide pool distortion caused by the CBS subdomain deletion. One possibility is that xanthine and guanine exert a yet unknown specific regulatory function which influences functional behavior of the enzymes implicated in purine nucleotide metabolism. Alternatively, if this result were to be interpreted as a sign of MP101 starvation for guanylate nucleotides, with the purine nucleotide pool distortion being a direct result thereof, it would have to be accepted that this GTP starvation is exceptionally unusual: it is not reflected in the GTP pool size (but could be reflected by the slower GTP turnover rate) and does not depend on the absolute IMPDH activity in the cell. While such a phenomenon is not impossible, much additional evidence would be needed to prove its validity. Indeed, the slower rate of GTP turnover in the guaBΔCBS mutant contrasts with the almost unchanged intracellular GTP concentration and may represent another example of the paradox expressed by P.W. Hochachka, among others, that essentially all metabolite concentrations are remarkably homeostatic over large changes in pathway fluxes (Hochachka, 1999). The apparent relevance of this paradigm to purine nucleotide metabolism in E. coli suggests that the metabolic regulation of this pathway may be more complex than is usually perceived.
The radioactive tracing experiments indicate that the swelling of the GTP pool observed in the guaBΔCBS mutant upon addition of guanine to the growth media may partially result from an increased flux through the IMPDH reaction, compared with the wild type strain (see Fig. 1). This could happen, for instance, if the CBS domain were the site for binding of an as yet unidentified negative allosteric regulator of the IMPDH activity in the conditions of a high guanylate nucleotide pool. Deletion of the subdomain in this case would render IMPDHΔCBS constitutively active and deplete the ATP pool by an enhanced consumption by IMPDH of IMP, its common substrate with AMPsS (Fig. 1), resulting in a lower flux of IMP toward AMP synthesis. However attractive owing to its simplicity, this interpretation is not easily reconciled with a number of observations. First, if the sole function of the CBS domain were to allosterically inhibit IMPDH in the conditions of a high guanylate pool, a wild type gene would not be able to complement the phenotype caused by the mutant IMPDH protein unless a dominant positive effect occurred in a “mixed” tetramer. Second, blocking the conversion of IMP into GMP by decoyinine only partially prevented the [ATP] decrease, suggesting that mechanisms in addition to the alteration of GMP synthesis are involved. Third, no signs of IMP deficiency were seen in the [14C(U)]-hypoxanthine tracing experiment (Fig. 6B). Finally, the total accumulation of [14C(U)]-hypoxanthine into nucleotides is 2 fold lower in the mutant; therefore the lower rate of adenylate labeling cannot be explained by a change in IMP partitioning caused by an enhanced competition by IMPDH for IMP, and must represent a more direct effect of the mutation on the AMP branch of IMP metabolism. We conclude that although a specific negative regulation of IMPDH catalytic function by the subdomain is possible, such regulation alone seems unable to account for all of the observed kinetic phenomena. Though we failed to observe a substantial effect of large ATP concentrations on IMPDH activity in crude E. coli extracts, our results do not rule out the possibility of the subdomain being involved in the allosteric regulation of IMPDH and it remains an important question whether there might be a yet undiscovered ligand which interacts with the subdomain to affect the IMPDH functional behavior. We note that GTP has been found to interact with IMPDH with an apparent Kd of 72.9 μM and induce IMPDH aggregation, although the functional significance of these observations is unclear (Ji et al., 2006).
