Abstract
There are widespread, genetically determined differences in gene expression. However, methods that compare transcript levels between individuals are subject to trans-acting effects and environmental differences. By looking at allele-specific expression in the F1 progeny of inbred mice, we can directly test for allelic imbalance (AI), which must be due to cis-acting variants in the parental strains. We tested over one hundred genes for AI between C57Bl/6J and A/J alleles in F1 mice, including a validation set of 23 genes enriched for cis-acting variants and a second set of 92 genes whose orthologs were previously examined for AI in humans. We assayed an average of two transcribed SNPs per gene in liver, spleen, and brain from three male and three female F1 mice. In the set of 92 genes, we observed 33 genes (36%) with significant AI including genes with AI that was specific to certain tissues or transcripts. We also observed extensive tissue-specific AI, with 11 out of 92 genes (12%) having differences in AI between tissues. Interestingly, several genes with alternate transcripts have transcript-specific AI. Finally, we observed that the presence of AI in human genes was correlated to the presence of AI in the mouse orthologs (one-tailed P = 0.003), suggesting that certain genes may be more tolerant of cis-acting variation across species.
Coding variation consisting of polymorphisms or mutations in the amino acid coding sequences of genes has been the traditional focus of genetic studies. Because the genetic code is known, it is straightforward to predict the location of a coding variant. Some examples of strong associations between coding variants and common diseases have been reported (Altshuler et al. 2000; Florez et al. 2004; Klein et al. 2005; Duerr et al. 2006; Sladek et al. 2007). Several noncoding variants have been also strongly associated to complex traits suggesting that they should not be ignored in genetic studies (Ueda et al. 2003; Graham et al. 2006; Grant et al. 2006; McPherson et al. 2007; Saxena et al. 2007).
Gene expression level as determined by microarrays has been shown to be a heritable trait in humans (Cheung et al. 2003). To determine the genetic variants that influence gene expression, the transcript level of each gene is considered a complex genetic trait, and linkage or association analysis is employed to map variants that contribute to transcript level variation (Cheung and Spielman 2002; Schadt et al. 2003). Previous studies in humans and model organisms have reported estimates that 20%–40% of genes with expression level differences are associated (or linked) to a nearby genetic marker (Brem et al. 2002; Yvert et al. 2003; Monks et al. 2004; Morley et al. 2004). However, guidelines based on distance do not definitively prove that a variant is cis or trans acting.
Measuring allele-specific expression levels allows one to definitively show if a gene is subject to cis-acting variants. Allele imbalance (AI) is determined using the alleles of a polymorphism in the transcribed region of the gene (i.e., variants in the coding and untranslated regions) as markers for the transcripts from each chromosome. Relative abundance of the alleles of this transcribed polymorphism can be determined in mRNA using many genotyping platforms (Knight 2004). If one allele of this transcribed polymorphism is overrepresented in the mRNA compared to the other allele, then that gene harbors at least one cis-acting variant. This cis-acting variant is likely not the transcribed polymorphism itself but might be located in a transcription factor binding site or region that determines transcript stability.
Several studies in humans and mice suggest that cis-acting variants are common in the mammalian genome. Genes expressed in human lymphoblast cells have been tested for AI, and 18%–53% of the genes tested showed allelic imbalance in at least one individual (Yan et al. 2002; Pastinen et al. 2004; Pant et al. 2006). In a study of 69 genes in F1 mice, four genes had strong evidence of AI (Cowles et al. 2002).
It is currently unknown whether AI is more often tissue-specific or a global phenomenon observed across tissues. In a study of human fetal liver and kidney, high correlation in AI between the two tissues was observed in the AI results, although only seven genes were tested for similarity between tissues (Lo et al. 2003). In contrast, out of four genes with AI in F1 mice, two showed tissue-specific AI (Cowles et al. 2002). These studies of tissue-specific AI have been small and more data are required to determine whether cis-acting variants commonly act in a tissue-specific manner.
We performed allelic imbalance assays in F1 mice. Because their parents are inbred, all F1 mice are heterozygous at single nucleotide polymorphisms (SNPs) where the parental strains have different alleles. Therefore, using existing databases, it is straightforward to identify transcribed SNPs that differ between the two parental strains, C57Bl/6J (B6) and A/J in this case. We have tested 115 genes for AI in C57Bl/6J × A/J F1 mice. We have tested genes previously analyzed for AI in mice (Cowles et al. 2002) and genes with expression differences between A/J and C57Bl/6J determined by microarray analysis. We have also analyzed a set of genes that had been tested for AI in human white blood cells (Pant et al. 2006). We have tested all of the genes in three organs: brain, liver, and spleen, and many genes (12%) show tissue-specific AI. Interestingly, we have observed AI differences between transcribed SNPs in the same genes. Many of these SNPs fall on reported alternate transcripts, confirming that different transcripts can be regulated by different cis-acting variants. Finally, we have found that genes that show AI in humans are more likely to show AI in mice.
Results
Validation of an assay for detecting allelic imbalance on a pilot set of genes
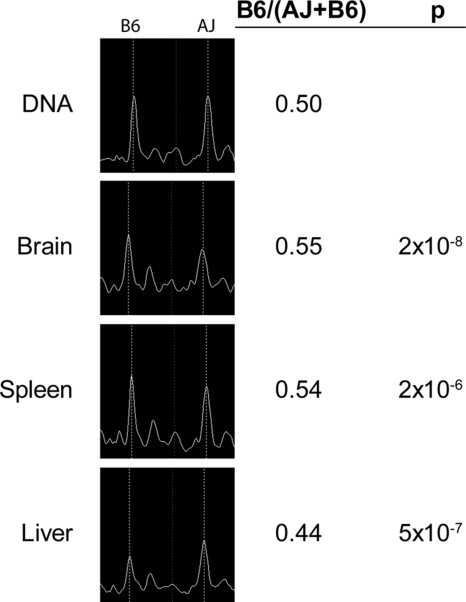
We used the Sequenom MassArray genotyping platform to detect allelic imbalance. We designed assays to transcribed SNPs in genes of interest where A/J and C57Bl/6J differed, and genotyped these assays on cDNA samples and genomic DNA for normalization as described in the Methods. An overview of our approach and results is presented in Figure 1. To validate our method for detecting allelic imbalance, we first tested a gene known to be imprinted, Grb10. Genetic imprinting can lead to AI, but the transcription imbalance of the two alleles is due to epigenetic influences inherited paternally or maternally and not dependent on genetic variants. In the case of imprinting, one would expect the opposite strain allele to be overrepresented in mice from a reciprocal cross where the strains of the parents were switched. Our control gene, Grb10, is expressed from the maternally inherited allele in all tissues except for brain, where it is expressed from the paternally inherited allele only (Hikichi et al. 2003). We observed overrepresentation of the B6 allele in liver and spleen from F1 mice, which have a B6 mother, and overrepresentation of the A/J allele in liver and spleen of RF1 mice which have an A/J mother. The reverse was observed in brain, as expected from the published data (Supplemental Fig. 1A). Therefore, from these data, we concluded that our method could detect allelic imbalance.
