Abstract
Studies in invertebrate model organisms have been a driving force in aging research, leading to the identification of many genes that influence life span. Few of these genes have been examined in the context of mammalian aging, however, and it remains an open question as to whether and to what extent the pathways that modulate longevity are conserved across different eukaryotic species. Using a comparative functional genomics approach, we have performed the first quantitative analysis of the degree to which longevity genes are conserved between two highly divergent eukaryotic species, the yeast Saccharomyces cerevisiae and the nematode Caenorhabditis elegans. Here, we report the replicative life span phenotypes for single-gene deletions of the yeast orthologs of worm aging genes. We find that 15% of these yeast deletions are long-lived. In contrast, only 3.4% of a random set of deletion mutants are long-lived—a statistically significant difference. These data suggest that genes that modulate aging have been conserved not only in sequence, but also in function, over a billion years of evolution. Among the longevity determining ortholog pairs, we note a substantial enrichment for genes involved in an evolutionarily conserved pathway linking nutrient sensing and protein translation. In addition, we have identified several conserved aging genes that may represent novel longevity pathways. Together, these findings indicate that the genetic component of life span determination is significantly conserved between divergent eukaryotic species, and suggest pathways that are likely to play a similar role in mammalian aging.
Aging is characterized by the progressive loss of function and fecundity over the lifetime of an organism. Although the rate can vary widely, nearly all metazoans do undergo an aging process whose general features include genomic instability, senescence, and sterility, as well as an age-dependent increase in mortality rate. Multiple models for why organisms age have been put forth by evolutionary biologists over the last century. Medawar proposed the mutation accumulation (MA) theory, suggesting that alleles with deleterious effects late in life would not be selected against and would, therefore, accumulate over many generations (Medawar 1946). Williams expanded on this idea, in his pleiotropy theory (commonly referred to as antagonistic pleiotropy [AP]), proposing that some alleles that increase mortality late in life might actually be favored by natural selection due to their beneficial effects early in life (Williams 1957). Kirkwood has further developed this notion of age-related trade-offs with the “disposable soma” theory, suggesting that in favorable environmental conditions organisms allocate the majority of their resources to reproduction and metabolism, rather than repair and maintenance, thus allowing damage to accumulate with time (Kirkwood 1977). Although several studies over the past decade have tried to distinguish between these models, it is still unclear which theory best describes the evolution of aging (Partridge and Gems 2006; Promislow et al. 2006; Martin 2007).
One feature in common among these evolutionary theories is that aging is not likely to be determined by a genetic program, because there is a minimal selective advantage associated with long-term post-reproductive survival of the individual. Life span in divergent eukaryotic species with distinct habitats is likely to be influenced by different factors. In the natural environment, survival of many organisms is heavily dependent on extrinsic factors such as predation rather than age-associated mortality. Thus, we might expect that most mechanisms associated with aging should be “private” (Martin et al. 1996)—that is, unique to each species. However, many studies have suggested that at least some mechanisms associated with aging, such as insulin/IGF1 signaling (Holzenberger et al. 2004) appear to be “public,” with similar patterns seen across diverse species. If aging is not programmed, why should organisms separated by hundreds of millions of years of evolution age by similar mechanisms? And perhaps, more importantly, to what extent are the various mechanisms of aging truly conserved over evolutionary time?
Empirical data demonstrate that aging is genetically modulated. Single gene mutations that significantly decrease mortality have been identified in each of the major model organisms used for gerontological research. Indeed, several hundred genes have been reported that influence longevity in yeast, worms, or flies (Guarente and Kenyon 2000; Warner 2003; Kaeberlein 2006). However, the genetic pathways that modulate aging in one organism may differ substantially from those in another. A handful of orthologous genes have been described as life span determinants in multiple organisms, supporting the idea that there are “public” mechanisms of aging. These include the Sir2-family proteins, insulin/IGF-like receptor proteins, and the target of rapamycin (TOR) kinase family (Kaeberlein 2006). No studies have quantified the degree to which genetic modifiers of aging are evolutionarily conserved. Using two divergent eukaryotic species, the unicellular budding yeast Saccharomyces cerevisiae and the multicellular nematode Caenorhabditis elegans, evolutionarily separated by ∼1.5 billion years (Wang et al. 1999), here, we quantitatively address this question by asking whether yeast orthologs of known worm aging genes are more likely to modulate yeast aging than randomly selected yeast genes.
