Abstract
We compared 12 different cell populations, including embryonic stem cells before and during differentiation into embryoid bodies as well as various types of normal and tumor cells to determine if pluripotent versus differentiated cell types use different mechanisms to establish their transcriptome. We first identified genes that were not expressed in the 12 different cell populations and then determined which of them were regulated by histone methylation, DNA methylation, at the step of productive elongation, or by the inability to establish a preinitiation complex. For these experiments, we performed chromatin immunoprecipitation using antibodies to H3me3K27, H3me3K9, 5-methyl-cytosine, and POLR2A. We found that (1) the percentage of low expressed genes bound by POLR2A, H3me3K27, H3me3K9, or 5-methyl-cytosine is similar in all 12 cell types, regardless of differentiation or neoplastic state; (2) a gene is generally repressed by only one mechanism; and (3) distinct classes of genes are repressed by certain mechanisms. We further characterized two transitioning cell populations, 3T3 cells progressing from G0/G1 into S phase and mES cells differentiating into embryoid bodies. We found that the transient regulation through the cell cycle was achieved predominantly by changes in the recruitment of the general transcriptional machinery or by post-POLR2A recruitment mechanisms. In contrast, changes in chromatin silencing were critical for the permanent changes in gene expression in cells undergoing differentiation.
It has been estimated that mature mRNAs corresponding to 30%–45% of the known genes can be detected in any given human or mouse cell type (Su et al. 2004). Although each cell has a distinct transcriptome, certain mRNAs are widely expressed across different cell types. In particular, genes encoding proteins involved in housekeeping functions such as DNA replication, mRNA processing, or protein translation show little cell type-specific expression (Schadt et al. 2004). On the other hand, genes that encode proteins that confer highly specific phenotypes (such as detoxification enzymes in hepatocytes or self-renewal transcription factors in stem cells) are expressed in only a few cell types. Therefore, the establishment and maintenance of a highly differentiated cell phenotype requires not only the activation of a small set of specific genes in a given cell, but also the repression of a larger set of genes that confer characteristics of other types of differentiated cells. Thus, an understanding of the mechanisms by which genes are kept in an off state is critical to our understanding of development and differentiation.
Gene expression can be regulated at different steps, including transcription initiation or elongation and mRNA processing, transport, stability, or translation (Orphanides et al. 1996; Uptain et al. 1997; Orphanides and Reinberg 2002; Sims III et al. 2004; Li et al. 2007; Komili and Silver 2008). One major control step is at the level of transcript production, which can be regulated by changes in the accessibility of the promoter region (due to changes in chromatin structure), changes in the amount of general transcription factors such as RNA polymerase II (RNAPII) that are recruited to the core promoter region (which is often due to changes in the abundance or activity of cell type-specific DNA binding transcription factors), and by changes in the efficiency and/or effectiveness of the transition from initiation to elongation. The latter mechanism, i.e., the control of transcription elongation by RNA polymerase II via release of a promoter-proximal paused polymerase, has recently been shown to be a rate-limiting step for a substantial fraction of yeast, fly, and mammalian genes (Ren et al. 2000; Radonjic et al. 2005; Guenther et al. 2007; Lis 2007; Muse et al. 2007; Tamkun 2007; Zeitlinger et al. 2007).
One might consider that highly differentiated cell types (such as liver tissue) may specialize in “long-term” repression mechanisms that could keep a large set of nonessential (for that particular cell type) genes off in a permanent manner. Highly stable repression can be achieved by certain chromatin modifications. Specifically, trimethylation of lysine 9 or lysine 27 of histone H3 (H3me3K9 or H3me3K27) are marks for silenced chromatin (Kouzarides 2007; Li et al. 2007). The silenced chromatin state can be maintained by the interaction of proteins such as HP1 and Polycomb with H3me3K9 and H3me3K27, respectively. In contrast, cells that have not yet committed to a specific differentiated phenotype (such as embryonic stem cells) may utilize more transient mechanisms such as the recruitment of the general transcriptional machinery to regulate gene expression. This would allow a rapid evolution of the transcriptome to occur once a differentiation pathway was initiated and a critical site-specific factor was expressed. To determine if pluripotent and differentiated cell types use different mechanisms to establish their transcriptome, we have compared 12 different cell populations, including embryonic stem cells before and during differentiation into embryoid bodies and various types of normal and tumor cells, to determine if the frequency of utilization of the different mechanisms changes as cells progress toward (e.g., during formation of embryoid bodies) or revert from (e.g., during neoplastic transformation) a differentiated phenotype (see Supplemental Fig. S1).
Results
We have chosen to compare the mechanisms that are used to prevent transcriptional activation in 12 different human or mouse cell populations (Table 1). We have chosen a variety of normal (liver tissue and primary fibroblasts from foot, lung, and foreskin) and cancer (liver tumor tissue, the Huh7 liver hepatoma cell line, the MCF7 breast adenocarcinoma cell line, and the Ntera2 testicular carcinoma cells) human cell types, to investigate whether different mechanisms predominate in differentiated cells from various tissues or in normal versus tumor cells. We have also chosen four different mouse cell populations, including mouse embryonic stem (mES) cells, differentiating embryoid bodies (EBs) derived from the mES cells, and 3T3 fibroblasts in G0/G1 or S phase of the cell cycle. These latter analyses will allow insight concerning transcriptional regulation during important cellular transitions (i.e., the transition from pluripotency to a differentiated phenotype and the transition from quiescent to proliferating cells). We also note that the cell populations can be divided into embryonic-like cell types (mES cells, EBs, 3T3 embryonal fibroblasts, and Ntera2 embryonal carcinomas) and adult cells (foot fibroblasts, cell lines derived from breast and liver cancer patients, and liver tissue samples).
Table 1.
Cell populations used in this study
To obtain cell cycle stage-specific 3T3 cells (Krek and DeCaprio 1995), the cells were grown in low concentrations of serum for 72 h (for the G0/G1 population) then stimulated to begin proliferation using normal concentrations of serum and harvested 16 h later (for the S phase cells). The percentage of cells in G0/G1, S, and G2/M phases was monitored using a fluorescence-activated cell sorter (Supplemental Fig. S2). The pluripotent state of the mES cells and their transition to EBs were monitored by Western blot (Supplemental Fig. S3) using antibodies to the stem cell-specific transcription factor POU5F1 (Scholer et al. 1990; Nichols et al. 1998; Niwa et al. 2000). The normal and tumor liver tissue samples were used directly (not grown in culture); the primary human fibroblasts were isolated, purified, and grown as previously described (Rinn et al. 2007), and the cancer cell lines were analyzed as asynchronously growing cell culture populations.
Identification of low expressed genes (LEGs)
Our first step in characterizing the mechanisms that are used to prevent transcriptional activation was to identify the sets of genes that are not expressed in each of the 12 different cell populations (see Supplemental Fig. S1 for an overview of the experimental setup). For these experiments, we prepared RNA from each cell population and analyzed RNA levels using Illumina mouse or human expression arrays (see Supplemental Table S1 for a list of all the RNA arrays used in this study). Using the criteria defined by the Illumina analysis program (see Methods), we first identified all genes that were in the “expressed” category (termed HEGs for highly expressed genes; P-value = 0). Next, we divided the remaining genes into two categories: genes that are not expressed (termed LEGs for low expressed genes; P-value between 1 and 0.05) and genes whose expression levels are between the HEGs and the LEGs (termed MEGs for middle expressed genes; P-value between 0.05 and 0). By distinguishing genes that are highly repressed (LEGs) from those that are on the boundary between the expressed HEGS and the nonexpressed LEGs (MEGs), we could investigate the extent to which the different repression mechanisms are utilized in the different categories.