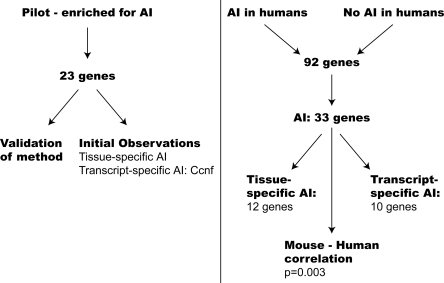
Figure 1.
Summary of genes tested and major findings.
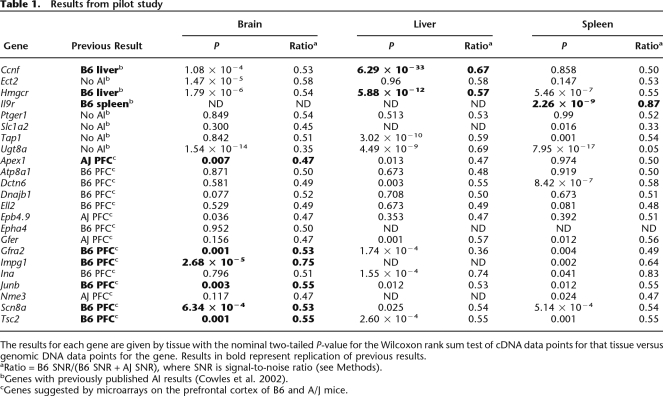
As part of our AI assay validation, we tested 23 genes that were likely enriched for the presence of AI. For these 23 genes, previous data existed to suggest the presence or absence of allelic imbalance (Supplemental Table 1). These included genes reported to have cis-acting variation, genes reported to not have cis-acting differences (Cowles et al. 2002), and genes suggested by microarray data on prefrontal cortex tissue from inbred B6 and A/J mice to be subject to cis-acting regulation (A. Kirby and M.J. Daly, unpubl.). We tested for allelic imbalance by comparing the distribution of allelic ratios between F1 genomic DNA replicates and F1 cDNA data points as described in the Methods. For all three positive controls (Ccnf, Il9r, and Hmgcr), our data agree with the published literature (Cowles et al. 2002; Table 1). For the genes suggested by microarrays, six of the genes have AI in brain in the predicted direction (Table 1).
Table 1.
Results from pilot study
The results for each gene are given by tissue with the nominal two-tailed P-value for the Wilcoxon rank sum test of cDNA data points for that tissue versus genomic DNA data points for the gene. Results in bold represent replication of previous results.
aRatio = B6 SNR/(B6 SNR + AJ SNR), where SNR is signal-to-noise ratio (see Methods).
bGenes with previously published AI results (Cowles et al. 2002).
cGenes suggested by microarrays on the prefrontal cortex of B6 and A/J mice.
Testing additional genes for allelic imbalance
In addition to the 23 genes described above, we tested 92 more genes for the presence of AI (Supplemental Table 1). These genes are a subset of 1983 genes whose orthologs were studied for AI in 12 human lymphoblast cell lines; 731 genes that showed AI in at least one cell line and 652 genes that showed no AI across all cell lines (Pant et al. 2006). We chose this subset of 92 genes because these genes had at least two transcribed SNPs with different alleles in A/J and C57Bl/6J mice. We assayed an average of two SNPs per gene in these 92 genes (45 with cis-acting variants in humans, 47 without).
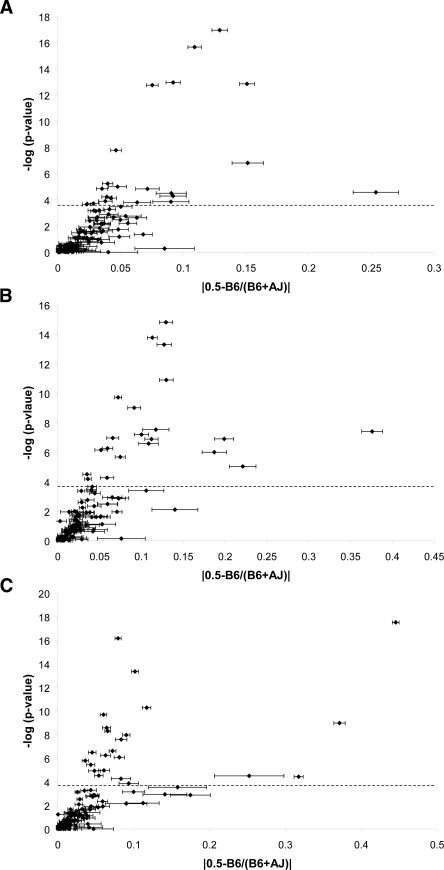
Ten of these 92 (11%) genes had significant evidence for AI in F1 mice (Table 2). Using standard statistical approaches (Benjamini and Hochberg 1995), we estimate the false discovery rate (FDR) for these 10 genes to be 0.05. We also estimated the rate at which we could replicate these results empirically. The results for nine of these genes were replicated (Bonferroni-corrected P < 0.05) or trended in the expected direction (nominal one-tailed P < 0.05) in mice from a reciprocal F1 cross (RF1 mice). We therefore estimate a nonreplication rate of 10% (one of 10 genes) for this analysis (see Methods). None of the genes tested showed evidence of genetic imprinting in the RF1 analysis (Supplemental Table 2). We also examined the relationship between statistical significance of AI and magnitude of AI. As expected, we observe correlation (R2 = 0.42–0.48) between these metrics for all three tissues tested (Fig. 2).