Results
Based on a survey of the aging literature, we have compiled a set of 276 C. elegans genes for which increased life span results from decreased gene expression or altered protein function (Supplemental Table S1) (see SAGE KE database at http://uwaging.org/genesdb/index.php) (Dillin et al. 2002; Lee et al. 2003; Boehm and Slack 2005; Hamilton et al. 2005; Hansen et al. 2005, 2007; Ventura et al. 2005; Winkelbauer et al. 2005; Henderson et al. 2006; Schafer et al. 2006; Curran and Ruvkun 2007; Pan et al. 2007; Syntichaki et al. 2007a). Using the basic local alignment search tool for proteins, or BLASTp, we searched the yeast proteome for proteins with high sequence similarity to each of the proteins encoded by the 276 known worm aging genes. A set of putative yeast orthologs was defined based on a stringent reciprocal best hit (RBH) criterion (described in Methods). Due to an ancient duplication event in the yeast genome (Wolfe and Shields 1997), we allowed for 1:2 worm:yeast ortholog sets by including both the best hit and the next best hit, when BLASTing worm against yeast, if the BLASTp scores were within 10% of each other. From this analysis, we identified 103 yeast orthologs that were associated with 78 of the 276 worm aging genes (Table 1; Supplemental Table S2). The 103 yeast genes that code for these proteins are defined as the RBH set.
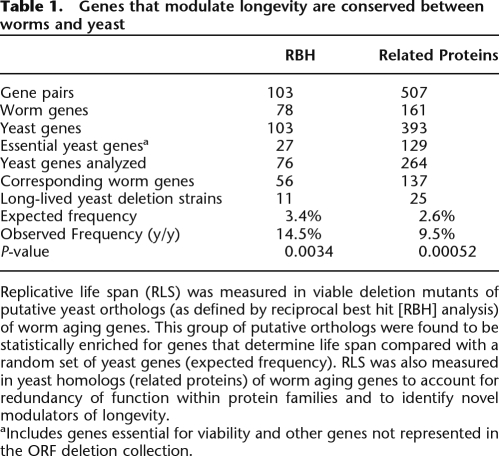
Table 1.
Genes that modulate longevity are conserved between worms and yeast
Replicative life span (RLS) was measured in viable deletion mutants of putative yeast orthologs (as defined by reciprocal best hit [RBH] analysis) of worm aging genes. This group of putative orthologs were found to be statistically enriched for genes that determine life span compared with a random set of yeast genes (expected frequency). RLS was also measured in yeast homologs (related proteins) of worm aging genes to account for redundancy of function within protein families and to identify novel modulators of longevity.
aIncludes genes essential for viability and other genes not represented in the ORF deletion collection.
To account for possible redundancy within protein families, a second set of yeast homologs to the worm aging genes was identified based on less-stringent homology criteria (described in Methods). This set of related proteins (RPs) is likely to contain nearly all of the true ortholog pairs, but also includes several proteins related by sequence similarity, but with different biological functions. A total of 393 yeast genes (including the 103 RBH genes) are contained in the RP set (Table 1; Supplemental Table S2).
We determined the replicative life span (RLS) for single-gene deletion mutants corresponding to genes contained in the RP set (Supplemental Table S3). Of the 393 genes in the RP set, 264 are viable as deletion alleles and are present in the yeast ORF deletion collection (Winzeler et al. 1999). RLS was measured for each of these 264 single-gene deletion mutants from the MATα (isogenic to BY4742) deletion collection. Deletion strains that tested as significantly long-lived in the initial analysis were verified by independently measuring the RLS of the corresponding deletion mutant from the MATa (isogenic to BY4741) deletion collection. This approach of initial testing in one mating type, followed by verification in the opposite mating type, is the same method used in the genome-wide screen for aging-related genes in yeast (Kaeberlein et al. 2005).
Based on the previously reported partial genome-wide analysis of RLS in yeast (Kaeberlein et al. 2005), we estimate that the random chance of a yeast gene deletion mutant being long-lived is 2.3% (13 long-lived strains from a pool of 564 randomly chosen deletion mutants). To account for the possibility that evolutionarily ancient and conserved genes are more likely to be associated with aging, we determined the frequency of aging-related genes in a subset of yeast genes with conserved worm orthologs. Based on RBH criteria (see Methods), we determined that 146 yeast genes in the random pool of 564 have putative worm orthologs. In this subset of conserved genes, 3.4% (5/146) of the deletion mutants were long-lived. Thus, the expected frequency of aging-related genes in a random set of yeast genes with worm orthologs is 3.4%.
Of the 76 genes examined in the RBH set, 11 deletion mutants were found to be significantly long-lived (Table 2; Fig. 1; Supplemental Fig. S1). After controlling for the potential effects of evolutionary conservation, we see a 4.3-fold enrichment for aging-related genes in yeast genes orthologous to worm genes associated with aging (G-test: χ12 = 8.57, P = 0.0034). The 76 yeast genes in the RBH set correspond to 56 worm genes. Twenty of the 56 worm aging genes had two yeast orthologs. By chance alone, these 20 genes are approximately twice as likely to have at least one yeast ortholog associated with aging. Even after controlling for the potential confounding effects of duplicate orthologs (see Methods), we still find that our set of yeast orthologs was enriched for genes associated with aging (G-test: χ12 = 5.37, P = 0.021). Therefore, we propose that yeast orthologs of worm aging genes are more likely to regulate yeast aging than yeast orthologs of randomly chosen worm genes, and we infer from these findings that the pathways modulating longevity are partially conserved between these two divergent species.