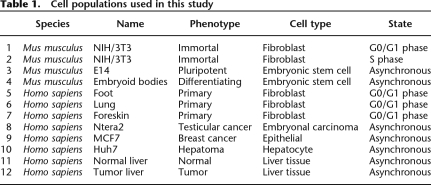
We found that, similar to previous studies (Su et al. 2004), ∼25%–35% of the genes represented on the arrays were expressed in each of the 12 cell types and that ∼40%–60% of the genes fell into the LEGs category (Fig. 1A). The percentage of genes that were in the LEGs category appeared to be slightly higher (with fewer genes in the MEGs category) in the mouse cells than in the human cells. However, this could be a consequence of slightly different compositions of the gene sets on the mouse versus the human arrays. We found that the sets of genes that were in the HEGs or the LEGs categories showed a very high overlap in all the cell populations (Fig. 1B). Interestingly, the overlap of the HEGs or the LEGs sets was very similar even in nonrelated cell populations (see Supplemental Table S8 for the RNA expression data for all 12 cell populations). For example, 81% of the Ntera2 (testicular embryonal carcinoma) LEGs were in the set of MCF7 (breast tumor) LEGs and 82% of the Huh7 (liver tumor cells) LEGs were in the set of MCF7 LEGs. Similarly, 74% of the Ntera HEGs were in the set of MCF7 HEGs and 72% of the Huh7 HEGs were in the MCF7 HEGs. These results suggest that, in general, many of the same genes are expressed in all cell types (e.g., housekeeping genes, genes required for cell cycle progression, etc.) and that many genes are repressed in all cell types (e.g., genes that are highly tissue-specific or developmentally regulated). However, the category of MEGs showed the most variable expression when comparing the different cell types, suggesting that separate analysis of the repression mechanisms of the MEGs and LEGs may be informative.
Figure 1.
Transcriptome analysis of 12 cell populations. (A) Identification of LEGs, MEGs, and HEGs. Illumina BeadChip arrays were used to analyze RNA from the 12 cell populations. The RNAs were then classified into low expressed genes (0.05 ≤ P-value ≤ 1), middle expressed genes (0 < P-value < 0.05), and highly expressed genes (P-value = 0). Percentages of the LEGs, MEGs, and HEGs are shown relatively to the total number of genes on the Illumina platform for all 12 cell populations. (B) Comparison of different cell populations. All possible pairwise comparisons between the genes in each category (the sets of LEGs, MEGs, and HEGs) were made, and then the percentage overlap was calculated relative to the average number of genes in each category. The upper and lower quartiles of the box plots are the 75th and 25th percentiles, respectively. The whisker top and bottom are 90th and 10th percentiles, respectively.
Identification of genes repressed by five different mechanisms
The second step in characterizing the mechanisms that are used to prevent transcriptional activation was to identify the sets of genes that are regulated by silenced chromatin structure, at the step of productive elongation, or by the inability to establish a preinitiation complex. Correlation of H3me3K27, H3me3K9, or DNA methylation with silenced chromatin is well established (Klose and Bird 2006; Squazzo et al. 2006; Barski et al. 2007; Kouzarides 2007) and therefore we analyzed these three silencing marks to determine their relative use in the different cell populations. These three silencing marks are mediated by distinct chromatin-modifying complexes. For example, EZH1 and EZH2 can cause methylation of H3 at lysine 27, EHMT1, EHMT2, SUV39H1/2, and SETDB1 can cause methylation of H3 at lysine 9, and DNMT1, TRDMT1, and DNMT3A/B can methylate DNA at CpG dinucleotides (Kirmizis et al. 2004; Klose and Bird 2006; Squazzo et al. 2006; Kouzarides 2007). Because different complexes create these three different types of silenced chromatin, we consider them to be three separate mechanisms of repression. The concept of gene regulation mediated by the transition of a bound to an elongating RNA polymerase II has also been previously established (Krumm et al. 1993; Lis 2007). For example, MYC can activate transcription by a post-RNAPII recruitment mechanism (Eberhardy and Farnham 2001). Also, recent genome-scale studies have identified a set of promoters that are bound by POLR2A but do not have detectable transcripts (Kim et al. 2005; Guenther et al. 2007). It is possible that this set of genes could have been regulated by transcript degradation. If transcription proceeded through the entire gene but the transcript was very unstable, H3K36me3 would be detected throughout the gene. To test this possibility, one group (Guenther et al. 2007) analyzed chromatin marks in the body of the gene. They showed that H3K36me3 was found on the body of the genes that had bound POLR2A and detectable transcripts but not on the body of genes that had bound POLR2A and no detectable transcripts, thus confirming the existence of a set of genes that have POLR2A bound to their promoters but do not produce full-length transcripts. For the purpose of our study, such genes are defined here as being regulated at the step of productive elongation; we define this as a fourth mechanism of repression. Finally, many of the promoters that are not silenced by repressed chromatin structure or regulated by a bound (but not elongating) POLR2A are likely regulated by the lack of a specific promoter- or enhancer-binding protein that is required to recruit RNA polymerase II and the general transcriptional machinery; we define the lack of critical factor as a fifth mechanism of repression.
To identify these different sets of promoters, we performed chromatin immunoprecipitation coupled with DNA microarrays (ChIP-chip) using antibodies to H3me3K27, H3me3K9, 5-methyl-cytosine (5-meC), and to the hypophosphorylated form of the largest subunit of RNA polymerase II in each of the 12 cell populations (see Supplemental Table S1 for a list of all the ChIP-chip arrays used in this study). Each ChIP sample was tested by PCR using primers specific for positive and negative controls; a list of promoters and primers used for positive and negative controls for each ChIP sample is provided in Supplemental Table S1. After confirmation that the ChIP assay was successful, amplicons were prepared and tested using the same positive and negative control primer sets (see Supplemental Fig. S4; see also Acevedo et al. 2007, 2008; O'Geen et al. 2007). After demonstrating that the amplicons were accurate representations of the starting ChIP samples, they were labeled and hybridized to mouse or human promoter arrays (see Supplemental Table S1 for a description of the type of promoter array used for each sample). The enrichment values for each promoter were determined (see Methods) and the top ranked set of 2000 promoters for each antibody and cell population combination was selected.