Table 2.
Genes with significant global AI
Results from Wilcoxon rank sum tests between cDNA data points and genomic DNA replicates are presented for each gene. Nominal P-values are given for F1 and RF1 mice. The threshold of significance in the RF1 data was P < 0.005, which is corrected for 10 tests. One-tailed P < 0.1 in the RF1 mice was considered a nonsignificant trend.
aRatio = B6 SNR/(B6 SNR + AJ SNR).
Figure 2.
Magnitude of allelic imbalance. The statistical significance from Wilcoxon rank sum tests (-log of the P-value) is plotted against the magnitude of AI (absolute value of the difference between 0.5 [no AI] and the average ratio B6/[AJ + B6]) for each of the 115 genes in each tissue. The dotted line indicates an empirical P = 0.05. Error bars are the standard error of the mean; genes with large standard errors have SNP differences in AI. (A) brain; (B) liver; (C) spleen.
Gender-specific allelic imbalance
We tested for AI in male and female mice separately. Specifically, we searched for genes where F1 mice of one gender showed significant AI but the other gender did not. None of the genes we examined met the criteria for gender-specific AI (Supplemental Table 3), suggesting that cis-acting variants with gender-specific effects are not common.
Tissue-specific allelic imbalance
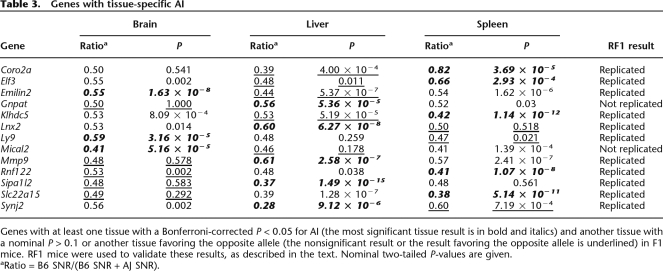
In the pilot study of 23 genes, we observed that several of the genes had tissue-specific AI (see Methods for criteria used to declare tissue specificity). For Ccnf, Ect2, Tap1, and Ina, evidence of AI was observed in one or two tissues, and no evidence of AI was observed in the remaining tissue(s). For Ugt8a and Gfra2, we observed evidence for AI favoring opposite alleles in different tissues. In spleen, there was significant overexpression of the A/J allele of Ugt8a, but in liver the B6 allele of this gene was overexpressed (Table 1). Motivated by the surprisingly high frequency of tissue-specific AI in the pilot gene set, we examined the set of 92 additional genes for tissue-specific AI. We found that 13 genes out of 92 (14%) had significant AI in at least one tissue and either no AI (10 genes) or AI favoring the opposite allele (seen in three genes: Emilin2, Klhdc5, and Synj2) in at least one of the remaining two tissues (Fig. 3; Table 2). We estimated the FDR for this analysis to be 0.05 (Benjamini and Hochberg 1995), and also empirically determined the rate of nonreplication in RF1 mice. We strongly replicated the tissue-specific AI results for 11 out of the 13 genes (Table 2), with an estimated nonreplication rate of 19% (see Methods).
Figure 3.
Example of tissue-specific allelic imbalance in Emilin2. Representative spectra of a transcribed SNP assayed in F1 genomic DNA and cDNA from three tissues. The peak locations for the A/J and B6 alleles are marked. For this gene, the B6 allele peak is higher than the A/J allele peak in brain and spleen, but the A/J allele peak is higher than the B6 peak in liver.
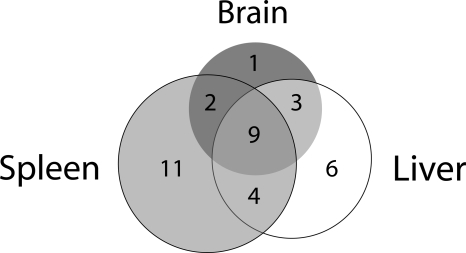
There was partial overlap in the genes that show AI in each of the three tissues (Fig. 4). In total, 15 out of 92 genes had evidence of AI in brain (16%), 22 out of 92 genes in liver (24%), and 26 out of 92 in spleen (28%). Each tissue had at least one gene where AI was specific to that tissue, with one, six, and 11 genes having AI specific to brain, liver, and spleen, respectively. For the remaining genes, the allelic imbalance results were generally consistent across at least two tissues (Fig. 4).
Figure 4.
Overlap of genes with allelic imbalance between tissues. For this analysis, only the 33 genes with any evidence of AI were considered. Three genes are represented twice in the diagram because two of the tissues had evidence for allelic imbalance favoring alleles from opposite strains.
Transcript-specific allelic imbalance
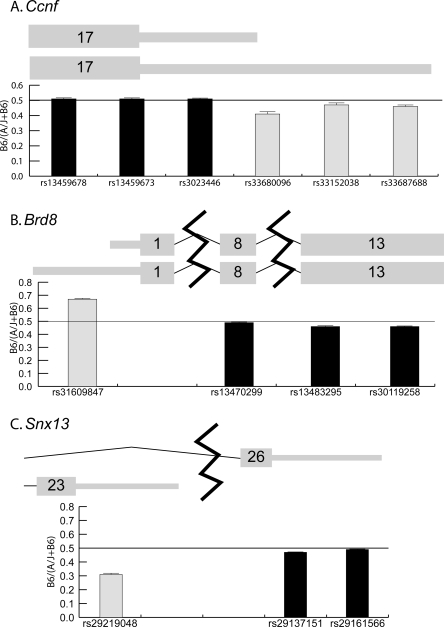
For the original set of 23 genes, we attempted to test as many reported SNPs in each gene as possible. For most genes, the expression results are consistent across the gene for all SNPs tested. However, for Ccnf, three of the eight SNPs assayed showed overrepresentation of the A/J allele in spleen, while the remainder showed no AI (Supplemental Table 5). These three SNPs were located near the end of the 3′ untranslated region (UTR) (Fig. 5). This result suggested that Ccnf transcripts of different length might be present and regulated differentially by a cis-acting variant, similar to the case of IRF5 in humans (Graham et al. 2006, 2007a, b). Data from the UCSC Genome Browser (genome. ucsc.edu) confirmed that this gene has multiple reported transcripts. In fact, the three SNPs in Ccnf with evidence of AI are located on the 3′ UTR of one transcript but not the other (Fig. 5). We confirmed these results using quantitative PCR in A/J and B6 animals (data not shown).