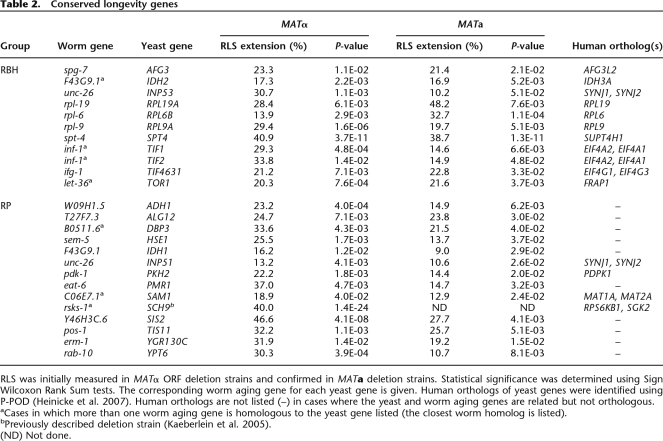
Table 2.
Conserved longevity genes
RLS was initially measured in MATα ORF deletion strains and confirmed in MATa deletion strains. Statistical significance was determined using Sign Wilcoxon Rank Sum tests. The corresponding worm aging gene for each yeast gene is given. Human orthologs of yeast genes were identified using P-POD (Heinicke et al. 2007). Human orthologs are not listed (–) in cases where the yeast and worm aging genes are related but not orthologous.
aCases in which more than one worm aging gene is homologous to the yeast gene listed (the closest worm homolog is listed).
bPreviously described deletion strain (Kaeberlein et al. 2005).
(ND) Not done.
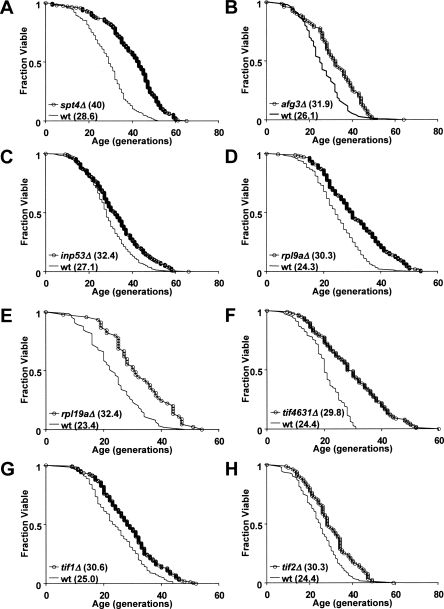
Figure 1.
Novel modulators of yeast replicative life span (RLS). Replicative life span is significantly extended by deletion of 11 of 76 yeast ORFs in the RBH set, including the transcription elongation factor SPT4 (A), ATP-dependent metalloprotease AFG3 (B), inositol polyphosphate 5-phosphatase INP53 (C), ribosomal proteins RPL9A (D), and RPL19A (E), and translation initiation factors TIF4631 (F), TIF1 (G), and TIF2 (H). Deletion alleles of TOR1, RPL6B, and IDH2 have been previously shown to increase RLS (Kaeberlein et al. 2005). Mortality curves were generated from life span data in both the MATα and MATa mating types; the mean life span for each deletion strain and the experiment-matched wild-type (wt) control is given in parentheses in the key of each panel.
Analysis of the single-gene deletion mutants corresponding to the larger set of yeast RP’s was carried out as described above for the RBH set. Of the 264 deletion strains examined, 25 were found to have a significant increase in RLS (9.5%), a 3.7-fold enrichment over the random set (G-test: χ12 = 12.03, P = 0.00052) (Table 2; Supplemental Fig. S1). The reduced frequency of long-lived strains among the RP set relative to the RBH set likely reflects the increased number of yeast genes that are not true orthologs of worm aging genes.
As a totally unbiased approach, we repeated this analysis examining only the yeast orthologs of worm genes identified to modulate life span in the RNAi screens (Dillin et al. 2002; Lee et al. 2003; Hamilton et al. 2005; Hansen et al. 2005; Curran and Ruvkun 2007). Of 196 worm genes, 52 of these have a yeast ortholog using RBH criteria and 111 have one or more related protein(s) in the RP set. In the RBH set, six of 46 nonessential yeast orthologs (or 13.0%) were found to modulate aging, a statistically significant enrichment (G-test: χ12 = 5.17, P = 0.023). Of the 182 nonessential yeast genes in the RP set, 18 are involved in life span determination (9.9%; G-test: χ12 = 1.1, P = 0.00086). Thus, we see a statistically significant enrichment of aging-related genes in both the RBH set and RP set when we limit our analysis to the worm genes identified in unbiased RNAi screens.
