Abstract
Ciliates are the only unicellular eukaryotes known to separate germinal and somatic functions. Diploid but silent micronuclei transmit the genetic information to the next sexual generation. Polyploid macronuclei express the genetic information from a streamlined version of the genome but are replaced at each sexual generation. The macronuclear genome of Paramecium tetraurelia was recently sequenced by a shotgun approach, providing access to the gene repertoire. The 72-Mb assembly represents a consensus sequence for the somatic DNA, which is produced after sexual events by reproducible rearrangements of the zygotic genome involving elimination of repeated sequences, precise excision of unique-copy internal eliminated sequences (IES), and amplification of the cellular genes to high copy number. We report use of the shotgun sequencing data (>106 reads representing 13× coverage of a completely homozygous clone) to evaluate variability in the somatic DNA produced by these developmental genome rearrangements. Although DNA amplification appears uniform, both of the DNA elimination processes produce sequence heterogeneity. The variability that arises from IES excision allowed identification of hundreds of putative new IESs, compared to 42 that were previously known, and revealed cases of erroneous excision of segments of coding sequences. We demonstrate that IESs in coding regions are under selective pressure to introduce premature termination of translation in case of excision failure.
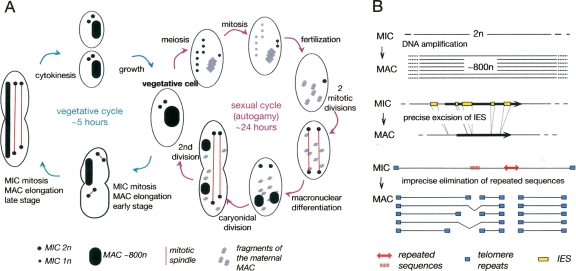
Paramecium is a unicellular eukaryote that belongs to the ciliate clade. One peculiar feature of ciliates is that, like multicellular eukaryotes, they separate germinal and somatic functions, in the form of two kinds of nuclei. A diploid germline micronucleus (MIC) undergoes meiosis to transmit the genetic information to the next sexual generation. A polyploid somatic macronucleus (MAC) is responsible for gene expression but develops anew at each sexual generation through reproducible rearrangements of the zygotic genome (for reviews, see Prescott 1994; Bétermier 2004; see Fig. 1A for a summary of the Paramecium life cycle).
Figure 1.
Nuclear dimorphism and the paramecium life cycle. (A) Life cycle. Left, vegetative cycle. During vegetative growth, paramecia divide by binary fission. The two micronuclei (MIC) undergo mitosis in the absence of nuclear envelope breakdown, while the macronucleus (MAC) elongates and divides by an amitotic process. Right, sexual cycle. P. aurelia species have two mating types, and sexually reactive cells can conjugate with a partner of the opposite mating type. In the absence of an appropriate partner, an auto-fertilization process (autogamy) occurs, illustrated here. Autogamy begins with meiosis of the two MIC to yield eight haploid products, seven of which degenerate. The eighth haploid gametic nucleus copies itself by mitosis and the two identical haploid nuclei fuse to from a completely homozygous diploid zygotic nucleus. Two post-zygotic mitotic divisions yield four diploid germline nuclei, which migrate to positions at the anterior and posterior of the cell. The two nuclei at the cell posterior differentiate into new MACs, the two nuclei at the cell anterior are the new MICs. During the first, caryonidal, cell division the macronuclear anlage do not divide. One is distributed to each daughter cell as they continue to endoreplicate DNA to attain the final MAC copy number of ∼800 n. After a second cell division in which both the MICs and the MAC divide, the sexual progeny enter the vegetative phase. Note that the progressively fragmented maternal MAC is present throughout meiosis, fertilization, and MAC differentiation and remains transcriptionally active. The fragments are lost by dilution in the course of the first cell divisions. The same events occur during conjugation; however, there is reciprocal exchange of haploid gametic MICs, so that the diploid zygotic nucleus produced by fertilization in each conjugating cell is heterozygous at all loci. Both sexual processes can be induced by standard laboratory protocols (Sonneborn 1974). (B) Genome reorganization. During MAC differentiation, DNA is amplified to a final copy number of ∼800 n. Different classes of repeated sequences, such as transposable elements and minisatellite repeats, schematized here, are eliminated by an imprecise mechanism that leads to chromosome fragmentation and de novo telomere addition, but which in some cases can be resolved by variable internal deletions. Internal Eliminated Sequences (IES) are short (<1 kb) unique-copy elements that interrupt both coding and noncoding sequences. IESs are precisely excised between 5′-TA-3′ dinucleotides at each end of the IES. A single TA remains in the MAC DNA. For a detailed review, see Bétermier (2004).
In Paramecium tetraurelia, the developmental genome rearrangements (Fig. 1B) consist of DNA amplification to a final copy number of ∼800 n and DNA elimination via two pathways. The first DNA elimination pathway is responsible for the removal of some 60,000 short, unique-copy elements (IES, for Internal Eliminated Sequence) that interrupt both coding and non-coding sequences. IESs are bound by 5′-TA-3′ dinucleotides and their perfectly precise excision is accomplished by a mechanism that produces double-stranded breaks followed by end joining (Gratias and Bétermier 2003). One TA dinucleotide remains in the chromosome after excision of the element.