Figure 5.
Examples of transcript-specific allelic imbalance. Representations of the exon and UTR structures are plotted with the thicker bars symbolizing coding sequence and the thinner bars symbolizing UTR sequence. Exon numbers are given in the bars for each exon. These diagrams are based on information from the UCSC Genome Browser (genome.ucsc.edu). Below the representation of each gene, a graph of B6/(A/J + B6) is plotted for SNPs that fall within the exons shown. The horizontal line across each graph marks a 50:50 ratio of B6 to A/J alleles. (A) Ccnf has two transcripts that differ in 3′ UTR length; data shown are from spleen. (B) Brd8 has two transcripts that differ in 5′ UTR length; data shown are the average of all tissues. (C) Snx13 has two transcripts with different last exons and 3′ UTRs; data shown are the average of all tissues.
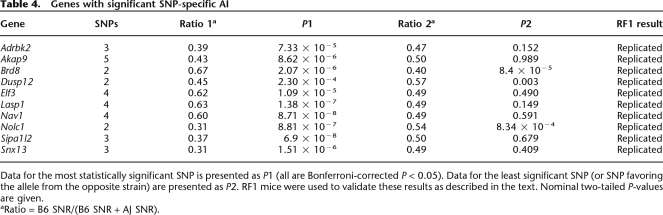
Based on the results in Ccnf, we tested for transcript-specific AI in the set 92 additional genes (see Methods). AI was determined for each SNP tested in a given gene (Supplemental Table 5). Ten genes out of 92 (11%) show significant differences in AI between SNPs (Table 4, see below). The estimated FDR for this analysis is 0.05 (Benjamini and Hochberg 1995). All of these differences were observed in the RF1 mice, and we estimated a nonreplication rate of 7% for these results (see Methods). Two genes had especially strong evidence of transcript-specific allelic imbalance. Specifically, Brd8 has two reported transcripts with different 5′ UTR lengths, and a SNP that lay only on the longer transcript showed a different pattern of AI from the remaining SNPs (Fig. 5; Supplemental Fig. 2). Snx13 has a shorter alternate transcript with a different last exon and 3′ UTR. A SNP in this alternate last exon showed a different pattern of AI from other SNPs in the gene (Fig. 5). We confirmed these transcript-specific observations by retesting the critical SNPs, by confirming these results in an analysis of mixed parental cDNA (Supplemental Table 5), and with qPCR on cDNA from the parental strains (data not shown).
Table 4.
Genes with significant SNP-specific AI
Data for the most statistically significant SNP is presented as P1 (all are Bonferroni-corrected P < 0.05). Data for the least significant SNP (or SNP favoring the allele from the opposite strain) are presented as P2. RF1 mice were used to validate these results as described in the text. Nominal two-tailed P-values are given.
aRatio = B6 SNR/(B6 SNR + AJ SNR).
Correlation between allelic imbalance in mice and humans
Because this set of 92 genes was drawn from genes that had been tested for AI in 12 human white blood cell samples (Pant et al. 2006), we were able to test whether the presence of AI in mice was correlated with the presence of AI in humans. Out of the 33 genes with strong evidence for AI in mice, 19 showed AI in humans and 14 did not, while of the 27 genes with no evidence of AI in mice, six showed AI in humans and 21 did not (one-tailed P = 0.003). These data suggest that genes that show AI in humans are more likely to have evidence of AI in mice.
Discussion
We have tested 115 genes for AI in C57Bl/6J × A/J F1 mice. Each of these genes harbors at least one transcribed SNP that differs between C57Bl/6J (B6) and A/J, which we used as a marker for transcripts from the chromosomes inherited from the two strains. We did not observe substantial gender-specific AI, but tissue-specific AI was prevalent. For some genes with several transcripts, certain transcripts had different levels of AI than others. Interestingly, genes with AI in humans were more likely to show AI in mice, and genes with no evidence of AI in humans were less likely to show AI in mice. These results constitute one of the largest surveys of cis-acting regulatory variation in mice to date.
We initially studied two groups of genes that were likely enriched for AI: genes from Cowles et al. (2002) and genes suggested by microarray analysis of prefrontal cortex tissue of A/J and B6 mice (A. Kirby and M. Daly, unpubl.). In this pilot study, we attempted to replicate some of these earlier results. We were able to replicate tissue-specific AI observed in three genes from Cowles et al. (2002) (Table 1). However, for many of the genes reported to not have cis-acting differences in Cowles et al. (2002) we observed strong AI. The reason for this discrepancy is unclear, although it might be explained by technical differences between the two assays or that we tested multiple SNPs in most genes whereas the original study only tested a single SNP per gene. It is unlikely that the AI we observed in these genes are false positive results, since we reproduced the same result in RF1 mice (Supplemental Tables 2, 4). We observed significant AI in the direction predicted from microarray experiments for six out of 15 genes. We did not observe AI in all of these 15 genes, which may be explained by trans-acting factors in the microarray data, or that the microarrays were performed on prefrontal cortex tissue whereas we tested whole brain.
The 115 genes that we examined for the presence of allelic imbalance were not randomly selected. Twenty-three of these genes were highly enriched for genes with AI, and these genes were not included in our estimates of the prevalence of AI or tissue-specific AI. For about half of the other 92 genes in our study, there was prior evidence of AI in human white blood cells, and we acknowledge that inclusion of these genes could introduce a small bias in our estimates of the extent of AI in mice, given the correlation that we discovered between genes showing AI in mice and human. However, we have no reason to believe that this set of genes would be biased in estimating the prevalence of gender- or tissue-specific AI within those genes that show AI.