Diverse cell types have very similar modes of transcriptional repression
Having identified the list of expressed and nonexpressed genes and determined the enrichment values for POLR2A, H3me3K27, H3me3K9, and 5-meC for each cell type, the next step was to combine the RNA expression data and the ChIP-chip data to derive a single list of genes. Because different platforms were used for RNA (Illumina) and ChIP-chip (NimbleGen) analyses, only genes represented on both platforms could be used for the combined analyses. Fortunately, between 13,000 and 16,000 genes (depending on the exact promoter platforms used for a particular experiment) could be analyzed for both expression and ChIP-chip studies. After deriving the combined expression and ChIP data set for a particular cell type, the next step was to determine what percentage of the LEGs or MEGs was represented in the top 2000 POLR2A, H3me3K27, H3me3K9, and 5-meC data sets and what percentage was not found in any of the top 2000 data sets (similar analyses using more and less stringent cutoffs produced similar results as the top 2000 data sets; data not shown). The percentages at which the top 2000 targets for the different types of repression marks are represented in the LEGs and MEGs categories for all 12 cell populations are shown in Supplemental Table S2. We note that restricting the set of targets to the top 2000 ranked promoters may eliminate or add some weak targets (depending upon the number of targets for each mark). However, the set of top 2000 targets is a generous set for most marks, with the enrichment value of the bottom target in the set falling below twofold (1.0 log2) for 11 of the 12 cell lines for 5-meC and for nine of the 12 cell lines for both H3me3K9 and H3me3K27 (see Supplemental Table S3 for the enrichment values of the promoters in the top 2000 targets that fell into the LEGs, MEGs, and HEGs categories for each ChIP-chip experiment). Thus, we would not classify the promoters lower than 2000 on the ranked lists as being targets in most of the cell lines. However, it is possible that the number of LEGs in the 5-meC category for MCF7 cells, in the H3me3K9 category in mESc, EB, and Huh7, and in the H3me3K27 category for Huh7, foreskin, and lung may increase modestly if the target set was extended.
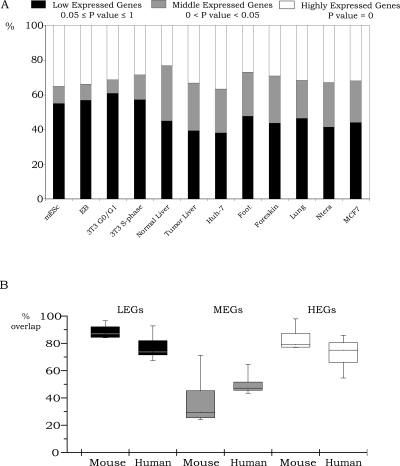
The first conclusion that can be drawn from these data is that 30%–60% of the LEGs and MEGs are not bound by POLR2A, H3me3K27, H3me3K9, or 5-meC (see Supplemental Table S2). This suggests that these promoters could be inactive due to the lack of a positively acting transcription factor that is needed to recruit the general transcription factors. Alternatively, other chromatin modifications besides the ones tested in this study may be responsible for repression. For example, methylation of lysine 20 on histone H4 has been associated with certain repressed regions, as has dimethylation of lysine 9 of histone H3 (Kouzarides 2007). Also, histones can be phosphorylated, ubiquitinated, and methylated on argines (Kouzarides 2007). It is possible that one or more of these modifications could play a role in transcriptional repression. Second, we note that the rank order by which POLR2A, H3me3K27, H3me3K9, and 5-meC are bound to LEGs or MEGs is fairly similar in all 12 cell types (H3me3K27 > H3me3K9 ≈ 5-meC > POLR2A), suggesting that, regardless of cell type, repression is achieved in a similar way (Fig. 2A,B). For example, H3me3K27 is the most common repression mark used in all 12 cell populations and H3me3K9 and 5-meC are used at similar percentages for eight of the 12 cell populations. However, we do note that H3me3K9 is used more frequently than 5-meC in the liver tissue samples and that we did not identify promoters regulating the LEGs or MEGs that were repressed by DNA methylation in the mES cells and EBs. We have confirmed that the 5-meC ChIP samples from the mES cells and EBs were of high quality by performing PCR analysis of the H19 region (see Supplemental Fig. S4) and by performing the ChIP-chip experiments several times (V.M. Komashko and P.J. Farnham, unpubl.). It is possible that, in the mES cells and EBs, some of the genes in the LEGs or MEGs categories were repressed by DNA methylation of genomic regions not represented on the promoter arrays. Future studies employing whole genome analyses of DNA methylation may identify such genes. Third, we note that a bound POLR2A is associated with a small percentage of the LEGs (4%–7%) but with a much larger percentage of the MEGs (11%–23%) in all cell types. Fourth, we note that, in general, fewer MEGs than LEGs are bound by H3me3K27 (Fig. 2C). Also, as expected, we found that a much larger percentage of HEGs than LEGs were bound by POLR2A, which confirms the general correlation of POLR2A recruitment and RNA expression. Fifth, we found that the percentage of genes bound by H3me3K9 or 5-meC was not correlated with expression (i.e., the percentages are similar for LEGs, MEGs, and HEGs). Although H3me3K9 and 5-meC are generally thought to be associated with silenced chromatin, several recent studies have shown the presence of these marks and/or associated proteins on actively transcribed genes (Vakoc et al. 2005; Wiencke et al. 2007; Yasui et al. 2007). Our results, along with the previous studies, suggest that H3me3K9 or 5-meC may require combination with other modifications to specify activation or repression.
Figure 2.
Comparison of repression mechanisms for 12 cell types. The percentage of LEGs (A) or MEGs (B) bound by H3me3K27, POLR2A, H3me3K9, or 5-meC is shown for each of the 12 cell populations. In panel C, box and whisker plots demonstrate the utilization of the four different marks in all 12 cell populations as shown for LEGs, MEGs, and HEGs. The upper and lower quartiles of the box plots are the 75th and 25th percentiles, respectively. The whisker top and bottom are 90th and 10th percentiles, respectively.
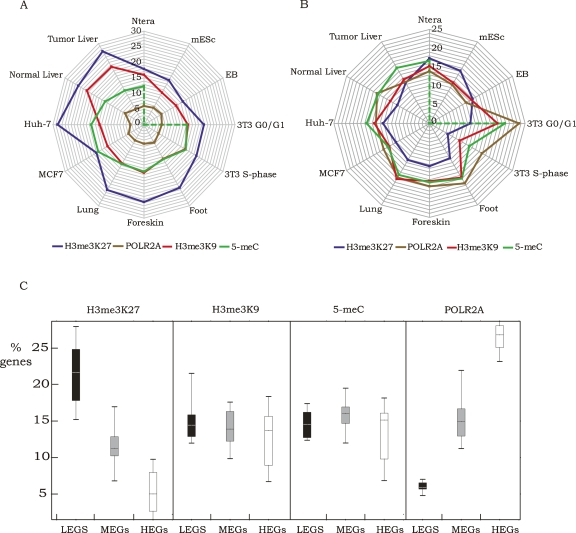
To determine if the repression mechanisms are redundant (i.e., a gene is kept inactive by several different mechanisms) or distinct (i.e., a gene is kept inactive by only one mechanism), the promoters comprising the sets of LEGs and MEGs bound by POLR2A, H3me3K27, H3me3K9, and 5-meC were compared for each cell type. For this analysis, the promoters were classified into sets that were bound by only one of the four marks, by two marks (shown individually for each of the six combinations), or by any three or four marks. As shown in Figure 3 (LEGs) and Supplemental Figure S5 (MEGs), in general a gene is repressed by only one mechanism and the same mechanism is often used in the different cell types. For example, HOXD3 is bound only by H3me3K27 in the primary foot, lung, and foreskin fibroblasts, in the Ntera2 and Huh7 cell lines, and in tumor liver tissue. However, it is bound by both H3me3K9 and H3me3K27 in the normal liver tissue and only by 5-meC in MCF7 cells (enrichment values for all promoters for all marks can be found in Supplemental Table S6 for the human cells and Supplemental Table S7 for the mouse cells; see Supplemental Table S9 for the HOXD3-specific information that was extracted from Supplemental Table S6).