The second DNA elimination pathway removes larger regions that often contain transposable elements (TE) and other repeated sequences, by an imprecise mechanism similar to that responsible for transposon silencing in eukaryotes. This mechanism involves short non-coding RNAs that probably target heterochromatin formation through histone methylation (for review, see Meyer and Chalker 2007). The heterochromatin is lost during MAC development. This DNA elimination pathway often leads to chromosome fragmentation. The new chromosome ends are healed by the addition of telomeric repeats, consisting of 200- to 300-nt random mixtures of G4T2 and G3T3 hexamers (Baroin et al. 1987).
Although these developmentally programmed genome rearrangements are highly reproducible, there is evidence that they generate some MAC chromosome heterogeneity within clonal cell populations, as shown by characterization of a few loci linked to the telomeric A and G surface antigen genes, in P. tetraurelia and Paramecium primaurelia, respectively. The elimination of repeated sequences, which usually leads to chromosome fragmentation, can also be resolved by variable internal deletions, as characterized in detail for one DNA elimination region in P. primaurelia (Fig. 1B; Le Mouël et al. 2003). Since a few rounds of endoreplication of the diploid zygotic genome precede DNA elimination, both chromosome fragmentation and variable internal deletions occur at this locus, even within a single homozygous cell. Similarly, the use of four different telomere addition regions, separated from each other by ∼10 kb, generates variability downstream from the A surface antigen gene in P. tetraurelia (Forney and Blackburn 1988; Amar and Dubrana 2004).
Patterns of MAC rearrangements may also vary between clonal cell populations. Indeed, variant MAC rearrangement patterns can be maintained across sexual generations, in the presence of a completely wild-type MIC genome (Epstein and Forney 1984; Meyer 1992; Duharcourt et al. 1995). The non-Mendelian inheritance of the rearrangement patterns can now be explained by “genome scanning” during development: The maternal MAC DNA is compared with the MIC DNA by a homology-dependent mechanism related to RNA interference (Mochizuki and Gorovsky 2004; Nowacki et al. 2005). The comparison ensures that only sequences present in the maternal MAC will be amplified and maintained in the new zygotic MAC (for review, see Meyer and Chalker 2007).
Given the strong heritability of the patterns of MAC rearrangements, variation in these patterns can give hold to the action of selection. To understand the constraints that the program of developmental genome rearrangements exerts on the evolution of the genome, it is essential to study these variations. P. tetraurelia somatic DNA was recently sequenced to a depth of ∼13× and assembled to provide a 72-Mb draft of the MAC genome (Aury et al. 2006). Annotation and analysis of the gene repertoire revealed that the very large number of protein-coding genes (nearly 40,000) is the consequence of at least three successive events of whole genome duplication (WGD) in the Paramecium lineage (Aury et al. 2006). In the present study, we have taken advantage of the whole genome shotgun (WGS) sequencing data (final assembly and >106 sequencing reads) to evaluate heterogeneity among the MAC chromosomes. It is important to note that this variability cannot be the result of allelic variations, since the genome sequence was obtained from entirely homozygous cells. We analyzed the heterogeneity produced by each of the three developmental processes: DNA amplification, IES excision, and elimination of TE and other repeated sequences. DNA amplification appears to be uniform across the genome. Conversely, the processes of TE elimination and IES excision are an important source of variability. Thanks to this heterogeneity, we have been able to identify hundreds of putative new IESs (whereas only 42 IESs were previously known in P. tetraurelia). This allows us to demonstrate that IESs located within coding regions are under selective pressure to introduce premature termination codons (PTC) in case of excision failure.
Besides somatic chromosomal rearrangements, we also analyzed rearrangements that have occurred in the germline, over evolutionary time. For this purpose, we exploit information from the recent WGD that was detected in the Paramecium lineage (Aury et al. 2006) that can be analyzed by alignment at the nucleotide level. We show that the rate of chromosomal rearrangements is remarkably low in Paramecium. Finally, the data allow us to infer a simple relationship between MAC chromosomes and the MIC chromosomes from which they derive.
Results
DNA amplification during macronuclear development
The 72-Mb draft assembly of P. tetraurelia MAC DNA consists of 697 scaffolds of which 188 are >47 kb, the minimum size observed for MAC chromosomes by pulsed field gel electrophoresis. These chromosome-sized scaffolds contain 96% of the genome assembly, and at least 60% of them represent complete MAC chromosomes since reads with telomere repeats map to both scaffold ends (Aury et al. 2006). In order to evaluate whether copy number is uniform across the macronucleus as expected if MAC-destined sequences are amplified to the same extent, we mapped the raw sequence reads (trimmed for quality, see Methods) to the assembly to determine the number of times each base of the assembly is present in the reads.