In the 92 genes, we do not observe any evidence for strong gender-specific AI. Gender-specific strain expression differences have been observed with microarrays (Yang et al. 2006), and we observe gender-specific expression differences between strains in seven genes using mixed A/J and B6 cDNA (Supplemental Table 3). These data suggest that gender-specific trans-acting variants and environmental influences are more common than cis-acting variants with effects in only one gender.
This study is the largest study to date of AI across different tissues. Data from smaller studies of AI in mice and humans have been inconclusive on the extent of tissue-specific AI (Cowles et al. 2002; Lo et al. 2003), although genes with tissue-specific expression patterns have been mapped to expression quantitative trait loci (eQTL) in mice (Yang et al. 2006). Interestingly, three genes in our study (Synj2, Emilin2, and Klhdc5) have highly significant AI favoring alleles from opposite strains in different tissues (Fig. 3; Table 3). One possible explanation for these data is the presence of multiple cis-acting variants including one or more variant(s) in tissue-specific regulatory elements that influence expression in only one tissue.
Table 3.
Genes with tissue-specific AI
Genes with at least one tissue with a Bonferroni-corrected P < 0.05 for AI (the most significant tissue result is in bold and italics) and another tissue with a nominal P > 0.1 or another tissue favoring the opposite allele (the nonsignificant result or the result favoring the opposite allele is underlined) in F1 mice. RF1 mice were used to validate these results, as described in the text. Nominal two-tailed P-values are given.
aRatio = B6 SNR/(B6 SNR + AJ SNR).
Interestingly, several genes in our study show evidence of cis-acting variation that only affects one transcript. Recently, a similar result was described for IRF5 in humans. In this gene, a SNP in the 3′ UTR creates a new polyadenylation signal, leading to a more stable shorter transcript (Graham et al. 2006, 2007a, b). We have studied three mouse genes in detail that show differences in AI between SNPs along their transcripts (Fig. 5), and these genes all have multiple reported transcripts (genome. ucsc.edu). In these three genes, we observe different patterns of transcript-specific AI (Fig. 5). More generally, we observed significant, reproducible differences in AI between SNPs in the same gene for 10 out of 60 genes for which we assayed more than one SNP. These data suggest that this phenomenon is relatively common in the mouse genome. We cannot suggest a mechanism for any of these genes, but differences in transcript stability due to different polyadenylation site usage and differences in microRNA binding efficiency are among the possible explanations; SNPs could also affect the efficiency of transcript export from the nucleus, the binding of transcript-specific regulators, or the overall rates of transcription of each transcript. Our results emphasize the potential importance of measuring the level of each transcript for a given gene when trying to understand the impact of cis-acting variants.
This is the first study to compare the presence of allelic imbalance across species. There are several caveats to this experiment: first, the tissues tested in mouse and human were different; second, the number of tested genes was small, so the power to detect an association is low; and third, the number of human samples was small with limited power to detect AI. Despite these caveats, we did observe a nominally significant association between genes with AI in both species (P = 0.003). Purifying selection has recently been described for cis-eQTL in yeast (Ronald and Akey 2007), which lends support to our findings that certain genes may be more tolerant of regulatory variation.
In our survey of allelic imbalance in mice, we observed many genes with tissue-specific AI, and our data suggest that the choice of tissue for studies of the genetics of gene expression should be considered carefully. We particularly recommend the creation of a human tissue bank, so that studies of the genetics of gene expression could be performed across a wide range of tissue types. We have also observed that different transcripts in the same gene can have differing AI, suggesting that expression should ideally be assessed at the transcript level. Determining the causal variants for the AI phenotypes observed would lead to new insights into the genetic control of gene expression.
Methods
Mice
Mice were bred and reared and organs were extracted by the Jackson Laboratory. Brains, livers, and spleens were obtained for three male and three female C57Bl/6J × A/J F1 mice, three male and three female A/J × C57Bl/6J F1 mice, three male and three female C57Bl/6J (B6) mice, and three male and three female A/J mice. Organs were flash frozen following extraction. All mice were 4–6 wk of age and were fed a standard chow diet.
DNA and RNA isolation
Genomic DNA was isolated from the F1, C57Bl/6J, and A/J mice using the DNeasy kit (Qiagen). To isolate RNA, frozen tissue was homogenized in Qiazol (Qiagen) using a mechanical rotor/stator homogenizer. Total RNA was purified using the RNeasy kit (Qiagen). mRNA was isolated from total RNA using the Oligotex kit (Qiagen). mRNA samples were treated with 10 units of DNaseI at 37°C for 30 min. Then, 2.5 mM EDTA was added to stabilize the RNA and the sample was incubated at 75°C for 10 min to denature the DNaseI.
cDNA synthesis
To synthesize cDNA, 100 ng of mRNA, 150 ng random hexamers, and 1 mM dNTPs were incubated at 65°C for 5 min and 4°C for 1 min. Next, 1× RT buffer, 5 mM MgCl2, 10 mM DTT, 120 units of RNaseOut, and 600 units of Superscript III (Invitrogen) were added to make a 60 μL reaction volume and the sample was incubated at 25°C for 10 min, 50°C for 50 min, and 85°C for 5 min. Controls lacking reverse transcriptase were included to indicate the presence of genomic DNA contamination. Synthesized cDNA samples were treated with 5 units of RNaseH at 37°C for 20 min.