In this study, we have identified 25 genes that modulate yeast RLS, of which 22 have not been previously reported (Table 2; Fig. 1; Supplemental Fig. S1). Using the Princeton Protein Orthology Database (P-POD, http://ortholog.princeton.edu/findorthofamily.html), we were able to identify clear human orthologs for at least 15 of the 25 worm–yeast gene pairs, including all genes in the RBH set (Table 2) (Heinicke et al. 2007). The novel genes in the RBH set found to influence yeast aging include a transcription elongation factor (SPT4), an ATP-dependent metalloprotease (AFG3), an inositol polyphosphate 5-phosphatase (INP53), two ribosomal proteins (RPL9A and RPL19A), and three translation initiation factors (TIF4631, TIF1, and TIF2) (Fig. 1). Strikingly, seven of the 11 genes in the RBH set with a conserved role in longevity are associated with protein translation.
To determine whether the ortholog pairs identified as conserved aging genes in this study are overrepresented in specific functional classes, we performed gene ontology analysis on the worm aging genes, their yeast orthologs in the RBH set, and the 11 conserved aging genes within the RBH set (see Methods; Supplemental Table S4). Focusing on the cellular component category, we noted that ribosome-associated genes are significantly enriched in each case, with 6/11 of the conserved aging genes in the RBH set annotated with this gene ontology term (Fig. 2A). In the biological process category, these ribosome-associated genes are annotated as being involved in translation initiation, ribosome biogenesis, and protein biosynthesis or translation (Supplemental Table S4). Although mitochondrial-associated genes are highly enriched in the set of worm aging genes and the corresponding yeast orthologs, the conserved aging genes in the RBH set are not statistically enriched for mitochondrial-associated genes (Fig. 2A).
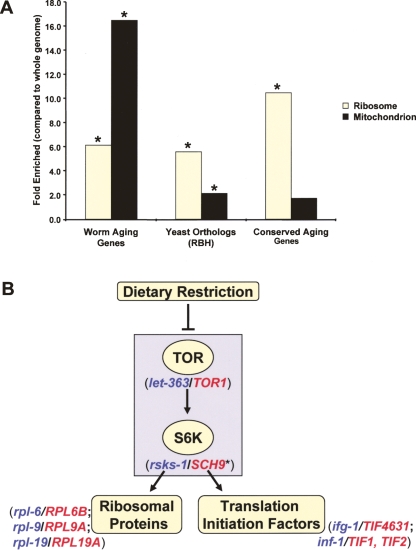
Figure 2.
Conserved aging genes are enriched for ribosome-associated genes. (A) Gene ontology analysis of the 276 worm aging genes, their yeast putative yeast orthologs in the reciprocal best-hit set (RBH, gene total = 103), and the 11 conserved aging genes (Table 2) revealed a significant enrichment in the ribosome cellular component when compared with the entire worm or yeast genome ([*] P <0.05, after Bonferroni Correction for multiple comparisons). Mitochondrial genes are highly enriched in worm aging genes and their corresponding yeast homologs and orthologs, but there is no significant enrichment for mitochondrial genes in the group of conserved longevity genes identified in this study. Essential genes were included in the gene ontology analysis, but replicative life span was not measured for these (RBH/ribosome = 8; RBH/mitochondrion = 8). (B) Conserved aging genes highlight the role of the TOR signaling pathway in modulating longevity. Orthologous gene pairs are shown in parentheses (worm genes are in blue and yeast genes are in red). The nutrient sensing kinases are enclosed in the gray box. Many downstream targets of TOR signaling, including ribosomal proteins and translation initiation factors, play a conserved role in life span determination. (*SCH9 was recently shown to have S6K activity (Urban et al. 2007), but also has homology with worm aging genes in the Akt family of kinases, such as akt-1 and sgk-1.)
Genetic and functional evidence suggests that decreased TOR or S6K activity may be one mechanism by which dietary restriction extends life span (Kennedy et al. 2007). There are many downstream targets of TOR signaling, including translation, autophagy, and the stress response (Wullschleger et al. 2006). In this study, we have identified translation initiation factors as highly conserved longevity modifiers, consistent with the hypothesis that these factors modulate longevity in a linear pathway with TOR and DR (Fig. 2B). Because our analysis was limited to loss-of-function mutations, however, it is unlikely that we would identify genes involved in autophagy or the stress response using this approach. Thus, we do not exclude the possibility that TOR signaling modulates longevity in divergent eukaryotes through regulation of both translation and other downstream targets.