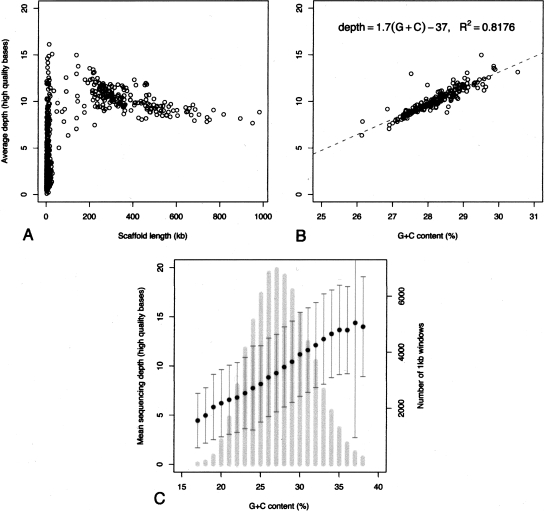
Figure 2A shows a scatter plot of average sequencing depth as a function of scaffold length for each of the 697 scaffolds. The average sequencing depth for the 188 chromosome-sized (>47 kb) scaffolds varies from 5× to 15×. Most of the small (<47 kb) scaffolds, which together represent only 4% of the assembly, have lower sequencing depth. We were able to map 46 of them to gaps in the larger scaffolds. We also remapped reads with telomere repeats (see Methods) to the small scaffolds. None of the small scaffolds resembles a complete chromosome; however, 83 have multiple remapped telomere reads and could represent chromosome ends (data not shown). We conclude that the majority of small scaffolds probably correspond to gaps or ends of the large scaffolds; they will not be taken into consideration in what follows.
Figure 2.
Depth of sequencing coverage and G+C content of the MAC genome assembly. (A) Scatter plot of average sequence depth in each of the 697 scaffolds of the assembly as a function of the size in nucleotides of the scaffold. The average sequencing depth is defined as the average number of reads that cover each nucleotide of the scaffold. (B) Scatter plot of the average sequencing depth of each of the 188 chromosome-sized scaffolds (>47 kb) as a function of the G+C content of the scaffold. The points were fit by linear regression, R2 = 0.82, P < 10−4. (C) Average sequencing depth and G+C content were calculated in 1-kb nonoverlapping windows for the 188 chromosome-sized scaffolds. Primary Y-axis: average of the sequencing depths for all of the 1-kb windows with the same G+C content (bins of 1%), plus or minus the SD, represented by black dots and dark-gray error bars, is plotted as a function of G+C content. Secondary Y-axis: histogram of G+C content, calculated using the 1-kb nonoverlapping windows (light-gray vertical bars).
A representation of average sequencing depth as a function of G+C content for each scaffold reveals a strong correlation between these parameters (Fig. 2B). The scaffolds vary by ∼10% in their G+C content, and the lower the G+C content, the lower the sequencing depth. To further test this correlation, we calculated average sequencing depth and G+C content in nonoverlapping 1-kb windows for the 188 chromosome-sized scaffolds (Fig. 2C). The 1-kb windows with lower G+C content than the genome's 28% average have below average sequencing depth, while the windows with higher G+C content also have above average sequencing depth, confirming the correlation. Since Paramecium has a very A+T-rich genome, the observed correlation can probably be explained by the fact that A+T-rich inserts are unstable in Escherichia coli. The higher the A+T content of a region, the poorer its representation in the shotgun sequencing libraries.
Thus all of the chromosome-sized scaffolds present approximately the same average sequencing depth, once corrected for their variation in G+C content. This implies that the amplification of the macronuclear genome is uniform, despite the existence of underamplified regions within chromosomes, for example adjacent to fragmentation/internal deletion sites. We cannot exclude the existence of underamplified chromosomes. However, independent data argue against this possibility. First, all previously characterized Paramecium genes are present in the assembly (data not shown). Second, the 78,110 ESTs generated in the course of the sequencing project (to the exclusion of ribosomal RNA and mitochondrial contaminants) all map unambiguously to the assembly (Aury et al. 2006). We conclude that the expressed portion of the Paramecium genome is amplified and maintained at uniform copy number in the macronucleus.
Low rate of chromosomal rearrangements during evolution
A striking characteristic of the P. tetraurelia genome is that 51% of the genes duplicated at the most recent WGD are still present in two copies. Alignment of each of the 12,026 pairs of paralogs revealed a distribution of amino acid identities comparable to that of mouse-human orthologs, with a peak near 95% identity. Although the synonymous substitution rates (Ks) of the paralogs are close to saturation, the nonsynonymous substitution rates (Ka) are very small, indicating that strong purifying selection maintains the amino acid sequences. Phylogenetic analysis showed that the most recent WGD occurred just before the speciation events that gave rise to the Paramecium aurelia complex of 15 sibling species (Coleman 2005; Aury et al. 2006).