Allelic Imbalance genotyping
All genotyping was performed using the mass spectrometry-based MassArray platform (Sequenom) (Gabriel et al. 2002; Altshuler et al. 2005). Primers and probes were designed using SpectroDesigner (Sequenom). Assays were multiplexed up to a maximum of five using the homologous Mass Extension (hME) protocol (Gabriel et al. 2002; Campbell et al. 2007). The iPLEX protocol was used to multiplex assays up to a maximum of 25 as follows. PCR was performed in 6 μL with 10 ng of genomic or cDNA, 0.6 pmol of each primer, 0.5 mM dNTPs, and 0.1 units of Taq DNA polymerase (Qiagen) in 1.25× PCR buffer (Qiagen) and 1.625 mM MgCl2. PCR conditions were 94°C for 15 min; 45 cycles of 94°C for 20 sec, 56°C for 20 sec, 72°C for 1 min; 72°C for 3 min. Extra dNTPs were inactivated using 0.3 U of shrimp alkaline phosphatase at 37°C for 20 min, and the enzyme was denatured at 85°C for 5 min. Single base extension was performed with 7.15–12.51 pmol of probe based on probe mass for each assay, 0.2 mM termination mix, 0.22× extension buffer, and 0.041 μL extension enzyme (Sequenom). Single base extension conditions were 94°C for 30 sec; 40 cycles of 94°C for 5 sec, 5 cycles of 52°C for 5 sec and 80°C for 5 sec; 72°C for 3 min. The iPLEX and hME assays yielded similar AI results when tested on the same set of genes.
Mouse homologs of human genes in Pant et al. (2006)
Using the human gene symbols provided in Pant et al. (2006), we performed database searches to identify the mouse homologs. We used the homolog table from Mouse Genome Informatics (MGI; www.informatics.jax.org). For human genes that were not listed in this table, we used Gene and Homologene from the National Center for Biotechnology Information (NCBI; www. ncbi.nlm.nih.gov) and the Ensembl database (www.ensembl.org) to determine the mouse homologs (Supplemental Table 1).
SNPs
SNPs were obtained from Ensembl, which has curated all mouse SNPs in dbSNP as well as SNPs discovered by the Sanger Institute. SNPs were also obtained from a database at the Broad Institute maintained by the mouse HapMap project. To design the AI genotyping assays, sequences for transcribed SNPs were trimmed so that both PCR primers would be in the same exon as the SNP. These design constraints allow the same assay to be run on genomic DNA and cDNA. SNP sites were avoided in the design of PCR primers and single base extension probes.
Data normalization and quality control
Each AI assay was run on three cDNA replicates for each F1 mouse and each organ. Six replicates of F1 genomic DNA were also run for each assay. Signal-to-noise (SNR) estimation was done automatically in the software provided by Sequenom. We chose SNR (as opposed to other measures, such as peak heights) because in pilot experiments this was the most reliable indicator of the relative proportion of alleles. For each SNP, the two alleles are present in equal amounts in the genomic DNA sample, and the SNRs for the two alleles should also be equal. Therefore, we used the genomic DNA replicates to calculate a correction factor to account for any technical biases or unknown copy number variants leading to unequal SNRs between the C57Bl/6J and A/J alleles. Specifically, the difference in SNR between the C57Bl/6J allele and A/J allele in the genomic DNA data was divided by the A/J allele SNR, and the mean of this correction factor was calculated across the six DNA replicates. The A/J SNRs were multiplied by the correction factor, and this value was added to the original A/J SNR value to obtain a normalized A/J SNR. The final AI ratio was calculated as the B6 SNR divided by the sum of the B6 and normalized A/J SNRs. Equal representation of the two alleles yielded a ratio of 0.5. Values between 0.5 and 1 indicate that the B6 allele is overrepresented and values between 0 and 0.5 indicate that the A/J allele is overrepresented.
Assays were required to pass several quality control (QC) measurements in order to be analyzed further. At least four of the six genomic DNA replicates had to be correctly called heterozygous by the Sequenom software indicating a high accuracy of genotyping, and the standard deviation of the AI ratio for the working genomic DNA replicates had to be <0.06. For each gender and tissue group (e.g., livers of male mice or spleens of female mice), there were a maximum of nine data points (three mice times three replicates). The group was only analyzed for AI if at least five data points had data (raw B6 SNR + raw A/J SNR >15) and the standard deviation of the replicates was <0.1. Mouse-to-mouse variability was tested for each group, and groups with significant mouse-to-mouse variability (P < 0.005) were excluded from further analyses. Each group was considered for QC individually, so that genes that were not expressed in one group would not lead to the failure of AI assays in the other groups.
It is unlikely that our AI results are due to random single-copy gene inactivation. We only analyzed data that were consistent across three F1 mice. In addition, for all genes with AI, we required that this result was reproduced in three RF1 mice from the reciprocal cross. Also, because we did not observe any reproducible gender-specific differences in AI, all AI results are consistent across six F1 mice (three male and three female) and six RF1 mice.
Statistical analysis
All statistical tests were coded and performed in R (www. r-project.org). For all tests, genomic DNA replicate data points were compared to cDNA data points using Wilcoxon rank sum tests. We observed that most of the variability in our assay was from experimental replicates and not mouse-to-mouse variability for data from the same tissue, gender, and SNP. Therefore, we used the experimental replicates as the cDNA data points. Genes were first evaluated for overall AI by performing Wilcoxon rank sum tests between all cDNA data points for the gene and all genomic DNA data points. A Bonferroni correction was applied to this data set for 92 tests. Genes were considered to show AI if the corrected P < 0.05.
There were three classes of analysis with multiple groups in each: gender analysis with male and female groups; tissue analysis with brain, liver, and spleen groups; and SNP analysis with groups for each transcribed SNP tested in a gene. The data in each class for a gene were evaluated for normality, and more deviation from normality was observed than expected by chance. Therefore, we have chosen to use nonparametric Wilcoxon rank sum tests in the analysis. We have performed the analysis using t-tests as well and observed similar results suggesting there is not a substantial loss of power when using the Wilcoxon rank sum tests (data not shown). In each class, two-tailed Wilcoxon rank sum tests were performed between the data points of each group and the genomic DNA data points to evaluate the presence of AI in that group. For example, to test for AI in the males for a gene, Wilcoxon tests between all cDNA data points from male mice for that gene and all genomic DNA data points for that gene were performed. In each class, the P-values obtained from the Wilcoxon tests were corrected for the number of tests performed using the Bonferroni method. For gender, the number of tests was two (one for each gender) times the number of genes, or 184 tests. For tissue, the number of tests was three (one for each organ) times the number of genes, or 276 tests. For SNPs, the number of tests was the number of SNPs in genes containing more than one SNP, which was equal to 169 tests. A gene was considered to have gender, organ, or SNP specific differences if one of the groups showed AI (corrected P < 0.05) and another group did not (nominal P > 0.1) or two of the groups showed overexpression of opposite alleles. We used these criteria to avoid declaring differences in AI for genes where all groups in a class (e.g., all tissues) show a trend favoring the same allele, since results like these are likely driven by statistical fluctuation and not true biological differences.