The activities of AMPsS and GMPR in crude dialyzed cell extracts were found to be two- to three times lower in the guaBΔCBS mutant compared with the wild type strain. As indicated above, the reduced GMPR activity is consistent with our observations of a slowed GTP turnover rate and of the guanine-dependent swelling of the GTP pool, whereas the reduced AMPsS activity contrasts with the increased ATP pool in the mutant, grown on minimal media. However, caution should be exercised in the interpretation of such results, since measurements of enzyme activities in crude extracts typically reflect the absolute cellular enzyme concentrations; they provide less information as to what the specific activities of these enzymes are in the crowded cellular milieu with potentially large concentrations of small-molecule effectors and enhanced protein-protein interactions. We note that the lack of a strong correlation between enzyme levels and nucleotide pools has been previously described (Kelln et al., 1975). At this point, we are unable to conclude whether the changes in AMPsS, IMPDH and GMPR activities in the cell extract are the primary effect of the guaBΔCBS mutation or represent a secondary, perhaps even compensatory, change. Interestingly, a pattern of purine nucleotide pool distortion that is similar to the one in MP101 was observed by C. Petersen in a GMPR null mutant (E. coli gsk-3 guaC2079) after addition of guanosine (Petersen, 1999); this supports our conclusion that the decreased GMPR activity and guanylate nucleotide turnover rate play an important role in the mutation-induced nucleotide pool changes. Notably, GMPR is a (α/β)8-barrel homotetramer with the overall topology similar to IMPDH (Li et al., 2006). It is therefore plausible that IMPDH may interact with GMPR to regulate the activity of either enzyme.
Despite decades of research, our understanding of how cellular ATP pools are regulated has remained limited for all organisms (Ataullakhanov and Vitvitsky, 2002). The existence of efficient (and probably redundant) mechanisms to keep the ATP/GTP ratio within a very narrow range is obvious but the nature of these mechanisms remains to be elucidated (Zalkin and Dixon, 1992). Two primary levels of control clearly cooperate in maintaining the purine nucleotide pool sizes in bacteria (Zalkin and Nygaard, 1996) and eukaryotes (Zalkin and Dixon, 1992): transcriptional control and enzyme-level regulation of purine biosynthetic enzymes. Although rarely stated in the literature, the implicit model is that enzyme-level regulation works as the “first line of defense” and is able to rapidly balance specific fluxes in the purine biosynthesis and interconversion circuits in response to the appearance in the media of one or more salvageable purine compounds (Petersen, 1999; Yamaoka et al., 2001; Zalkin and Nygaard, 1996). Transcriptional regulation of purine nucleotide biosynthesis, mediated in prokaryotes primarily by PurR transcriptional repressor (Zalkin and Nygaard, 1996), is a slower mechanism; as suggested in this study by the lack of a strong correlation between the enzyme levels and the nucleotide pools/fluxes, it is less capable of “fine tuning” purine metabolism. Notably, while transcriptional regulation has been described in detail in E. coli and some other prokaryotes, few in vivo experiments on enzyme-level regulation have been reported. For instance, it remains unclear if the classical in vitro competitive inhibition of IMPDH by GMP (Gilbert et al., 1979) is of physiological significance. The results of the experiment shown in Fig. 4 indicate that GMP is not a strong inhibitor of E. coli enzyme, even at concentrations that are ca. 10 fold greater than physiological.
The near-absolute evolutionary conservation of the CBS subdomain in the IMPDH structure suggests preservation of function; hence, extrapolation of our results to human cells indicates that, unlike previously thought, changes in retinal nucleotide pools are to be expected in the RP10 pathogenesis. However, the normal phenotype of a guaB+/guaBΔCBS “heterozygous” mutant contrasts with the disease development in RP10 heterozygous patients (Bowne et al., 2002) suggesting caution in the extrapolation of our data to the disease pathogenesis. We provide no specific data regarding physiological significance of the reported nucleic acid binding by the subdomain (McLean et al., 2004; Mortimer and Hedstrom, 2005). However, the lack of an effect of the guaBΔCBS mutation on growth characteristics suggests that no severe changes in RNA metabolism have occurred in the mutant strain under the conditions tested.