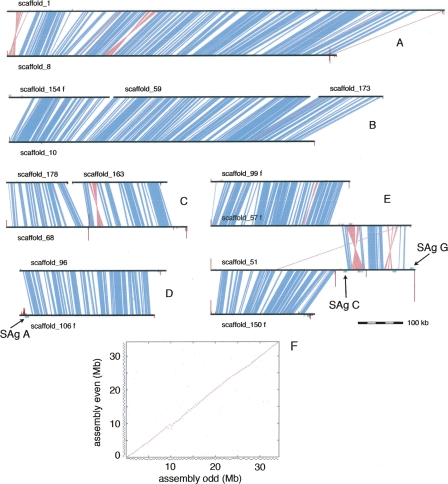
We carried out an all-against-all nucleotide comparison of the 188 chromosome-sized scaffolds in order to obtain a picture of the recent WGD at the nucleotide sequence level. Segments of >80% nucleotide identity, corresponding to ∼30% of the nucleotides in the assembly, cover most of the ORFs that are related by the recent WGD as well as some noncoding sequences that may include gene regulatory regions or noncoding RNA. The segments were grouped into syntenic blocks and the blocks were clustered using a transitive algorithm. We then drew all of the clusters (examples in Fig. 3; all of the drawings and a synteny viewer are available at http://paramecium.cgm.cnrs-gif.fr/tool/synteny). The drawings show the scaffolds (horizontal black lines) joined by segments of nucleotide alignment in blue (or pink for inverted regions), decorated by remapped telomere repeats (vertical maroon lines) and variable surface antigen genes (turquoise boxes). The drawings guided us in separation of the scaffolds into two half genomes. A dot plot comparison of the two half genomes presents a nearly continuous diagonal (Fig. 3F).
Figure 3.
Pairs of chromosomes revealed by internal nucleotide comparison of the MAC genome assembly. Examples of the clusters obtained from the internal nucleotide comparison (see Methods). The majority of clusters show pairs of complete MAC chromosomes as in A and D. The cluster in B corresponds to two complete MAC chromosomes, but one of them consists of three scaffolds separated by two sequencing gaps. The cluster in C illustrates possible assemblies of three polymorphic chromosomes that cover a single region; scaffold 68 is a consensus of two shorter chromosomes and one long one, resulting from fragmentation or internal deletion upon DNA elimination, while scaffolds 178 and 163 represent chromosomes created by fragmentation. The cluster in E shows a translocation that has occurred since the recent WGD. Note that scaffold 51 contains two internal sites with remapped telomeric repeats. The leftmost site corresponds to the MIC elimination region that was sequenced in P. primaurelia (Le Mouël et al. 2003). Horizontal black lines, scaffolds; blue polygons, segments of >82% nucleotide identity; pink polygons, inverted segments of >82% nucleotide identity; vertical maroon lines are proportional in height to the number of remapped reads that contain telomere repeats (i.e., at least three repeats of CCC[CA]AA with no more than one mismatch), the repeats were masked for the alignment of the reads against the assembly; turquoise boxes, remapped surface antigen genes. (F) Dot plot internal comparison of the MAC genome assembly. The genome was divided into two arbitrary half genomes for the dot plot, using the drawings of chromosome clusters related by the recent WGD. The small staggered lines along the outside of each axis represent the individual scaffolds.
All regions of the genome are paired, and 48 of the 73 clusters consist of a single pair of scaffolds, each of which is a complete MAC chromosome as indicated by correctly oriented telomere repeats at either end. Four additional clusters also consist of a pair of scaffolds, but at least one of the four scaffold ends is not marked by telomere repeats. The Paramecium genome appears to have undergone remarkably few large-scale rearrangements since the recent WGD, which is at an evolutionary distance, in terms of synonymous substitution rates, roughly equivalent to that of the divergence of rodents and primates from their common ancestor (Aury et al. 2006). Only six simple translocations (example, Fig. 3E) and one reciprocal translocation were found, along with 76 local inversions. This appears to be in striking contrast with the much higher rate of rearrangements in other taxa, as illustrated by the comparison of mouse and human chromosomes (Waterston et al. 2002) or by the analysis of the recent WGD in Arabidopsis (Blanc et al. 2003).
Heterogeneity of MAC chromosome fragmentation
Most of the sequence reads containing telomere repeats map, as expected, to the ends of assembly scaffolds. However, additional sites where telomere repeats have been mapped are located within assembly scaffolds. Interestingly, one of these sites, found on scaffold 51 (Fig. 3E), is located in a region orthologous to a locus that has been extensively analyzed in P. primaurelia: Characterization of both MAC and MIC DNA showed that there is heterogeneity in the fragmentation of MAC chromosomes at that locus (Caron 1992; Le Mouël et al. 2003). The polymorphic MAC chromosomes result from elimination of a 21-kb region, containing an inactive copy of the Tc1/mariner-like Tennessee TE and minisatellite sequences, leading to chromosome fragmentation as well as variable internal deletions during MAC development. This suggests that most telomere repeats remapped to internal sites within assembly scaffolds result from heterogeneity generated by the process of imprecise elimination of heterochromatic MIC DNA regions (as schematized in Fig. 1B).