Figure 3.
Promoters are generally repressed by a single mechanism. Shown are pie charts indicating the percentage of LEGs that are bound by only one of the repression marks, by each of the possible different combinations of two marks, or by any three or four marks; also shown is the percentage of LEGs that are not bound by any of the repression marks.
Distinct classes of genes are repressed by certain mechanisms
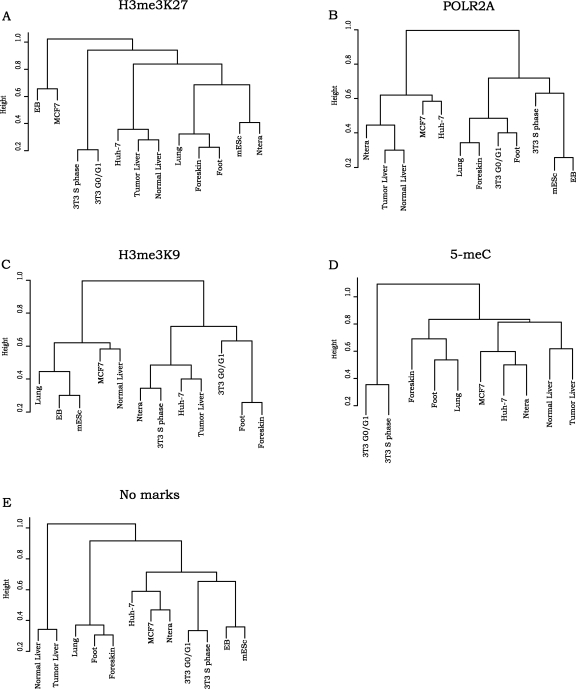
To determine if distinct classes of genes are repressed by different mechanisms in a given cell type and, if so, if the classes are similar or different for the 12 different cell populations, we used the DAVID program (Dennis et al. 2003) to identify significantly enriched gene ontology categories in the LEGs and MEGs that also corresponded to the top 2000 targets bound by POLR2A, H3me3K27, H3me3K9, or 5-meC and those inactive promoters that are not bound by any of the tested marks. For these analyses, all categories of genes having at least 10 members and an enrichment P-value <0.01 were selected. All of the resultant gene ontology categories for an individual repression class in each cell population were then used for cluster analyses (Fig. 4; see Methods for details). We found that similar cell types often clustered next to each other, suggesting that related cell types repress certain types of genes using a common repression mechanism. For example, comparison of the H3me3K27-bound sets (Fig. 4A) from the 12 cell populations showed that fibroblasts collected from distal parts of the body (foot and foreskin) were closer to each other than to the fibroblasts that were collected from internal organs such as lung. These results confirm identity relationships of different fibroblasts previously shown using RNA expression analyses (Rinn et al. 2006). We also found that mES cells clustered close to the Ntera2 embryonal cancer cell line, which we have previously shown to have a similar chromatin structure as ES cells (Squazzo et al. 2006), and that, as expected, the liver tissues were close to each other regardless of normal or tumor state (Acevedo et al. 2008). Comparison of the gene ontology categories of the POLR2A-bound sets of LEGs (Fig. 4B) demonstrated separations of the cell lines into two groups: normal and cancer, which may suggest different regulation at the step of productive elongation. We also noticed that mouse 3T3 fibroblasts and human foot, foreskin, and lung fibroblasts, all of which had been synchronized in G0 phase prior to analysis, were in the same cluster in the POLR2A-bound set of LEGs. However, the S phase 3T3 fibroblasts were not found in the same cluster. This suggests that a stalled RNA polymerase regulates specific gene categories in certain stages of the cell cycle in a variety of cell types in different species. Clustering of the gene ontology categories obtained for the 5-meC data of the LEGs (Fig. 4D) resembles the cluster obtained with H3me3K27-bound set, showing similar cell type relationships. In contrast, we did not find any strong cell type or cell state relationships in the H3me3K9-bound clusters (Fig. 4C).
Figure 4.
Cluster analysis of gene ontology categories in the sets of LEGS repressed by different mechanisms. The sets of LEGs for each cell population that were bound by H3me3K27 (A), POLR2A (B), H3me3K9 (C), 5-meC (D), or no marks (E) were analyzed using the DAVID gene ontology program. The functional annotations that passed the cutoff criteria (see Methods) for each cell population were compared by transforming the lists into a binary data set (if a functional category was present, then its P-value was assigned to 1; if it was absent, then its P-value was assigned to 0). We then performed hierarchical cluster analysis and plotted the dendograms in R (http://www.R-project.org) using hclust function.
We next refined the gene ontology lists by removing noninformative terms (such as multi-gene family, alternative splicing, and direct protein sequencing) and condensing similar categories (such as transcription factor and transcriptional regulator). After this, the 20 most significantly enriched categories for the POLR2A, H3me3K27, H3me3K9, or 5-meC sets for each of the 12 cell populations were selected. We found that the most significantly enriched categories for a given mark were similar in the different cell types. Thus, the condensed lists were comprised of less than the possible 240 categories; there were 34 categories for H3me3K27 (Supplemental Fig. S6A), 22 categories for POLR2A (Supplemental Fig. S6B), 52 categories for H3me3K9 (Supplemental Fig. S6C), 53 categories for 5-meC (Supplemental Fig. S6D), and 47 categories for the “no marks” set (Supplemental Fig. S6E). We note that DNA binding proteins are found in the top 20 sets of gene ontology terms of the H3me3K27-, H3me3K9-, POLR2A-, and 5-meC-bound promoters in all 12 cell types. In contrast, the genes that are not active but which are not bound by any of the repression marks are not significantly enriched in DNA binding proteins. We also found that certain categories were enriched in both of the H3me3K27 and H3me3K9 lists, which is not surprising considering that about 1/3 of the promoters bound by H3me3K27 are also bound by H3me3K9 (Fig. 3).