In conclusion, our study of a guanine-prototrophic E. coli mutant expressing IMPDHΔCBS provides the first in vivo insights into the biological function of the CBS subdomain. It is clear that the subdomain is involved in the global regulation of the intracellular purine nucleotide pools and turnover rates in E. coli by what seems to be a coordination of the activities of the enzymes of two branches of IMP metabolism. In particular, the two critical parameters that seem to be affected by the deletion of the subdomain are the IMPDH-GMPS-GMPR cyclic flux and the rates of adenylate nucleotide synthesis and, possibly, degradation. Thus, the presence of the subdomain in IMPDH structure is essential for maintaining the physiological concentrations of adenylate and guanylate nucleotides and, accordingly, the ATP/GTP ratio in E. coli. The unexpected finding that ATP rather than GTP pools are grossly affected by the mutation both in the minimal media and upon guanine addition suggests the existence of a novel mechanism of specific regulation of ATP homeostasis. Further studies on the mechanism of purine nucleotide metabolism regulation by the CBS subdomain of IMPDH are warranted.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
The E. coli MP101 strain (guaBΔCBS) was constructed using the two-step recombineering technology as described (Datsenko and Wanner, 2000; Ellis et al., 2001; Yu et al., 2000). E. coli BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) was used as the parent strain (Datsenko and Wanner, 2000). Briefly, the part of the guaB sequence encoding the CBS pair (amino acid residues 93–215) was replaced with a kanamycin resistance gene flanked by FRT recombination sequences and 42 nt homologies to the chromosomal guaB sequence. The resulting KanR strain was auxotrophic for guanine. A second round of recombineering restored guanine prototrophy by removal of the antibiotic resistance cassette leaving an in-frame 24 amino acid “scar” sequence in place of the subdomain as shown in Fig. S1.
Single copy number plasmids bearing guaBwt and guaBΔCBS genes under the control of the native promoter were created using the Copy Control PCR Cloning Kit (Epicentre). A pair of primers (5’-GTGAGCGAGATCAAATTCTAAATCAGCAGG and 5’-TCAGGAGCCCAGACGGTAGTTC) was used to amplify the guaB locus of E. coli BW25113 and E. coli MP101 strains, including the 5’ regulatory/promoter region. The resulting PCR products were blunt-end cloned in the pCC1 plasmid, yielding the pGUAB plasmid containing wild type guaB and pGUA4 plasmid containing guaBΔCBS. Both plasmids were able to restore guanine prototrophy to a guaB null mutant (data not shown).
A pair of primers (5’-ACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGACAATATTTATT AACCACTCTGGTCGAG and 5’-TCAGGAGCCCAGACGGTAGTTC) was used to amplify the guaBΔCBS gene and place it under control of the lacUV5 constitutive promoter as shown in Fig. S2. The lacUV5 promoter sequence was fused to the native guaBΔCBS 5’-UTR and cloned in the pCR2.1-TOPO vector, yielding the pGUA22 plasmid. The re-circularized pCR2.1-TOPO plasmid, termed pCR2.1-TOPO-E. was used as an “empty” vector control.
Measurements of nucleotide pools
Nucleotide pools were measured using 33Pi labeling of exponentially-growing E. coli cells followed by thin-layer chromatography (TLC) of the cell extract (Bochner and Ames, 1982). A fresh overnight cell culture grown in the “MOPS medium” (Neidhardt et al., 1974) was pelleted and resuspended in 1 ml of fresh media to a final OD600 of 0.3. For strains containing pGUAB and pGUA4 plasmids the medium was supplemented with 12.5 μg/ml chloramphenicol. Up to 30 μCi/ml of 33Pi (40-158 Ci/mg) was added and the cell culture was grown at 37°C to an OD600 of 0.5 – 0.7. Where indicated, 30 μg/ml of a purine base or 60 μg/ml of a nucleoside was added. Immediately before, and at various times after, base/nucleoside addition portions of the cell culture were mixed with ice-cold formic acid to extract nucleotides (150 μl culture was mixed with 7 μl 11 N formic acid). Two-dimensional TLC on PEI-cellulose plates (Selecto Scientific; solvent Ta, first dimension: 0.75 M Tris, 0.45 M HCl; solvent Sb, second dimension: 74 g (NH4)2SO4, 0.4 g (NH4)HSO4, 4 g Na2EDTA, 100 ml H2O) was used for nucleotide separation as described (Bochner and Ames, 1982). Measurements of nucleoside triphosphates in time-course experiments were performed using the unidimensional TLC system of Jensen et al. (Jensen et al., 1979), consisting of PEI-cellulose plates developed in 0.85 M KH2PO4 adjusted to pH 3.4 by addition of 0.85 M H3PO4. The radiolabeled nucleotide spots were detected by phosphorimaging and quantified using the MultiGauge software (Fujifilm). To calculate the absolute nucleotide concentrations, the cellular ATP content was measured using the Enliten® luciferase ATP assay system (Promega). The volume of E. coli was assumed to be 0.8285 μ3 as previously described (Harvey et al., 1967). Concentrations of the rest of radiolabeled compounds were calculated by normalization of their TLC spot intensities to the intensity of the ATP spot (Bochner and Ames, 1982). All time-course experiments were performed in duplicate or triplicate. The results of a representative experiment are shown.