The comparison of paralogous scaffolds resulting from the recent WGD supports this interpretation. We found 14 cases where a single scaffold is paired with two or more scaffolds. Possible explanations are (1) incomplete assembly, i.e., one chromosome is covered by two or more scaffolds that were not joined during assembly because of sequencing gaps or (2) a DNA elimination region responsible for chromosome fragmentation has appeared or disappeared since the recent WGD or (3) heterogeneity of chromosome fragmentation. The example shown in Figure 3B can be explained by sequencing gaps between scaffolds 154, 59, and 173 that together constitute a single MAC chromosome, since only the extremities of scaffolds 154 and 173 have correctly oriented, remapped telomere repeats indicative of MAC chromosome ends. However, the cluster consisting of scaffolds 178, 163, and 68 (Fig. 3C) clearly corresponds to the third possibility. Three observations support this interpretation. First, all three scaffolds in this cluster appear to be complete MAC chromosomes capped by telomeres. Second, many remapped telomere repeats are visible at the center of scaffold 68, between the 5′ part of the scaffold that aligns with scaffold 178 and the 3′ part that aligns with scaffold 163. Third, we were able to find two BAC clones that span the gap between scaffolds 178 and 163, indicating the existence of molecules corresponding to a large chromosome as expected if the DNA elimination event can be resolved by internal deletions. Hence, scaffold 68 probably represents three molecules, a large chromosome equivalent to the assembled scaffold 68, and two smaller versions, as would arise if DNA elimination led to both chromosome fragmentation (the two smaller chromosomes) and internal deletions (the large chromosome). Three other clusters present similar topology (clusters 5, 11, 35; http://paramecium.cgm.cnrs-gif.fr/tool/synteny).
We conclude that scaffolds with telomere repeats remapped to internal sites are very likely to represent the consensus assembly for a heterogeneous set of MAC chromosomes generated by variable outcomes of MIC DNA elimination. We can unambiguously identify 10 such sites (clusters 5, 11, 35, 42, 47) and additional sites may be present in clusters 28, 33, 40, and 44. Since the reads present only 13× redundancy compared to the 800 haploid copies of the genome that are present in the MAC, this is a minimal estimate of the number of MIC regions responsible for generating MAC chromosome heterogeneity.
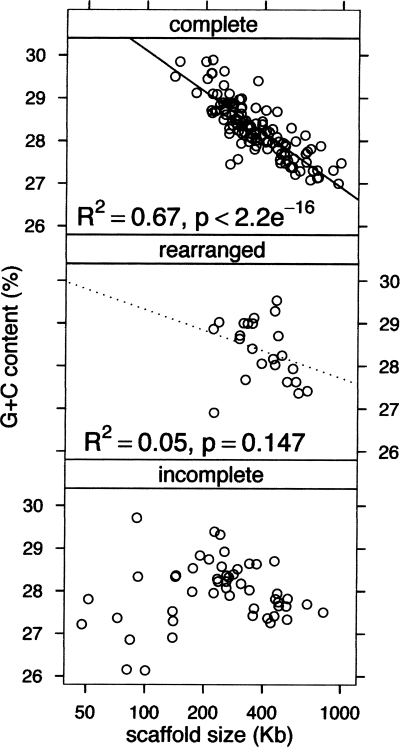
G+C content and chromosome length
MAC chromosomes are small and numerous. The 114 scaffolds that were validated as being complete chromosomes (see Methods) vary greatly in size, from 138 to 982 kb. These scaffolds also vary in G+C content, by ∼10% (cf. Fig. 2B). Strikingly, the scaffolds that are complete chromosomes display a significant inverse correlation between size and G+C content (R2 = 0.67, P < 2 × 10−16; Fig. 4). Confirmation of this strong negative correlation was obtained by examination of the G+C content of noncoding DNA on the chromosomes (introns: R2 = 0.565, P < 2 × 10−16; intergenic regions: R2 = 0.418, P = 4.8 × 10−15) and of the third base of four codon amino acids (R2 = 0.712, P < 2 × 10−16). The correlation—not to mention the quality of the assembly—is all the more impressive given the lower sequencing depth of A+T-rich sequences (cf. Fig. 2C). The correlation is weaker for the group of complete scaffolds from clusters with a translocation, i.e., the group of chromosomes that may have changed size recently owing to large-scale rearrangements. Scaffolds that are chromosome fragments show no correlation at all (Fig. 4).
Figure 4.
G+C content is inversely proportional to chromosome size. Scatter plots of G+C content of scaffolds as a function of scaffold size. The data for complete MAC chromosomes can be fit by a linear regression model. The “rearranged” scaffolds are complete chromosomes from clusters with translocations since the recent WGD as shown in Figure 3E; not all of the scaffolds in this group have changed size since the WGD; however, inclusion of this group with the other complete chromosomes adds several significant outliers to the data and reduces the value of the correlation coefficient R2.