An additional minimum threshold for the magnitude of AI was applied to the genes with statistically significant results as defined by the corrected P-values above. Genes were considered to show AI if the ratio of SNR for the B6 allele to the sum of SNRs for both alleles was ≤0.44 or ≥0.55. These values were determined based on an empirical null distribution as follows. For each working assay with six genomic DNA data points (226 SNPs), we assigned three data points as genomic DNA control and the remaining three genomic DNA data points as “experimental” values. We then calculated the mean ratio for the “experimental” data points in each assay and examined the distribution of means across all the assays; 95% of the assays had means between 0.44 and 0.55, so we used these values as our thresholds for the magnitude of AI.
To validate the AI results and to test whether any of these results were due to genetic imprinting, we tested the same genes in the brain, liver, and spleen of reciprocal A/J × C57Bl/6J F1 mice (RF1). These mice are genetically identical to the original F1 mice with the strains of the mother and father reversed. For global tests of AI, we compared all cDNA and genomic DNA values as was done in the F1 mice, To validate potential results from F1 mice that showed specificity in AI for gender, tissue, or SNPs, we created a list of the two most divergent groups for each potential instance of AI, and performed Wilcoxon rank sum tests between these same two groups in the RF1 data set. The P-values were Bonferroni corrected for the number of potential positive results in each class: 13 for tissue-specific AI, 10 for SNP-specific AI, and 10 for overall AI. RF1 data were considered to have replicated the F1 result if the Bonferroni-corrected P-value was <0.05 in RF1 mice and the allele overrepresented in the RF1 data was the same as in the F1 mice.
To determine empirically a nonreplication rate, we not only calculated the rate of negative findings in the RF1 mice but also estimated the number of apparent replications that we expected to see by chance in the RF1 mice. To estimate this latter quantity for genes with global AI, the 10 genes with the most nonsignificant P-values in the F1 data set for the Wilcoxon rank sum test between all cDNA data points and genomic DNA data points were analyzed in the RF1 mice, to mimic our analysis of the 10 genes with putative global AI in the F1 mice. None of these 10 genes was a false positive in the RF1 data set (Bonferroni-corrected P-value < 0.05). Because we observed that one out of 10 genes with significant global AI in F1 mice did not replicate in RF1 mice, we estimated an empirical rate of nonreplication of 10%.
To determine a rate of experimental nonreplication for tissue and SNP-specific AI, we performed a similar analysis, this time using genes that had no evidence for tissue or SNP-specific effects in the F1 mice. Genes were considered negative for tissue- or SNP-specific effects in F1 mice if the nominal P-values for AI were >0.1 for all tissues (or all SNPs), suggesting no AI in the gene, or if all tissues (or SNPs) had a Bonferroni-corrected P-value < 0.05 favoring the same allele, suggesting consistent AI. Wilcoxon rank sum tests were performed for these genes in the RF1 data between the two most similar groups in the F1 data set, in other words, those showing the least evidence of specificity, and P-values were Bonferroni corrected. Genes with corrected P-values < 0.05 and magnitude of AI >0.55 or <0.44 in the RF1 data set were considered false-positive results. For tissue-specific AI, we tested 24 genes that had no evidence of tissue-specific AI in F1 mice, and one (4.2%) of these genes showed apparent evidence of tissue-specific AI in RF1 mice. From this result, we considered that 4.2% (0.42 genes) of the 11 genes with reproduced tissue-specific AI (Table 4) could represent false positives, despite the apparent replication. Including the two genes with apparent evidence of tissue-specific AI in F1 but no replication in the RF1 mice, we estimate that the rate of nonreplication is 2.42 genes (0.42 + 2) out of 13, or 19%. For SNP-specific AI, there were 14 genes with no evidence of SNP differences in AI in the F1 mice. Of these 14 genes, one (7.1%) had significant apparent SNP-specific differences in RF1 mice despite no evidence in the F1 mice. All 10 genes that were positive of SNP differences in AI were replicated in RF1 mice, so we estimate the rate of nonreplication for SNP-specific AI differences in F1 mice as 7.1%.
To compare genes between mice and humans, we considered any genes with reproduced tissue- or SNP-specific AI as well as genes with AI across all data points as showing AI in mice. Genes were considered to not show AI in mice if the nominal P > 0.05 for AI in both F1 and RF1 mice. Thirty-two genes met neither criteria and were not included in the analysis. Results were similar using a χ2 test with one degree of freedom or a Fisher’s exact test.
Mixed cDNA studies
To assess gene expression in the parental mouse strains using Sequenom AI assays, we assayed an equal mix of C57Bl/6J and A/J cDNA for each gender and organ. cDNA from three B6 and three A/J mice was mixed in equal proportion to approximate a F1 mouse. The mixed cDNA for each gender and tissue was tested in the same AI assays described above with six replicates. To account for small differences in the amounts of B6 and A/J cDNA, all data points for the mixed cDNA from each group were averaged. Operating under the assumption that the mean should equal 0.5, we calculated the difference between 0.5 and the actual mean. Then, we added this value to all the data points in the group so the mean would be equal to 0.5. Analysis of the mixed cDNA sample then proceeded as described above.
Quantitative PCR
Quantitative PCR was performed using SYBR green (Applied Biosystems). Three to four replicates of cDNA from each mouse were tested, and three mice each of B6 and A/J were tested. In a 15 μL reaction volume, 7.5 μL of SYBR green master mix, 10 pmol of each PCR primer, and 5 ng of cDNA were added. Cycling conditions were 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. PCR primers were the same as used in the Sequenom AI assays.
Acknowledgments
We thank members of the Hirschhorn laboratory for advice on this project, especially Guillaume Lettre for helpful discussions. We thank Tracey Petryshen for the prefrontal cortex gene expression data. All mouse work and organ extraction were done at the Jackson Laboratories in Bar Harbor, Maine. This work has been supported by the Smith Family Pinnacle Program Project grant from the American Diabetes Association.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.068692.107.