14C-purine tracing
[14C(U)]-hypoxanthine and [8-14C]-guanine were purchased from Moravek Biochemicals. Ten OD600 units of exponentially growing E. coli cells were pelleted and resuspended in 5 ml MOPS media that had been pre-equilibrated at +15°C. 14C-labeled and unlabeled purines were immediately added as indicated. The cell suspension was then shaken in a +15°C water bath. At various times after addition of the radiotracer, 0.5 ml samples of the cell suspension were withdrawn and the cells collected by centrifugation at +4°C for 1 min in a tabletop centrifuge at the top speed; the cells were then mixed with 20 μl of ice-cold 2 N formic acid to extract nucleotides. Each experiment was conducted in 2 replicates; results of a representative experiment are shown.
33Pi pulse-chase experiments
Freshly grown cells were pelleted and resuspended to an OD600 of 0.1 in MOPS media containing 30 μCi/ml of 33Pi (40-158 Ci/mg). The culture was grown at 37°C in a shaking incubator. When the OD600 reached 0.4–0.5, the Pi concentration in the media was increased from 0.2 mM to 77.2 mM by addition of 21X unlabeled phosphate concentrate (1.18 M NaH2PO4, 0.441 M KH2PO4). Where indicated, 30 μg/ml guanine was also added at this point. Immediately before and at various times after addition of unlabeled Pi, nucleotides were extracted by combining aliquots of the culture with formic acid as described above. The rate of disappearance of the labeled nucleoside triphosphate pool was fit to a single exponential function, and the gross flux was calculated from the apparent first-order rate constant as described (Yuan et al., 2006).
Assays of enzyme activities in cell extract
Exponentially growing cells were harvested by centrifugation at an optical density of 0.5 – 0.6. Cells were washed with extraction buffer (100 mM Tris-HCl, pH 8.1, 2 mM Na2EDTA, 0.1 mM DTT, 30 μM PMSF) and resuspended in the same buffer. A single pass through a French press was used to disrupt the cells. The resulting extract was cleared by centrifugation and dialyzed against extraction buffer. Protein concentration was measured by the BioRad Bradford protein assay (Bradford, 1976). Enzyme activities were normalized to the protein concentration in each sample.
Assays of IMPDH activity were performed in 100 mM Tris-HCl, pH 8.1, 10 mM KCl, 0.1 mM DTT in the presence of 0.1 mM NAD, 0.1 mM IMP. Unless indicated otherwise, the production of XMP was monitored by the absorbance increase at 290 nm (ε = 5.4 mM-1 cm-1). Assays of IMPDH activity in the presence of nucleotide additives and in the IMPDH overexpression experiments were conducted using a TLC-based method with physiological concentrations of KCl, NAD and IMP (Neidhardt et al., 1987). The assay mix contained 50 mM HEPES, pH 7.0, 0.174 mM [8-14C]-IMP, 0.79 mM NAD, 150 mM KCl, 0.1 mM DTT, 2 μl cell extract (8 – 11 μg protein), and 0.25 – 10 mM additives, as indicated, in a final volume of 20 μl. The reaction was incubated at 37°C for 5 min, stopped by addition of 11 N formic acid to a final concentration of 0.5 M and centrifuged at top speed in a tabletop centrifuge for 5 min. One μl of the supernatant was spotted on a PEI-cellulose TLC plate and air-dried. The TLC plate was developed in 0.75 M Tris, 0.45 M HCl, 0.25 M LiCl.