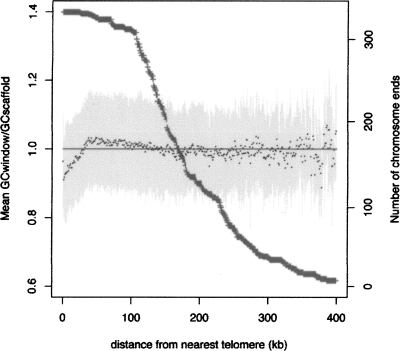
In order to explain the correlation, we looked for G+C variation within chromosomes. The only systematic variation we could detect was lower G+C content of the ∼30 kb at the chromosome ends (Fig. 5). This effect cannot account for the G+C variation between chromosomes: A region of fixed size with a high, rather than a low, G+C content would be required to explain the correlation. It is not surprising that subtelomeric sequences tend to have lower G+C content than the rest of a MAC chromosome, since these regions are primarily noncoding.
Figure 5.
G+C content variation along chromosomes. Mean G+C content (calculated in nonoverlapping 1-kb windows and normalized against the average G+C content of the scaffold) is plotted as a function of the distance from the nearest telomere (black points) ±SD (light-gray lines). Only chromosome ends with validated telomeres were used for the calculation. The number of chromosome ends decreases as the distance from the telomere increases (dark-gray curve and second Y-axis), because fewer and fewer scaffolds are of a size equal to or greater than twice the distance (X-axis).
Inverse correlations between chromosome size and G+C content have been found in the genomes of birds (International Chicken Genome Sequencing Consortium 2004), mammals (Mikkelsen et al. 2007), and yeast (Bradnam et al. 1999). Meiotic recombination rate provides the link between chromosome size and G+C content. There is a strong negative correlation between recombination rate and the size of chromosome arms in mammals, yeast, and birds (Kaback et al. 1992; Kaback 1996; Lander et al. 2001; Pardo-Manuel de Villena and Sapienza 2001; International Chicken Genome Sequencing Consortium 2004; Meunier and Duret 2004), and recombination can lead to an increase in G+C content via biased gene conversion (Galtier et al. 2001). Of course the negative correlation we have found concerns the fragmented MAC chromosomes and not the MIC chromosomes that undergo meiosis. The simplest explanation for this correlation is therefore that MAC chromosomes are proportional in size to the MIC chromosomes from which they are derived.
IES excision heterogeneity
Remarkably, some ciliates reconstitute their protein-coding genes at each sexual generation through the assembly of smaller gene segments present in the MIC genome. The most dramatic examples are the scrambled genes that must be reordered in hypotrich ciliates (Prescott 1994; Nowacki et al. 2008) and genes interrupted by multiple short internal sequences, the IESs, that must be spliced out in Paramecium (Bétermier 2004). A similar situation, though restricted to a small fraction of the genome, is found in vertebrates where immunoglobulin genes are rearranged during lymphocyte maturation (for review, see Jones and Gellert 2004).
The genome of P. tetraurelia is estimated to contain ∼60,000 IESs. IES boundaries are characterized by an absolutely conserved 5′-TA-3′ dinucleotide which is required for excision, while the 6 bp internal to the TA often resemble a loosely defined consensus (Klobutcher and Herrick 1995). The process of IES excision is initiated early during MAC development, after a few cycles of endoreplication of the diploid zygotic genome (Bétermier et al. 2000), but the excision machinery remains active at later times of MAC development, probably until sexual progeny enter the vegetative growth phase (Ku et al. 2000). In order to assess the level of heterogeneity generated among the ∼800 copies of the MAC chromosomes by such a massive recombination program, we have aligned all shotgun sequence reads with the genome assembly to search for short insertions or deletions (indels). A threshold indel size of 10 bp was chosen to be smaller than the smallest known IES (26 bp), but large enough to avoid any sequencing errors in the reads.
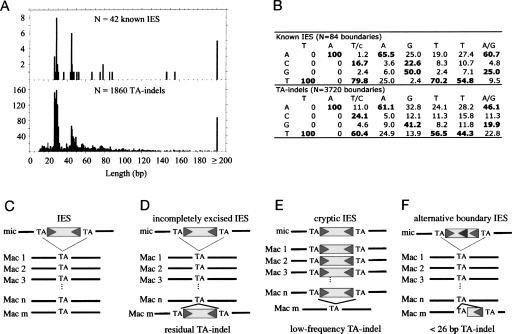
We identified 2169 indels (>10 bp) within high-quality alignments, among which 1860 (86%) are flanked by TA dinucleotides (hereafter named TA-indels). If indels were randomly distributed within sequences, we would have expected only 6% of them to be TA-indels (see Methods). Probable origins of TA-indels are (1) imprecise DNA elimination events between TA dinucleotides (Le Mouël et al. 2003), (2) rare events of IES excision using alternative boundaries (Dubrana and Amar 2000; Haynes et al. 2000), (3) the retention of IESs on a fraction of macronuclear copies, or (4) erroneous excision of sequences that are not IESs on a fraction of macronuclear copies. The TA-indels we found are unique-copy, AT-rich sequences (77.3% A+T vs. 71.6% for the whole genome), with a length distribution very similar to that of known IESs (Fig. 6A), characterized by two striking modes: a major mode at 28 bp and a minor one at 44 bp (Gratias and Bétermier 2001). The ends of the TA-indels closely resemble the degenerate IES consensus (Fig. 6B). For all of these reasons, we consider that the majority of the TA-indels probably result from the IES excision process.