Discussion
Invertebrate models have been a driving force in aging research, but the degree to which the genetic modifiers of longevity are conserved between different organisms has remained largely unexplored. We have compared two divergent eukaryotes (worms and yeast) and found that the ortholog of a gene with a known longevity role in worms is statistically more likely than a random gene to play a similar role in yeast.
For several reasons, we believe that the observed overlap between worm and yeast aging genes is likely to be an underestimate of the true conservation among longevity genes. First, our rigorous criteria for verification of long-lived yeast deletion mutants (see Methods) and known issues of aneuploidy in the yeast ORF deletion collection, both create a low false positive rate, but may fail to identify some deletion mutants with increased RLS. As evidence for this idea, we note several cases (15 in the RBH set) in which the deletion mutant is significantly long-lived in MATα but not MATa (Supplemental Table S3). While it is possible that these deletions regulate aging in a mating type-specific manner, more likely scenarios are that (1) the deletion would become significant in both mating types with analysis of a greater number of mother cells, or (2) the deletion strain was incorrectly annotated or has acquired a suppressor mutation in one mating type or the other. None of these deletions were included in the reported set of long-lived yeast strains. Second, there is an unknown false positive rate among genes reported to be long-lived in C. elegans. Third, the longevity-modulating role of more than half of the worm aging genes in our set of 276 was defined in large-scale RNAi screens, and the efficiency of RNAi-mediated knock down is known to vary. Therefore, many long-lived C. elegans mutants correspond to partial loss-of-function alleles, rather than null alleles. In contrast, all of the yeast data from this study were derived from single-gene deletion mutants that (with the notable exception of the ribosomal protein pairs) result in complete loss of protein activity. Finally, other methods of ortholog identification, such as the use of protein–protein interaction networks, may increase the number of ortholog pairs that have a conserved role in life-span determination. For example, previous studies have argued that SCH9 is the functional ortholog of akt-1 (Longo and Finch 2003), and more recently, SCH9 was shown to have ribosomal S6 kinase activity (Urban et al. 2007). Because of our stringent reciprocal best-hit requirement, however, SCH9 falls into the RP group in this study, although functional evidence indicates it is orthologous to a known C. elegans aging gene.
If aging is not the result of a genetic program, why should orthologs of the genes modulating aging in one organism regulate aging in another? Through careful examination of the pathways that have a conserved role in modulating longevity, we propose a possible explanation. Nutrient-responsive signaling pathways and translational control have been closely linked to life span extension by dietary restriction in a number of studies (Kaeberlein and Kennedy 2007), and many of the orthologs identified in our study map to these pathways. There is likely to be selective pressure on eukaryotic organisms to assess food availability and alter behavior accordingly in order to increase fitness. Life span modulation by dietary restriction, and genetic mimetics thereof, may be under selective pressure, indirectly, by virtue of their close apposition to organismal responses to food availability. Thus, “public” mechanisms of aging exist not to regulate aging, but to regulate the response of the organism to available nutrients, with aging as a pleiotropic response. Other pathways modulating aging may not be so tightly partnered to phenotypes under high selection, and therefore may be more variable across eukaryotic species. If true, these findings may help resolve the apparent paradox inherent in the argument that aging is not genetically programmed but is evolutionarily conserved.
Translation has been previously shown to decrease as a function of age (Rattan 1996; Syntichaki et al. 2007b). The regulation of protein translation may represent a “checkpoint” in the trade-off between reproduction and longevity. Decreased translation causes slow growth in yeast and decreased brood size in C. elegans (Chen et al. 2007; Pan et al. 2007), indicating that this process is necessary for maximal reproductive fitness. Although decreased translation and slow growth are not necessarily predictors of longevity in yeast (Steffen et al. 2008), there is an enrichment for genes involved in protein translation in the conserved aging genes (Fig. 2). Thus, our data support the idea that, in a favorable (nutrient-rich) environment, the activity of genes involved in growth and reproduction is limiting for life span. In unfavorable environmental conditions (such as DR), the activity of these genes is reduced, limiting the rate of growth and reproduction and enhancing longevity (and in some cases, e.g., daf-2 mutants, extending reproductive periods). In order to really understand how natural selection might act on these genes, we would need to compare the fitness of mutant strains against that of wild type under different environmental conditions. Thus, although our findings are consistent with antagonistic pleiotropy and the “disposable soma,” further studies will be required to understand how the activity of nutrient-sensing kinases and translation factors involved in growth and reproduction are limiting for life span in yeast, worms, and other model organisms.