Characterization of transitioning cell populations
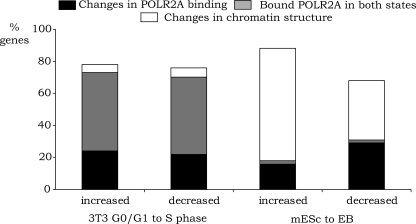
Although, in general, the set of genes expressed in G0/G1 cells is very similar to the set of genes expressed in S phase cells, the two cell populations have distinct roles in the cell cycle and thus must exhibit differential expression of certain genes. Similarly, the set of genes expressed in mES cells are very similar to those expressed in EB cells, but genes involved in pluripotency (such as Pou5f1) or genes that specify a specific differentiating phenotype (e.g., globin genes) are differentially expressed in the two populations. The transition between G0/G1 and S phase and the initiation of differentiation (as monitored by the down-regulation of Pou5f1) are both rapid events, with major changes (DNA replication or gain of a differentiated phenotype, respectively) occurring within a short period of time (Supplemental Figs. S2, S3). However, these two transitions are distinguished by the fact that cell cycle stages are transient cell states (requiring activation or repression of genes in a periodic manner) whereas the ES to EB transition is permanent (requiring long-term activation or repression of specific genes). We were interested to know if the gene expression changes that occur during these two cell state transitions are regulated similarly or if the transient nature of a cell cycle stage requires a different mode of regulation than is used to set up the permanent state of differentiation. Therefore, using the Illumina RNA array data, we performed differential expression analysis and identified four distinct sets of genes: those that are up-regulated when G0/G1 cells enter S phase, those that are down-regulated when G0/G1 cells enter S phase, those that are up-regulated when mES cells differentiate into EBs, and those that are down-regulated when mES cells differentiate into EBs (Supplemental Table S4). We then characterized the mechanisms that regulate the changes in RNA levels in the four cell populations (Fig. 5). For this analysis, we identified the subset of genes that showed expression changes and a >2000 position change in the target gene list for POLR2A, H3me3K27, H3me3K9, or 5-meC. For example, an up-regulated gene was required to move 2000 positions higher in the POLR2A list of ranked targets or 2000 positions lower in the H3me3K27, H3me3K9, or 5-meC list. Conversely, a down-regulated gene was required to move at least 2000 positions lower in the POLR2A list or 2000 positions higher in the H3me3K27, H3me3K9, or 5-meC list. In addition, we eliminated all genes that were not true targets of each mark (but were simply changing positions in the very bottom of the ranked lists) by requiring that the enrichment value be at least 0.7 (on a log2 scale) in the appropriate set. For example, for a gene classified as being up-regulated in S phase due to increases in POLR2A binding, the POLR2A enrichment value must be at least 0.7 (on a log2 scale) in S phase; similarly, for a gene classified as being down-regulated in S phase by decreases in POLR2A binding, the POLR2A enrichment value must be at least 0.7 (on a log2 scale) in G0/G1. We also identified the set of genes that showed changes in RNA levels during the G0 to S phase progression or after differentiation of the mES to EB but which were bound by high levels of POLR2A in both cell states. Interestingly, we found that half of the RNAs that were either up- or down-regulated during the G0/G1 to S phase transition had high levels of bound POLR2A in both cell states whereas very few genes regulated during the mES to EB transition were bound by POLR2A in both cell states (Fig. 5). Conversely, we found that a large number of genes regulated during the mES to EB transition showed changes in chromatin structure whereas very few genes regulated during the G0/G1 to S phase transition were regulated by chromatin structure.
Figure 5.
Mechanistic analysis of transitioning populations. The subsets of genes that showed expression changes and a >2000 position change in the target gene list for POLR2A, H3me3K27, H3me3K9, or 5-meC were identified by comparing G0/G1 to S phase 3T3 cells and mESc to EBs. For example, an up-regulated gene in S phase or EBs was required to move 2000 positions higher in the POLR2A list of ranked targets or 2000 positions lower in the H3me3K27, H3me3K9, or 5-meC list. Conversely, a down-regulated gene in S phase or EBs was required to move at least 2000 positions lower in the POLR2A list or 2000 positions higher in the H3me3K27, H3me3K9, or 5-meC list. In addition, we eliminated all genes that were not true targets of each mark (but were simply changing positions in the very bottom of the ranked lists) by requiring that the enrichment value be at least 0.7(on a log2 scale) in the appropriate set. For ease of display of the data, we combined the sets of promoters regulated by changes in H3me3K27, H3me3K9, or 5-meC and identified this set as “regulated by changes in chromatin structure.” We also identified the set of genes that showed changes in RNA levels during the G0 to S phase progression or after differentiation of the mES to EB but which were bound by high levels of POLR2A in both cell states (promoters had a >1.0 [on a log2 scale]), enrichment for POLR2A in both cell states, and lacked silencing marks in either cell state (enrichments <0.7 [on a log2 scale] for H3me3K27, H3me3K9, or 5-meC).
Discussion
We compared 12 different cell populations, including embryonic stem cells before and during differentiation into embryoid bodies and various types of normal and tumor cells to determine if pluripotent and differentiated cell types use different mechanisms to establish their transcriptome. We found that (1) the percentage of low expressed genes bound by POLR2A, H3me3K27, H3me3K9, or 5-meC is similar in all 12 cell types, regardless of differentiation or neoplastic state; (2) a gene is generally repressed by only one mechanism; and (3) distinct classes of genes are repressed by certain mechanisms. However, although the most significantly enriched GO categories are repressed by a specific mechanism in all cell types (see Supplemental Fig. 6A–E), other GO categories are repressed by a specific mechanism only in highly related cells (see Supplemental Fig. 4A–E). We further characterized two transitioning cell populations, mES cells differentiating into embryoid bodies and 3T3 cells progressing from G0/G1 into S phase. We found that the transient regulation through the cell cycle was achieved predominantly by changes in the recruitment of the general transcriptional machinery or by post-RNAPII recruitment mechanisms. In contrast, changes in chromatin silencing were critical for the permanent changes in gene expression in cells undergoing differentiation.
Although we characterized a diverse set of mouse and human cell populations, we found that the majority of the genes expressed in one cell type were also expressed in the other cell types. Many of these genes encode housekeeping proteins that regulate basic cellular processes. For example, many of the top 20 (as ranked by P-value) gene ontology categories of the commonly expressed HEGs are related to protein synthesis and protein transport. Although we did not further characterize this set of commonly expressed genes, they are likely to provide a rich source for identifying motifs and factors that are used to ensure expression of a gene in all cell types. To assist in such future endeavors, we have provided the set of commonly expressed genes in mouse and human cells as Supplemental Table S5. We also found that the majority of the genes that were not expressed in one of the cell populations were also not expressed in certain other of the cell populations. It is likely that many of the identified LEGs are cell type-specific or developmentally regulated genes that are expressed in a limited numbers of tissues and/or for a limited time. Interestingly, we found that, although the cell types analyzed were quite diverse, they each employed the five characterized mechanisms (lack of recruitment of the general transcriptional machinery, stimulation of a bound POLR2A, or silencing by the presence of H3me3K27, H3me3K9, or DNA methylation) at similar levels.