References
- Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Lindgren C.M., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Vohl M.C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Nemesh J., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Lane C.R., Schaffner S.F., Bolk S., Brewer C., Schaffner S.F., Bolk S., Brewer C., Bolk S., Brewer C., Brewer C., et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Chakravarti A., Collins F.S., Daly M.J., Donnelly P., Collins F.S., Daly M.J., Donnelly P., Daly M.J., Donnelly P., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Brem R.B., Yvert G., Clinton R., Kruglyak L., Yvert G., Clinton R., Kruglyak L., Clinton R., Kruglyak L., Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Campbell C.D., Lyon H.N., Nemesh J., Drake J.A., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Lyon H.N., Nemesh J., Drake J.A., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Nemesh J., Drake J.A., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Drake J.A., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., Cooper R.S., Ardlie K.G., Groop L.C., Ardlie K.G., Groop L.C., Groop L.C., et al. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: A possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes. 2007;56:1460–1467. doi: 10.2337/db06-1051. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Spielman R.S. The genetics of variation in gene expression. Nat. Genet. 2002;(Suppl.) 32:522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Jen K.Y., Morley M., Spielman R.S., Morley M., Spielman R.S., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Hirschhorn J.N., Altshuler D., Lander E.S., Hirschhorn J.N., Altshuler D., Lander E.S., Altshuler D., Lander E.S., Lander E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Abraham C., Regueiro M., Griffiths A., Regueiro M., Griffiths A., Griffiths A., et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez J.C., Burtt N., de Bakker P.I., Almgren P., Tuomi T., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Burtt N., de Bakker P.I., Almgren P., Tuomi T., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., de Bakker P.I., Almgren P., Tuomi T., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Almgren P., Tuomi T., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Tuomi T., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Holmkvist J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Gaudet D., Hudson T.J., Schaffner S.F., Daly M.J., Hudson T.J., Schaffner S.F., Daly M.J., Schaffner S.F., Daly M.J., Daly M.J., et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53:1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Higgins J., DeFelice M., Lochner A., Faggart M., DeFelice M., Lochner A., Faggart M., Lochner A., Faggart M., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Graham R.R., Kozyrev S.V., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Kozyrev S.V., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., Gonzalez Escribano M.F., Pons-Estel B., Pons-Estel B., et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- Graham R.R., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., Koeuth T., Ortmann W.A., Hom G., Ortmann W.A., Hom G., Hom G., et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. 2007a;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D.S., Manku H., Wagner S., Reid J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J., Manku H., Wagner S., Reid J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J., Wagner S., Reid J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J., Reid J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J., Gutin A., Lanchbury J.S., Vyse T.J., Lanchbury J.S., Vyse T.J., Vyse T.J. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum. Mol. Genet. 2007b;16:579–591. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Stefansson H., Emilsson V., Helgadottir A., Emilsson V., Helgadottir A., Helgadottir A., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Hikichi T., Kohda T., Kaneko-Ishino T., Ishino F., Kohda T., Kaneko-Ishino T., Ishino F., Kaneko-Ishino T., Ishino F., Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31:1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., SanGiovanni J.P., Mane S.M., Mayne S.T., Mane S.M., Mayne S.T., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J.C. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Yang H.H., Gere S., Buetow K.H., Lee M.P., Gere S., Buetow K.H., Lee M.P., Buetow K.H., Lee M.P., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Tybjaerg-Hansen A., Folsom A.R., Folsom A.R., et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks S.A., Leonardson A., Zhu H., Cundiff P., Pietrusiak P., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Leonardson A., Zhu H., Cundiff P., Pietrusiak P., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Zhu H., Cundiff P., Pietrusiak P., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Cundiff P., Pietrusiak P., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Pietrusiak P., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Edwards S., Phillips J.W., Sachs A., Schadt E.E., Phillips J.W., Sachs A., Schadt E.E., Sachs A., Schadt E.E., Schadt E.E. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G., Ewens K.G., Spielman R.S., Cheung V.G., Spielman R.S., Cheung V.G., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant P.V., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Ballinger D.G., Cox D.R., Frazer K.A., Cox D.R., Frazer K.A., Frazer K.A. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., Villeneuve A., Gaudin T., Brandstrom H., Gaudin T., Brandstrom H., Brandstrom H., et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- Ronald J., Akey J.M., Akey J.M. The evolution of gene expression QTL in Saccharomyces cerevisiae. PLoS One. 2007;2:e678. doi: 10.1371/journal.pone.0000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Hirschhorn J.N., Daly M.J., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., Milligan S.B., Lamb J.R., Cavet G., Lamb J.R., Cavet G., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Boutin P., Vincent D., Belisle A., Hadjadj S., Vincent D., Belisle A., Hadjadj S., Belisle A., Hadjadj S., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Ueda H., Howson J.M., Esposito L., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Howson J.M., Esposito L., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Esposito L., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Heward J., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Snook H., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Chamberlain G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Rainbow D.B., Hunter K.M., Smith A.N., Di Genova G., Hunter K.M., Smith A.N., Di Genova G., Smith A.N., Di Genova G., Di Genova G., et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Vogelstein B., Kinzler K.W., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Ingram-Drake L., Drake T.A., Lusis A.J., Drake T.A., Lusis A.J., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G., Brem R.B., Whittle J., Akey J.M., Foss E., Smith E.N., Mackelprang R., Kruglyak L., Brem R.B., Whittle J., Akey J.M., Foss E., Smith E.N., Mackelprang R., Kruglyak L., Whittle J., Akey J.M., Foss E., Smith E.N., Mackelprang R., Kruglyak L., Akey J.M., Foss E., Smith E.N., Mackelprang R., Kruglyak L., Foss E., Smith E.N., Mackelprang R., Kruglyak L., Smith E.N., Mackelprang R., Kruglyak L., Mackelprang R., Kruglyak L., Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]