Figure 6.
Sequence analysis of TA-indels and comparison with known IESs. (A) Length distribution of TA-indels and known IESs in P. tetraurelia. (B) Consensus sequence and frequency matrix of the 8-bp motif at the boundaries of TA-indels and of known IESs. Nucleotide frequencies that are higher than the genome average are indicated in bold. (C–F) Putative relationships between IESs and TA-indels. The IES is drawn as a box flanked by two TA repeats; triangles indicate the additional six nucleotides that define the consensus for the terminal inverted repeats. IESs that are excised with 100% efficiency are totally absent from the macronucleus, and therefore cannot be detected as TA-indels in our analyses (C). TA-indels may correspond to IESs for which some copies remain unexcised (D) or to cryptic IESs that are excised at low frequency (E). Some TA-indels (and notably those <26 bp) may correspond to IESs with alternative boundaries (F). Some TA-indels may also correspond to imprecise deletion events (data not shown).
Among the 1860 TA-indels, we identified 744 elements (40%) that are present in a sequence read but absent from the assembly (residual TA-indel), and 1116 elements (60%) that are present in the assembly but excised from a sequence read (low-frequency TA-indel). Given that the genome assembly represents the consensus sequence of the different copies of each macronuclear chromosome, residual TA-indels represent “incompletely excised” IESs (Fig. 6D). Conversely, low-frequency TA-indels may represent “cryptic” IESs (Fig. 6E), and it is striking that some are coding sequences, suggesting erroneous IES excision. In agreement with this suggestion, low-frequency TA-indel boundaries have a poorer resemblance to the 6-bp consensus. The 9% of TA-indels <26 bp (the shortest size reported for a Paramecium IES) could (1) correspond to IES fragments with alternative excision boundaries (Fig. 6F) or (2) result from imprecise elimination of repeated sequences, which can generate internal chromosome deletions between TA dinucleotides (Le Mouël et al. 2003). Indeed, for 22% of the TA-indels <26 bp, more than two macronuclear versions were present in the reads while this was the case for only 4% of the TA-indels ≥26 bp. We also detected 30 cases of a TA-indel nested within another TA-indel, a situation that has been reported for IESs (Duharcourt et al. 1998; Mayer et al. 1998).
Do the TA-indels correspond to MAC variability, or to contamination from MIC DNA? We cannot exclude the possibility that some residual TA-indels are MIC contaminants. However, we favor the hypothesis that the majority of the TA-indels reflect variability in the MAC DNA, for two reasons. First, low-frequency TA-indels are by definition present in most copies of the MAC DNA, and we consider that their excision from one MAC copy probably represents a cryptic event (but see Discussion), so these TA-indels cannot represent MIC contamination. Second, the ratio of MIC to MAC DNA in the cell is ∼1/200 (two diploid MIC, one MAC with ∼800 haploid copies); however, the DNA used to make the plasmid sequencing libraries was purified from MACs obtained by cell fractionation (see Methods). Judging by analysis of mitochondrial sequences, the enrichment in MAC DNA with respect to mitochondrial DNA is >1000-fold (Keller and Cohen 2000 and our unpublished results). The enrichment with respect to MIC DNA should be similar. But even if MIC contamination were 10 times greater than this, we would still find only ∼50 MIC reads in 106 reads (most of which would not contain an IES), clearly insufficient to account for several hundred residual TA-indels.
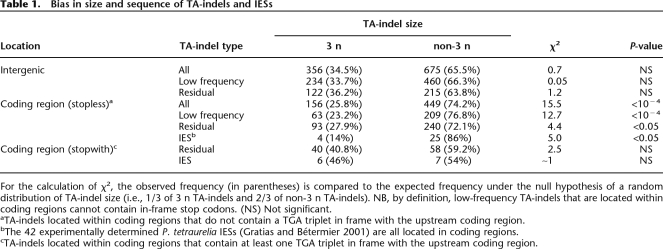
The TA-indels identified in this analysis occur everywhere in the genome: 56.7% of residual TA-indels are located in coding regions, 39.8% in intergenic regions, and 3.5% in introns (these regions constitute, respectively, 75.0%, 21.9%, and 3.2% of the genome). Low-frequency TA-indels are essentially intergenic (61.8%). Both classes of TA-indels are underrepresented in coding regions, which suggests that incompletely excised and, in particular, cryptic IESs are counter-selected in coding regions. Interestingly, when TA-indels are found within coding regions, they display a bias in their size distribution. As shown in Table 1, TA-indels that are a multiple of 3 (hereafter called 3 n TA-indels) occur at a significantly lower frequency than the expected 33%. However, this is only the case for the elements that lack a stop codon in phase with the upstream coding sequence (Table 1). In other words, within coding regions, there is a highly significant excess of TA-indels that lead to a translational frameshift and/or a premature stop codon. Since the 42 previously characterized P. tetraurelia IESs show exactly the same trend (Table 1), it appears that IESs and coding sequences may be subject to constraints that would favor premature translation termination in the case of an excision error.