The percentage of LEGs regulated by a bound POLR2A was, in general, fairly low for all cell populations (4%–7%) but a greater percentage of the MEGs were regulated by a bound POLR2A (11%–23%). Taken together, these results are similar to studies of gene regulation in Drosophila and human stem cells, which have suggested that 10%–50% of genes (depending on the method of estimation) are regulated by release of a bound RNA polymerase II (Ren et al. 2000; Radonjic et al. 2005; Guenther et al. 2007; Lis 2007; Muse et al. 2007; Tamkun 2007; Zeitlinger et al. 2007). These previous studies have shown that genes induced by environmental or developmental signals are often regulated by paused RNA polymerase II. Also, we have previously shown that the Cad promoter, which displays increased activity when quiescent cells are stimulated to reenter the cell cycle, is regulated by MYC via a post-RNAPII recruitment mechanism (Eberhardy and Farnham 2001). In other words, high levels of POLR2A are bound to the Cad promoter in both G0/G1 and S phases of the cell cycle. Similarly, a study of yeast genes showed that POLR2A is located at many inactive promoters in quiescence (Radonjic et al. 2005). To address whether genes that are regulated during the G0/G1 to S phase transition of mammalian cells are, in general, regulated by a post-RNAPII recruitment mechanism, we determined the number of genes whose expression was increased or decreased in S phase (as compared to G0/G1 phase), that had high levels of bound POLR2A in both states, and did not have silencing marks in either state. We found that ∼50% of the genes that were increased or decreased in S phase had high levels of bound POLR2A in both cell states. Thus, regulation of a pre-bound RNA polymerase II appears to be a key mechanism by which transcription is linked to cell cycle progression. In contrast, a similar analysis of the mES cell to EB transition revealed that ∼2% of the genes regulated during differentiation had a bound POLR2A in both cell states, but that many of the genes were regulated by changes in chromatin structure. Our finding that regulation of a bound RNA polymerase II is important in cell cycle-dependent regulation fits the biological pattern of cell cycle regulation. For example, genes that are down-regulated in S phase will need to be up-regulated in the G1 phase of the next cell cycle. Thus, using a stable repression mechanism such as silenced chromatin is not appropriate. In contrast, the genes that show altered expression when pluripotent mES cells are differentiated into EBs are undergoing permanent changes in gene expression that can be controlled for the long-term via chromatin modifications.
In conclusion, our studies suggest that transcriptional repression is regulated in very similar ways in a variety of different types. Specifically, most genes are repressed by only one mechanism, specific gene categories correlate with specific modes of repression, H3me3K27 is found at more repressed promoters than the other tested marks, and that many promoters may be inactive in a given cell type due to lack of a specific activator that can recruit the general transcriptional machinery.
Methods
Cell culture
Ntera2 cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS, 2 mM glutamine, and 1% Penicillin/Streptomycin. MCF7 cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS and 2 mM glutamine. Huh7 cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin. Mouse embryonic stem cells line E14 was obtained from BayGenomics (UC Davis). The cells were grown according to the protocol http://baygenomics.ucsf.edu/protocols/comp1/feederES.html in Glasgow Minimum Essential Medium supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 1× nonessential amino acids, 10% fetal bovine serum, a 1:1000 dilution of beta-mercaptoethanol stock solution (70 μL of beta-mercaptoethanol to 20 mL of deionized water), and 700 units per mL of leukocyte inhibitory factor (Chemicon) on gelatin-coated plates. Differentiation of embryonic bodies was induced by gentle shaking of mouse embryonic stem cells in suspension on bacterial plates in the absence of leukocyte inhibitory factor (Zhang et al. 2003). Differentiation medium: Iscove’s Modified Dulbecco’s Medium supplemented with 15% FBS, 2 mM glutamine, 1% of ascorbic acid solution (0.05 g in 10 mL H2O), 3 μL per mL of monothiolglycerol (26 μL per 2 mL). Embryonic bodies were collected on day 4.5 when expression of POU5F1 transcription factor was significantly reduced as determined by Western blot analysis (Supplemental Fig. S3). Mouse 3T3 fibroblasts were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% BCS and 4 mM glutamine. For synchronization in G0/G1 phase of the cell cycle 3T3 fibroblasts were grown until 60% confluent. For synchronization in S phase of the cell cycle 3T3 fibroblasts were grown until 30% confluent. To initiate the growth arrest cells were fed with Dulbecco’s Modified Eagle’s Medium supplemented with 0.5% BCS and 4 mM glutamine. After 48 h fresh starvation media was added and cells were incubated for 24 more hours for the total time of 72 h. After 72 h ∼86% of the cells showed 2 n DNA content by FACS analysis (Supplemental Fig. S2). To release from growing arrest, cells were directly fed with medium containing 10% FBS for 16 h. After 16 h ∼60% of the cells reached S phase by FACS analysis (Supplemental Fig. S2). The investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR1208–01 from the National Research Resources, National Institutes of Health. Human Primary fibroblast cultures were propagated in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS and 1% Penicillin/Streptomycin until 80% confluent. Fibroblasts were then synchronized in G0/G1 by the addition of media containing only 0.1% fetal bovine serum for 48 h (Rinn et al. 2006). Mouse ES cells and embryonic bodies were incubated at 37°C in a humidified 6% CO2 incubator. All other cells were incubated at 37°C in a humidified 5% CO2 incubator. Human normal adjacent tissues and tumor liver tissues were obtained from the National Disease Research Interchange NDRI (Philadelphia, PA) and from the University of Wisconsin, Madison Hospital (Madison, WI). Samples were surgically collected and preserved by flash-freezing in liquid nitrogen followed by storage at −70°C. The patient (OD04962) was a 58 yr-old African American male.
RNA expression arrays
Total RNA was prepared from 107−2 × 107 cells using RNeasy kit (Qiagen) following the manufacturer’s instructions. RNA quality was ensured using the Agilent Systems Bioanalyzer. Biotinylated RNA was prepared with the Illumina TotalPrep RNA Amplification kit (Ambion) and hybridized to Illumina Human Ref-8 v2 and Mouse Ref-8 Expression BeadChips. Human Ref-8 v2 Expression BeadChip contains 22,000 transcripts, Mouse Ref-8 Expression BeadChip contains 24,000 well annotated RefSeq transcripts. Hybridization signals were detected with Illumina BeadArray Reader. RNA labeling, amplification, and array hybridization were performed by the Expression Analysis Core at UC Davis.
ChIP assays and amplicons preparation
ChIP assays (200 μg/assay) were performed following the ChIP protocol provided at http://www.genomecenter.ucdavis.edu/farnham and http://genomecenter.ucdavis.edu/expression_analysis. ChIP assays with smaller amount of chromatin were performed following the protocol (Acevedo et al. 2007). ChIP assay with 5-Methylcytidine antibody for all cell lines and tissues except MCF7 was performed using ChIP-IT Express kit (Active Motif, Cat # 53008). For ChIP with 5-Methylcytidine antibody genomic DNA was extracted by shaking cells in digestion buffer (100 mM NaCl, 10 mM TrisCl, pH 8.0, 25 mM EDTA, pH 8.0, 0.5% SDS) and 0.1 mg/mL Proteinase K for 12–18 h at 50°C and purified using phenol-chlorophorm extraction method. Extracted DNA was sonicated to an average size of 800 bp, denatured at 95°C for 10 min, quickly chilled on ice and captured on magnetic beads following the protocol as described by the manufacturer. For MCF7 cell line the IP setup was done as described in the MicroChIP protocol, but instead of heat-denaturation NaOH was added to IP dilution buffer to the final concentration of 7 mM to separate DNA strands. The primary antibodies used in this study were as follows: mouse monoclonal RNA polymerase II 8WG16 IgG2a (Covance Cat # MMS-126R), two different rabbit polyclonal H3me3K9 IgGs (Abcam Cat # ab8898 and ab1186), rabbit polyclonal H3me3K27 IgG (Upstate Cell Signaling Cat # 07–449), mouse monoclonal 5-Methylcytidine (Eurogentec Cat # BI-MECY-0100). The secondary rabbit anti-mouse IgG was purchased from MP Biomedicals (Cat # 55436). The nonspecific rabbit IgG used as a negative control in the ChIP assays was purchased from Alpha Diagnostics (Cat # 20009–5). For PCR analysis of the ChIP samples (200 μg/assay) prior to amplicon generation, purified immunoprecipitates (QIAquick PCR purification kit, Qiagen) were dissolved in 50 μL of water. Standard PCR reactions using 2 μL of the immunoprecipitated DNA were performed. PCR products were separated by electrophoresis through 1.5% agarose gels and visualized using ethidium bromide. Amplicons were prepared by adapting the standard protocol for the whole genome amplification using the Sigma GenomePlex WGA2 kit as described in O'Geen et al. (2006). Briefly, the initial random fragmentation step was eliminated and DNA from an entire ChIP sample or from 10 ng of total chromatin was amplified. A detailed protocol for the WGA2 method is provided at http://genomics.ucdavis.edu/farnham and http://genomecenter.ucdavis.edu/expression_analysis. Precipitated DNA from ChIP with smaller starting material (4–30 μg/assay) was measured by Quant-iT PicoGreen assay (Invitrogen) (Acevedo et al. 2008). DNA was purified and amplified using the Sigma GenomePlex Single Cell WGA4 kit as described in Acevedo et al. (2007). PCR positive and negative primers used for ChIP samples and amplicons confirmation are described in Supplemental Table S1.