Worms and yeast are separated evolutionarily by a greater distance (∼1.5 billion years) than worms and mammals (∼1 billion years) (Wang et al. 1999); therefore, quantitative evidence for conservation of longevity pathways between these species would suggest similar overlap between invertebrates and mammals. Indeed, we find that at least 15 of the aging-related ortholog pairs between yeast and worms have clear human orthologs (Table 2). While this manuscript was in preparation, Kapahi and colleagues reported the identification of 24 developmental arrest genes that modulate adult life span in C. elegans, many of which also have mammalian orthologs (Chen et al. 2007). As novel aging genes are reported in C. elegans, we will continue to examine their yeast relatives to identify genes with a conserved role in life span determination. We believe that many of these genes, and the pathways within which they function, are likely to modulate aging in mammals, and that mammalian orthologs of these gene pairs are reasonable candidates as potential therapeutic targets for age-associated diseases.
Methods
Ortholog identification
For the purposes of this study, we have defined a worm aging gene as any worm gene whose decreased function has been reported to increase life span (Supplemental Table S1). Computational algorithms can identify putative ortholog pairs between yeast and worms; however, this determination is complicated by the fact that there are no definitive parameters for determining true orthology. Here, we used a custom program to analyze BLASTp matches between WS156 (longest splice form for each gene) and SGD1 protein sequences downloaded from Wormbase (www.wormbase.org) and the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org), respectively. All-by-all BLASTp searches of worm to yeast and yeast to worm were done using the -m 8 output option without filtering and with an E-value cutoff of 0.01. BLASTp hits with an E-value greater than 0.001 or a BLAST bit score below 40 were excluded from analysis in this study.
In order to identify yeast orthologs of the 276 reported worm aging genes, we have adopted a multiple stringency level strategy. The reciprocal best-hit (RBH) group contains protein pairs that meet a reciprocal best BLASTp match requirement with at least 20% sequence identity and 20% amino acid alignment. Reciprocal best hits were identified using blast bit scores as the measure of similarity. To allow for 1:2 orthologs, we have included in this group any yeast proteins with a BLASTp score within 10% of the best hit. The RBH group represents putative ortholog pairs and many, if not all of these ortholog pairs are true orthologs. However, it is likely that other true ortholog pairs are not represented here due to the stringent reciprocal best-hit requirement. The “related proteins” (RP) group contains the top six yeast homologs to each worm aging gene, with at least 20% sequence identity and 10% amino acid alignment. This group allows us to address the possibility of redundancy of function within protein families in the context of aging. It is important to note that the RBH group is completely overlapping (included) in the “related proteins” group. Approximately 1/3 of the orthologs and homologs in the “related proteins” group cannot be tested in the replicative life span assay, because these genes are essential, or the deletion strains are very sick (and thus not represented in the MATα haploid deletion collection) (Supplemental Table S2). Ortholog identification is complete for currently known worm aging genes; however, we will continue to periodically refine our putative ortholog sets as new worm aging genes are reported and as gene predictions are updated.
Yeast replicative life span analysis
Single-gene deletion mutants corresponding to all nonessential genes were analyzed for replicative aging. The life span of a minimum of 20 cells from each strain in both sets was determined in the MATα haploid collection. Sign Wilcoxon Rank Sum tests were performed to determine whether the strain exhibited an enhanced life span relative to experiment-matched control cells (BY4742) (Supplemental Table S3). One of three decisions was made at this point. If the deletion strain was statistically long-lived (P <0.05), 20 cells from the haploid MATa collection were examined to confirm that long life span was linked to the deletion in question. While the ORF deletion set has been immensely valuable to the yeast research community, there are examples of aneuploidy in a small fraction of deletions and growth suppressors have arisen in certain cases. In deletion strains where data from the 20 cells analyzed was ambiguous, we chose to examine the life span of more cells. In general, less than 100 cells were examined in total for these MATα strains, although in a few cases more cells were analyzed. If the life span of one of these deletions was long-lived after collection of more data, verification was performed in the MATa set. Finally, in cases where MATα deletion strains were unlikely to be long-lived, based on analysis of 20 cells, no further analysis was performed.
Statistical analysis of worm aging genes with duplicate yeast orthologs
Our statistical analysis of conserved aging genes included worm genes that had two yeast orthologs. If the probability that a randomly chosen yeast gene is associated with aging is given by ϕ, then worm genes with two yeast orthologs are approximately twice as likely to have one or more yeast orthologs associated with aging (1 − [1 − ϕ]2 ≈ 2ϕ, assuming ϕ is small). Thus, we may overestimate the degree to which having a worm ortholog associated with aging increases the likelihood that a yeast gene is associated with aging. To control for our allowance of 1:2 orthologs, we performed a more conservative analysis, calculating the probability that deletion of one of two putative yeast orthologs (corresponding to a single worm aging gene, and chosen at random) would result in a long-lived phenotype. Of the 56 worm aging genes in the RBH group, 36 have a single yeast ortholog (three of which are aging related) and 20 worm genes have duplicate yeast orthologs (seven of which have at least one yeast ortholog that is aging related). For one worm aging gene (inf-1), both yeast orthologs are associated with aging in yeast, so the probability that a randomly chosen ortholog is associated with aging is 1. For the other six worm genes that have duplicate orthologs (one of which is aging related), the probability is 1/2. So, based on the life span data collected, we would expect seven of the 56 worm aging genes (or 12.5%) to have a yeast ortholog that is associated with aging, compared with 3.4% of the random set. Thus, even taking duplicate orthologs into account, we find that yeast orthologs of worm aging genes are significantly enriched for genes that impact longevity (Fisher’s G-test, G = 5.37, P = 0.021).