Adenylosuccinate synthetase was assayed with the use of [8-14C]-IMP as a substrate. The reaction mix was a modification of the previously described (Juang et al., 1993) and contained 50 mM HEPES, pH 7.0, 8 mM magnesium acetate, 0.4 mM [8-14C]-IMP, 1.5 mM GTP, and 8 mM aspartate. The production of AMPs was monitored by separation of the reaction mix on PEI-cellulose TLC plates2.
The assay for GMPS activity was an adaptation of the published procedure (Patel et al., 1975). The 25-μl reaction mix contained 90 mM Tris HCl, pH 8.1, 0.1 mM DTT, 0.1 mM Na2EDTA, 0.3 mM [8-14C]-XMP (Moravek Biochemicals), 3 mM ATP, 4 mM MgCl2, 20 mM glutamine and up to 4 μl diluted E. coli extract (1 – 1.5 μg protein). The reaction was stopped by addition of 0.5 M formic acid. The production of GMP was monitored by separation of the reaction mix on PEI-cellulose TLC plates developed in 0.75 M Tris, 0.45 M HCl, 0.25 M LiCl.
GMPR activity was measured using a modification of the procedure by Nakamura et al. (Nakamura et al., 1992). [8-14C]-GMP was purchased from Moravek Biochemicals. The 25 μl reaction mix contained 100 mM Tris-HCl, pH 7.24, 1 mM Na2EDTA, 6.6 mM NADPH, 4 mM DTT, 0.2 mM [8-14C]-GMP (12 mCi/mmol) and 5 μl E. coli extract (35 – 50 μg protein). The reaction was stopped by addition of 0.5 M formic acid and centrifuged at top speed in a tabletop centrifuge for 5 min. Two μl of the supernatant was spotted on a PEI-cellulose TLC plate and air-dried. The TLC plate was then immersed in methanol for 10 min, air-dried, and developed in 0.85 M KH2PO4, pH 3.4 (Jensen et al., 1979) to separate reaction products. The radiolabeled IMP spot was detected by phosphorimaging and quantified using the MultiGauge software (Fujifilm).
Supplementary Material
Acknowledgments
We thank Drs. A. Hershko, E.K. Jaffe and J.R. Peterson for critical reading of this manuscript. We are grateful to Dr. A.T. Yeung, C. Meyers, M. Hussein, Dr. V. Schramm and Dr. M. Andrake for helpful technical discussions.
Footnotes
This work was supported by National Institutes of Health Grants GM072425, CA06927 and also supported by an appropriation from the Commonwealth of Pennsylvania. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute, or any other sponsoring organization.
The abbreviations used are: IMP, inosine 5’-monophosphate; XMP, xanthosine 5’-monophosphate; PRPP, 5-phosphoribosyl 1-pyrophosphate; (p)ppGpp, guanosine 3’-diphosphate 5’-triphosphate and guanosine 3’-diphosphate 5’-diphosphate; IMPDH, IMP dehydrogenase (guaB); GMPR, GMP reductase (guaC); GMPS, GMP synthetase (guaA); AMPsS, adenylosuccinate synthetase (purA); AMPs, adenylosuccinate; DTT, dithiothreitol; PMSF, phenylmethanesulphonylfluoride; TLC, thin layer chromatography.
Manuscript in preparation.