ChIP-chip assays
Amplicons were applied either to mouse promoter arrays or human promoter arrays (see www.nimblegen.com for details). The labeling and hybridization of DNA samples for ChIP-chip analysis were performed by NimbleGen Systems, Inc., or at UC Davis (Supplemental Table S1). Briefly, each DNA sample (1 μg) was denatured in the presence of 5′-Cy3- or Cy5-labeled random nonamers (TriLink Biotechnologies) and incubated with 100 units (exo-) Klenow fragment (NEB) and dNTP mix (6 mM each in TE buffer [10 mM Tris/1 mM EDTA, pH 7.4; Invitrogen]) for 2 h at 37°C. Reactions were terminated by addition of 0.5 M EDTA (pH 8.0), precipitated with isopropanol, and resuspended in water. Then, 12 μg of the Cy5-labeled ChIP sample and 12 μg of the Cy3-labeled total sample were mixed, dried down, and resuspended in 40 μL of NimbleGen Hybridization Buffer (NimbleGen Systems). After denaturation, hybridization was carried out in a MAUI Hybridization System (BioMicro Systems) for 18 h at 42°C. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation, and scanned at 5-μm resolution using the GenePix 4000B scanner (Axon Instruments). Fluorescence intensity raw data were obtained from scanned images of the oligonucleotide tiling arrays using NimbleScan 2.2 extraction software (NimbleGen Systems). For each spot on the array, log2 ratios of the Cy5-labeled test sample versus the Cy3-labeled reference sample were calculated. Then, the biweight mean of this log2 ratio was subtracted from each point; this procedure similar to mean-normalization of each channel.
We validated the ChIP-chip assays in several ways. First, we performed duplicate arrays for a subset (13 of the 48) of samples, for which we found a high correlation between arrays (R-value ranges from 0.8 to 0.97). Second, we examined the enrichment values on the arrays for our positive and negative control promoters (which we first analyzed by ChIP-PCR) to ensure that the positive controls were enriched and the negative controls were not enriched. Third, the reproducibility of our ChIP-chip assays was also validated by comparison of the different samples. As noted in the text, similar numbers of genes and the same gene ontology categories were identified for a particular mark in all 12 cell populations. Finally, we performed PCR assays of a subset of the targets identified on the arrays.
Data analysis
Hybridization signals were analyzed using BeadStudio Gene Expression Module v.3 (Illumina). All transcripts were normalized and ranked according to the detection P-value, which is calculated based on signals of negative controls. The detection P-value is calculated as 1-R/N, where R is the rank of the gene signal relative to negative controls and N is the number of negative controls. The detection P-value can be interpreted as probability of seeing a certain signal level without specific probe-target hybridization. For the low expressed genes (LEGs) we set a P-value threshold at 0.05 (0.05 ≤ P-value < 1), for the middle expressed genes (MEGs) we set a P-value threshold to be >0, but <0.05 (0 < P-value < 0.05). Genes with highest expression (HEGs) had a P-value equal to 0. In order to match Illumina RNA expression arrays and NimbleGen promoter arrays we obtained the unique Entrez Gene ID. We were able to obtain unique Gene IDs for 15,560 transcripts from the Human Ref-8 v2 platform and for 18,286 from the Mouse Ref-8 platform. It is theoretically possible that some of the RNAs identified as LEGs were actually expressed but were not detected on the Illumina arrays due to poor probe design. However, most genes were identified in the HEGs set in a least one cell type, indicating that the probes for most genes are good. Also, for our analyses, we only used the probes for the known genes (based on annotation from Entrez Gene ID and RefSeq genomes) that were present on both NimbleGen promoter and Illumina arrays. These genes have well characterized exon–intron structure, so the chance that a probe is designed to an intron or nontranscribed region is very small. However, there were 2481 genes that were classified as LEGs in all eight human cell types. Of these 2481 genes, the vast majority was not bound by POLR2A on the NimbleGen platform ChIP-chip arrays, supporting the hypothesis that these genes are not on in the cell types examined.
Several different promoter array designs were used in this study. Mouse amplicons samples were hybridized to the UCSC mm5 1.5 kb promoter array, which consists of a single array design containing 26,842 promoters. Human samples were hybridized either to UCSC hg17 1.5 kb promoter array, UCSC hg17 5 kb promoter array, UCSC hg18 4.4 kb promoter array, or UCSC hg18 RefSeq 2.5 kb promoter array. Human UCSC hg17 1.5 kb promoter array is a single array design containing 24,275 promoters. Human UCSC hg17 5 kb and hg18 4.4 promoter arrays consist of two individual arrays (promoter 1 and promoter 2), containing 27,295 and 23,047 promoters, respectively. Human UCSC hg18 RefSeq 2.5 kb promoter array is a single array design containing 24,659 well-characterized RefSeq genes. The exact design used for each experiment is indicated in Supplemental Table S1. Regions on the promoter arrays bound by the individual factors were determined using the Maxfour peak calling method (M.C. Bieda and P.J. Farnham, in prep.). Briefly, a value was assigned based on the highest mean of four consecutive probes in each promoter, then obtained results were combined if two-array designs were used.
RNA expression arrays and promoter arrays were matched based on unique Entrez Gene IDs. To obtain a number of genes in each category (e.g., low expressed genes, middle expressed genes, and high expressed genes) bound by H3me3K27, H3me3K9, RNAPII, or 5-meC we ranked promoter array data by the Maxfour values and determined how many genes belong to each expression category (e.g., low expressed genes, middle expressed genes, and high expressed genes) in the top 2000 genes for every factor. We then normalized the obtained value by the total number of genes in each expression category.
Functional annotations were performed using the program Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2007 (Dennis et al. 2003) (see also http://niaid.abcc.ncifcrf.gov/). The same parameters were used for all analyses presented in this study. These parameters were Gene Ontology (GO) Molecular Function term, level 2; InterPro name is the Protein Domains section; and SP_PIR_Keywords in the Functional Category section. The EASE Score Threshold was set at 0.01.