Gene ontology (GO) analysis
All GO terms associated with each of the 276 worm aging genes were downloaded from WormMart (www.wormbase.org) and analyzed by a custom program similar to SGD GO Term Finder, http://db.yeastgenome.org/cgi-bin/GO/goTermFinder.pl). GO terms are statistically enriched (using a cutoff of P <0.05 after Bonferroni correction) relative to the GO term frequency in the entire worm genome. Individual statistical tests were computed using Fisher’s exact test, which is based on the hypergeometric distribution as described at SGD (http://www.yeastgenome.org/help/goTermFinder.html).
GO term analysis of yeast genes was done using SGD GO Term Finder, including all ORFs in the background set and the default settings for annotation source, annotation type, and evidence codes. The P-value cutoff used in this analysis was P <0.05 (after Bonferroni correction).
Acknowledgments
This work was funded by an award from the Ellison Medical Foundation to B.K.K. and M.K. and by National Institutes of Health Grant R01 AG024287 to B.K.K. E.D.S. was supported by NIH training grant P30 AG013280. This work was performed while M.K. was an AFAR research grant recipient. D.E.L.P. was funded by an award from the Ellison Medical Foundation.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074724.107.
References
- Boehm M., Slack F., Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Chen D., Pan K.Z., Palter J.E., Kapahi P., Pan K.Z., Palter J.E., Kapahi P., Palter J.E., Kapahi P., Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S.P., Ruvkun G., Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1370/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Kamath R.S., Ahringer J., Kenyon C., Ahringer J., Kenyon C., Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Guarente L., Kenyon C., Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Liu W., Odell I., Ruvkun G., Lee S.S., Odell I., Ruvkun G., Lee S.S., Ruvkun G., Lee S.S., Lee S.S. A systematic RNAi screen for longevity genes in C. elegans. Genes & Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A.L., Dillin A., Kenyon C., Hsu A.L., Dillin A., Kenyon C., Dillin A., Kenyon C., Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of life span from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.J., Kenyon C., Taubert S., Crawford D., Libina N., Lee S.J., Kenyon C., Crawford D., Libina N., Lee S.J., Kenyon C., Libina N., Lee S.J., Kenyon C., Lee S.J., Kenyon C., Kenyon C. Life span extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Heinicke S., Livstone M.S., Lu C., Oughtred R., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K., Livstone M.S., Lu C., Oughtred R., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K., Lu C., Oughtred R., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K., Oughtred R., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K., Kang F., Angiuoli S.V., White O., Botstein D., Dolinski K., Angiuoli S.V., White O., Botstein D., Dolinski K., White O., Botstein D., Dolinski K., Botstein D., Dolinski K., Dolinski K. The Princeton Protein Orthology Database (P-POD): A comparative genomics analysis tool for biologists. PLoS ONE. 2007;2:e766. doi: 10.1371/journal.pone.0000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S.T., Bonafe M., Johnson T.E., Bonafe M., Johnson T.E., Johnson T.E. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Kappeler L., De Magalhaes Filho C., Kappeler L., De Magalhaes Filho C., De Magalhaes Filho C. IGF-1 signaling and aging. Exp. Gerontol. 2004;39:1761–1764. doi: 10.1016/j.exger.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M. Genome-wide approaches to understanding human ageing. Hum. Genomics. 2006;2:422–428. doi: 10.1186/1479-7364-2-6-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Kennedy B.K., Kennedy B.K. Protein translation, 2007. Aging Cell. 2007;6:731–734. doi: 10.1111/j.1474-9726.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R.W., Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Powers R.W., Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Steffen K.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Westman E.A., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Hu D., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Dang N., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Kerr E.O., Kirkland K.T., Fields S., Kennedy B.K., Kirkland K.T., Fields S., Kennedy B.K., Fields S., Kennedy B.K., Kennedy B.K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Steffen K.K., Kaeberlein M., Steffen K.K., Kaeberlein M., Kaeberlein M. Ruminations on dietary restriction and aging. Cell. Mol. Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood T.B. Evolution of ageing. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- Lee S.S., Lee R.Y., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G., Lee R.Y., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G., Kamath R.S., Ahringer J., Ruvkun G., Ahringer J., Ruvkun G., Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Longo V.D., Finch C.E., Finch C.E. Evolutionary medicine: From dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Martin G.M. Modalities of gene action predicted by the classical evolutionary biological theory of aging. Ann. N. Y. Acad. Sci. 2007;1100:14–20. doi: 10.1196/annals.1395.002. [DOI] [PubMed] [Google Scholar]