References
- Ataullakhanov FI, Vitvitsky VM. What determines the intracellular ATP concentration. Biosci Rep. 2002;22:501–511. doi: 10.1023/a:1022069718709. [DOI] [PubMed] [Google Scholar]
- Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E, Mahmood NA, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci U S A. 2006;103:10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Blanton SH, Cepko CL, Blackshaw S, Birch DG, et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:559–568. doi: 10.1093/hmg/11.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Mortimer SE, Hedstrom L, Zhu J, Spellicy CJ, et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47:34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Chapman AG, Atkinson DE. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microb Physiol. 1977;15:253–306. doi: 10.1016/s0065-2911(08)60318-5. [DOI] [PubMed] [Google Scholar]
- Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day P, Sharff A, Parra L, Cleasby A, Williams M, Horer S, et al. Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP. Acta Crystallogr D Biol Crystallogr. 2007;63:587–596. doi: 10.1107/S0907444907009110. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Petsko GA, Hedstrom L. Crystal structure of a ternary complex of Tritrichomonas foetus inosine 5’-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis. Biochemistry. 2002;41:13309–13317. doi: 10.1021/bi0203785. [DOI] [PubMed] [Google Scholar]
- Gilbert HJ, Lowe CR, Drabble WT. Inosine 5’-monophosphate dehydrogenase of Escherichia coli. Purification by affinity chromatography, subunit structure and inhibition by guanosine 5’-monophosphate. Biochem J. 1979;183:481–494. doi: 10.1042/bj1830481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- Harvey RJ, Marr AG, Painter PR. Kinetics of growth of individual cells of Escherichia coli and Azotobacter agilis. J Bacteriol. 1967;93:605–617. doi: 10.1128/jb.93.2.605-617.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hochachka PW. The metabolic implications of intracellular circulation. Proc Natl Acad Sci U S A. 1999;96:12233–12239. doi: 10.1073/pnas.96.22.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignoul S, Eggermont J. CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol. 2005;289:C1369–1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- Jensen KF. Apparent involvement of purines in the control of expression of Salmonella typhimurium pyr genes: analysis of a leaky guaB mutant resistant to pyrimidine analogs. J Bacteriol. 1979;138:731–738. doi: 10.1128/jb.138.3.731-738.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KF, Houlberg U, Nygaard P. Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-alpha-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal Biochem. 1979;98:254–263. doi: 10.1016/0003-2697(79)90138-6. [DOI] [PubMed] [Google Scholar]
- Jensen KF. Regulation of Salmonella typhimurium pyr gene expression: effect of changing both purine and pyrimidine nucleotide pools. J Gen Microbiol. 1989;135:805–815. doi: 10.1099/00221287-135-4-805. [DOI] [PubMed] [Google Scholar]
- Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxid Redox Signal. 2005;7:813–822. doi: 10.1089/ars.2005.7.813. [DOI] [PubMed] [Google Scholar]
- Ji Y, Gu J, Makhov AM, Griffith JD, Mitchell BS. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J Biol Chem. 2006;281:206–212. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- Juang RH, McCue KF, Ow DW. Two purine biosynthetic enzymes that are required for cadmium tolerance in Schizosaccharomyces pombe utilize cysteine sulfinate in vitro. Arch Biochem Biophys. 1993;304:392–401. doi: 10.1006/abbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Kelln RA, Kinahan JJ, Foltermann KF, O’Donovan GA. Pyrimidine biosynthetic enzymes of Salmonella typhimurium, repressed specifically by growth in the presence of cytidine. J Bacteriol. 1975;124:764–774. doi: 10.1128/jb.124.2.764-774.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan A, Aherne A, Palfi A, Humphries M, McKee A, Stitt A, et al. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(-/-) mice. Hum Mol Genet. 2002;11:547–557. doi: 10.1093/hmg/11.5.547. [DOI] [PubMed] [Google Scholar]