For the cluster analysis we combined obtained functional annotations with enrichment P-values for every factor in one data set. We transformed the data set into a binary one: If a functional category was present, then its P-value was assigned to 1; if it was absent, then its P-value was assigned to 0. We performed hierarchical cluster analysis and plotted the dendograms in R (http://www.R-project.org) using hclust function. The following parameters were used for clustering: distance metric = binary, agglomeration rule = Ward’s minimum variance. Heatmaps were prepared in R (see http://www.R-project.org) using the RColorBrewer palettes of R package version 1.0–1.
Acknowledgments
We thank John Rinn and Howard Chang (Stanford University) for the generous gift of human primary fibroblasts, all members of the Farnham laboratory, especially Mark Bieda and Victor Jin, for helpful discussions, and Katie Pollard (UC Davis) for help with the statistical analyses. This work was supported in part by Public Health Service grants CA45250, HG003129, and DK067889.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to Gene Expression Omnibus under accession no. GSE10504.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074609.107.
References
- Acevedo L.G., Iniguez A.L., Holster H.L., Zhang X., Green R., Farnham P.J., Iniguez A.L., Holster H.L., Zhang X., Green R., Farnham P.J., Holster H.L., Zhang X., Green R., Farnham P.J., Zhang X., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Genome-scale ChIP-chip analysis using 10,000 human cells. Biotechniques. 2007;43:791–797. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo L.G., Bieda M., Green R., Farnham P.J., Bieda M., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Analysis of the mechanisms mediating tumor specific changes in gene expression in human liver tumors. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-07-5590. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Wang Z., Wei G., Chepelev I., Zhao K., Wei G., Chepelev I., Zhao K., Chepelev I., Zhao K., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Dennis G.J., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R., Farnham P.J., Farnham P.J. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A., Levine S.S., Boyer L.A., Jaenisch R., Young R.A., Boyer L.A., Jaenisch R., Young R.A., Jaenisch R., Young R.A., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A., Bartley S.M., Kuzmichev A., Margueron R., Reinberg D., Green R., Farnham P.J., Bartley S.M., Kuzmichev A., Margueron R., Reinberg D., Green R., Farnham P.J., Kuzmichev A., Margueron R., Reinberg D., Green R., Farnham P.J., Margueron R., Reinberg D., Green R., Farnham P.J., Reinberg D., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Silencing of human polycomb target genes is associated with methylation of histone H3 lysine 27. Genes & Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Komili S., Silver P.A., Silver P.A. Coupling and coordination in gene expression processes: A systems biology view. Nat. Rev. Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krek W., DeCaprio J.A., DeCaprio J.A. Cell synchronization. Methods Enzymol. 1995;254:114–124. doi: 10.1016/0076-6879(95)54009-1. [DOI] [PubMed] [Google Scholar]
- Krumm A., Meulia T., Groudine M., Meulia T., Groudine M., Groudine M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays. 1993;15:659–665. doi: 10.1002/bies.950151005. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L., Carey M., Workman J.L., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lis J.T. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K., Grissom S.F., Zeitlinger J., Adelman K., Zeitlinger J., Adelman K., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A., Klewe-Nebenius D., Chambers I., Scholer H., Smith A., Chambers I., Scholer H., Smith A., Scholer H., Smith A., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G., Miyazaki J., Smith A.G., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- O'Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Blahnik K., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H., Squazzo S.L., Iyengar S., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J., Squazzo S.L., Iyengar S., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J., Iyengar S., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J., Rinn J.L., Chang H.Y., Green R., Farnham P.J., Chang H.Y., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange T., Reinberg D., Lagrange T., Reinberg D., Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Radonjic M., Andrau J.-C., Lijnzaad P., Kemmeren P., Kockelkorn T.T.J.P., van Leenen D., van Berkum N.L., Holstege F.C.P., Andrau J.-C., Lijnzaad P., Kemmeren P., Kockelkorn T.T.J.P., van Leenen D., van Berkum N.L., Holstege F.C.P., Lijnzaad P., Kemmeren P., Kockelkorn T.T.J.P., van Leenen D., van Berkum N.L., Holstege F.C.P., Kemmeren P., Kockelkorn T.T.J.P., van Leenen D., van Berkum N.L., Holstege F.C.P., Kockelkorn T.T.J.P., van Leenen D., van Berkum N.L., Holstege F.C.P., van Leenen D., van Berkum N.L., Holstege F.C.P., van Berkum N.L., Holstege F.C.P., Holstege F.C.P. Genome-wide analyses reveal RNA Polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rinn J.L., Bondre C., Gladstone H.B., Brown P.O., Chang H.Y., Bondre C., Gladstone H.B., Brown P.O., Chang H.Y., Gladstone H.B., Brown P.O., Chang H.Y., Brown P.O., Chang H.Y., Chang H.Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Helms J.A., Farnham P.J., Segal E., Farnham P.J., Segal E., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Edwards S.W., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Edwards S.W., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., GuhaThakurta D., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Holder D., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Ying L., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Svetnik V., Leonardson A., Hart K.W., Russell A., Li G., Leonardson A., Hart K.W., Russell A., Li G., Hart K.W., Russell A., Li G., Russell A., Li G., Li G., et al. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 2004;5:R73. doi: 10.1186/gb-2004-5-10-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer H.R., Ruppert S., Suzuki N., Chowdhury K., Gruss P., Ruppert S., Suzuki N., Chowdhury K., Gruss P., Suzuki N., Chowdhury K., Gruss P., Chowdhury K., Gruss P., Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Sims R.J., Mandal S.S., Reinberg D., Mandal S.S., Reinberg D., Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Squazzo S.L., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Green R., Margueron R., Reinberg D., Farnham P.J., Margueron R., Reinberg D., Farnham P.J., Reinberg D., Farnham P.J., Farnham P.J. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W. Stalled polymerases and transcriptional regulation. Nat. Genet. 2007;39:1421–1422. doi: 10.1038/ng1207-1421. [DOI] [PubMed] [Google Scholar]
- Uptain S.M., Kane C.M., Chamberlin M.J., Kane C.M., Chamberlin M.J., Chamberlin M.J. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A., Mandat S.A., Olenchock B.A., Blobel G.A., Olenchock B.A., Blobel G.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wiencke J.K., Zheng S., Morrison Z., Yeh R.-F., Zheng S., Morrison Z., Yeh R.-F., Morrison Z., Yeh R.-F., Yeh R.-F. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210895. [DOI] [PubMed] [Google Scholar]
- Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Nagarajan R.P., Thatcher K.N., Farnham P.J., LaSalle J.M., Thatcher K.N., Farnham P.J., LaSalle J.M., Farnham P.J., LaSalle J.M., LaSalle J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A., Nechaev S., Adelman K., Levine M., Young R.A., Adelman K., Levine M., Young R.A., Levine M., Young R.A., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.J., Chung Y.S., Eades B., Choi K., Chung Y.S., Eades B., Choi K., Eades B., Choi K., Choi K. Gene targeting strategies for the isolation of hematopoietic and endothelial precursors from differentiated ES cells. Methods Enzymol. 2003;365:186–202. doi: 10.1016/s0076-6879(03)65013-5. [DOI] [PubMed] [Google Scholar]