- Martin G.M., Austad S.N., Johnson T.E., Austad S.N., Johnson T.E., Johnson T.E. Genetic anlaysis of ageing: Role of oxidative damage and environmental stresses. Nat. Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- Medawar P.B. Old age and natural death. Modern Quart. 1946;2:30–49. [Google Scholar]
- Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P., Olsen A., Chen D., Lithgow G.J., Kapahi P., Chen D., Lithgow G.J., Kapahi P., Lithgow G.J., Kapahi P., Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Gems D., Gems D. Beyond the evolutionary theory of ageing, from functional genomics to evo-gero. Trends Ecol. Evol. 2006;21:334–340. doi: 10.1016/j.tree.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Promislow D.E.L., Fedorka K.M., Burger J.M.S., Fedorka K.M., Burger J.M.S., Burger J.M.S. Evolutionary biology of aging: Future directions. Elsevier; New York: 2006. [Google Scholar]
- Rattan S.I. Synthesis, modifications, and turnover of proteins during aging. Exp. Gerontol. 1996;31:33–47. doi: 10.1016/0531-5565(95)02022-5. [DOI] [PubMed] [Google Scholar]
- Schafer J.C., Winkelbauer M.E., Williams C.L., Haycraft C.J., Desmond R.A., Yoder B.K., Winkelbauer M.E., Williams C.L., Haycraft C.J., Desmond R.A., Yoder B.K., Williams C.L., Haycraft C.J., Desmond R.A., Yoder B.K., Haycraft C.J., Desmond R.A., Yoder B.K., Desmond R.A., Yoder B.K., Yoder B.K. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J. Cell Sci. 2006;119:4088–4100. doi: 10.1242/jcs.03187. [DOI] [PubMed] [Google Scholar]
- Steffen K.K., MacKay V.L., Kerr E.O., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., MacKay V.L., Kerr E.O., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Kerr E.O., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Tsuchiya M., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Hu D., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Fox L.A., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Dang N., Johnston E.D., Oakes J.A., Tchao B.N., Johnston E.D., Oakes J.A., Tchao B.N., Oakes J.A., Tchao B.N., Tchao B.N., et al. Yeast lifespan extension by depletion of 60S ribosomal subunits is mediated by Gnc4. Cell. 2008 doi: 10.1016/j.cell.2008.02.037. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N., Troulinaki K., Tavernarakis N., Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007a;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N., Troulinaki K., Tavernarakis N., Tavernarakis N. Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2007b;1119:289–295. doi: 10.1196/annals.1404.001. [DOI] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Wanke V., Anrather D., Ammerer G., Riezman H., Anrather D., Ammerer G., Riezman H., Ammerer G., Riezman H., Riezman H., et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Ventura N., Rea S., Henderson S.T., Condo I., Johnson T.E., Testi R., Rea S., Henderson S.T., Condo I., Johnson T.E., Testi R., Henderson S.T., Condo I., Johnson T.E., Testi R., Condo I., Johnson T.E., Testi R., Johnson T.E., Testi R., Testi R. Reduced expression of frataxin extends the life span of Caenorhabditis elegans. Aging Cell. 2005;4:109–112. doi: 10.1111/j.1474-9726.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- Wang D.Y., Kumar S., Hedges S.B., Kumar S., Hedges S.B., Hedges S.B. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. Biol. Sci. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H.R. Subfield history: Use of model organisms in the search for human aging genes. Sci. Aging Knowl. Environ. 2003;2003:RE1. doi: 10.1126/sageke.2003.6.re1. [DOI] [PubMed] [Google Scholar]
- Williams G.C. Pleiotropy, natural selection, and the evolution of senescence. Evolution Int. J. Org. Evolution. 1957;11:398–411. [Google Scholar]
- Winkelbauer M.E., Schafer J.C., Haycraft C.J., Swoboda P., Yoder B.K., Schafer J.C., Haycraft C.J., Swoboda P., Yoder B.K., Haycraft C.J., Swoboda P., Yoder B.K., Swoboda P., Yoder B.K., Yoder B.K. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., Bangham R., Benito R., Boeke J.D., Bussey H., Benito R., Boeke J.D., Bussey H., Boeke J.D., Bussey H., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Shields D.C., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N., Loewith R., Hall M.N., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]