- Krogh A. Progress in physiology. Am J Physiology. 1920;90:243–251. [Google Scholar]
- Leung HB, Schramm VL. Adenylate degradation in Escherichia coli. The role of AMP nucleosidase and properties of the purified enzyme. J Biol Chem. 1980;255:10867–10874. [PubMed] [Google Scholar]
- Li J, Wei Z, Zheng M, Gu X, Deng Y, Qiu R, et al. Crystal structure of human guanosine monophosphate reductase 2 (GMPR2) in complex with GMP. J Mol Biol. 2006;355:980–988. doi: 10.1016/j.jmb.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Mager J, Magasnik B. Guanosine 5’-phosphate reductase and its role in the interconversion of purine nucleotides. J Biol Chem. 1960;235:1474–1478. [PubMed] [Google Scholar]
- McLean JE, Hamaguchi N, Belenky P, Mortimer SE, Stanton M, Hedstrom L. Inosine 5’-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem J. 2004;379:243–251. doi: 10.1042/BJ20031585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger LJ, Zalkin H. Glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli. Purification and properties. J Biol Chem. 1979;254:3382–3392. [PubMed] [Google Scholar]
- Meyer S, Savaresi S, Forster IC, Dutzler R. Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5. Nat Struct Mol Biol. 2007;14:60–67. doi: 10.1038/nsmb1188. [DOI] [PubMed] [Google Scholar]
- Miller MD, Schwarzenbacher R, von Delft F, Abdubek P, Ambing E, Biorac T, et al. Crystal structure of a tandem cystathionine-beta-synthase (CBS) domain protein (TM0935) from Thermotoga maritima at 1.87 A resolution. Proteins. 2004;57:213–217. doi: 10.1002/prot.20024. [DOI] [PubMed] [Google Scholar]
- Mortimer SE, Hedstrom L. Autosomal dominant retinitis pigmentosa mutations in inosine 5’-monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem J. 2005;390:41–47. doi: 10.1042/BJ20042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Natsumeda Y, Nagai M, Shiotani T, Weber G. Direct assay method for guanosine 5’-monophosphate reductase activity. Anal Biochem. 1992;206:115–118. doi: 10.1016/s0003-2697(05)80019-3. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: ASM Press; 1987. [Google Scholar]
- Nimmesgern E, Black J, Futer O, Fulghum JR, Chambers SP, Brummel CL, et al. Biochemical analysis of the modular enzyme inosine 5’-monophosphate dehydrogenase. Protein Expr Purif. 1999;17:282–289. doi: 10.1006/prep.1999.1136. [DOI] [PubMed] [Google Scholar]
- Pankiewicz KW, Goldstein BM. Inosine monophosphate dehydrogenase A major therapeutic target. Washington, DC: Oxford University Press; 2003. [Google Scholar]
- Patel N, Moyed HS, Kane JF. Xanthosine-5’-phosphate amidotransferase from Escherichia coli. J Biol Chem. 1975;250:2609–2613. [PubMed] [Google Scholar]
- Petersen C. Inhibition of cellular growth by increased guanine nucleotide pools. Characterization of an Escherichia coli mutant with a guanosine kinase that is insensitive to feedback inhibition by GTP. J Biol Chem. 1999;274:5348–5356. doi: 10.1074/jbc.274.9.5348. [DOI] [PubMed] [Google Scholar]
- Roy-Burman S, Visser DW. Transport of purines and deoxyadenosine in Escherichia coli. J Biol Chem. 1975;250:9270–9275. [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayton MM, Rudolph FB, Fromm HJ. Regulation, genetics, and properties of adenylosuccinate synthetase: a review. Curr Top Cell Regul. 1983;22:103–141. doi: 10.1016/b978-0-12-152822-5.50008-7. [DOI] [PubMed] [Google Scholar]
- Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka T, Yano M, Kondo M, Sasaki H, Hino S, Katashima R, et al. Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J Biol Chem. 2001;276:21285–21291. doi: 10.1074/jbc.M011103200. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol. 2006;2:529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- Zalkin H, Dixon JE. De novo purine nucleotide biosynthesis. Prog Nucleic Acid Res Mol Biol. 1992;42:259–287. doi: 10.1016/s0079-6603(08)60578-4. [DOI] [PubMed] [Google Scholar]
- Zalkin H, Nygaard P. Biosynthesis of Purine Nucleotides. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996. pp. 561–579. [Google Scholar]
- Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, et al. Characteristics and crystal structure of bacterial inosine-5’-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.