Table 1.
Bias in size and sequence of TA-indels and IESs
For the calculation of χ2, the observed frequency (in parentheses) is compared to the expected frequency under the null hypothesis of a random distribution of TA-indel size (i.e., 1/3 of 3 n TA-indels and 2/3 of non-3 n TA-indels). NB, by definition, low-frequency TA-indels that are located within coding regions cannot contain in-frame stop codons. (NS) Not significant.
aTA-indels located within coding regions that do not contain a TGA triplet in frame with the upstream coding region.
bThe 42 experimentally determined P. tetraurelia IESs (Gratias and Bétermier 2001) are all located in coding regions.
cTA-indels located within coding regions that contain at least one TGA triplet in frame with the upstream coding region.
We also examined the properties of the genes that contain TA-indels, compared to genes that do not contain TA-indels, and found the former to be less highly expressed (average of 0.7 ESTs per gene as opposed to an average of two ESTs per gene), to be less often retained in two copies after the recent WGD (60% as opposed to 68%), and to evolve more rapidly (Ka of 0.14 as opposed to a Ka of 0.10, P < 0.0001). We conclude that more highly constrained genes either contain fewer IESs (and fewer cryptic IESs) or contain IESs that are more faithfully excised.
Discussion
We have used WGS data from the P. tetraurelia genome sequencing project (Aury et al. 2006) to evaluate variability in the somatic DNA produced by the programmed genome rearrangements that occur at each sexual generation in this organism.
Variability in the somatic genome
Analysis of sequencing depth across the assembly indicated that DNA is amplified to a uniform copy number during MAC development, as reported for the other holotrich ciliate with a sequenced somatic genome, Tetrahymena thermophila (Eisen et al. 2006). This is in sharp contrast with the situation in hypotrich ciliates such as Euplotes or Oxytricha, which have extensively fragmented somatic genomes. In these organisms, the copy number of the different MAC chromosomes, which usually contain a single gene, is variable owing to a process of differential amplification (Baird and Klobutcher 1991; La Terza et al. 1995; Donhoff and Klein 1996; Frels et al. 1996).
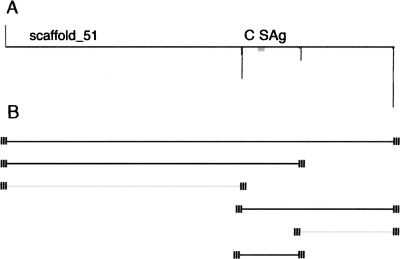
We found that the two DNA elimination processes (precise excision of IESs and imprecise elimination of repeated sequences) both contribute to substantial heterogeneity among MAC chromosomes. In other words, the genome assembly only represents the consensus sequence for MAC DNA. This conclusion is further supported by previously published experimental data. Phan et al. (1989) hybridized P. tetraurelia MAC chromosomes separated by pulsed field gel electrophoresis with probes specific for the A and C surface antigen (SAg) genes, which are present in a single copy per haploid genome. The A SAg probe hybridized with a single chromosome (cf. Fig. 3D). However, the C SAg probe hybridized with four chromosomes. This hybridization pattern is entirely consistent with the genome assembly, as shown schematically in Figure 7, because the C SAg maps to scaffold 51 (cf. Fig. 3E), which has two internal telomere sites. In other words, the unique MIC region that contains the C SAg gene is carried by four different MAC chromosomes as a result of alternative outcomes of DNA elimination at these sites. The drawings of the scaffolds with the remapped telomere reads show that at least eight other MAC chromosomes harbor such sites. This is a lower limit, however it is clear that not every fragmentation event can also be resolved by internal deletions, as demonstrated by extensive characterization of MAC chromosomes bearing the A SAg in P. tetraurelia (Epstein and Forney 1987; Forney and Blackburn 1988; Phan et al. 1984; Amar and Dubrana 2004). We have no data concerning sites where DNA elimination is resolved only by internal deletions, as they would not have been detected by the present analysis.
Figure 7.
Assembled scaffolds can represent several MAC chromosomes that derive from a unique region of MIC DNA. (A) The assembly scaffold 51 is shown; remapped telomere repeats appear as vertical lines and the C SAg (GenBank accession no., M65164; ParameciumDB accession no., GSPATG00017138001) as a gray box. Note that reads with telomeric repeats map not only to the ends of this scaffold, but also to two internal locations. (B) Predicted MAC chromosomes. The prediction that six MAC chromosomes derive from the same germline region is based on the assumption that each internal telomere site corresponds to a MIC region whose elimination can lead either to fragmentation of the chromosome or to internal deletions. The small striped boxes represent the telomeres at the ends of each MAC chromosome. Chromosomes that contain the C SAg are in black, and chromosomes that do not are in gray. The prediction is in agreement with previously published experimental data showing that four MAC chromosomes hybridize with a C SAg probe (Phan et al. 1989).
IES excision is precise in Paramecium, as required for gene expression and as shown by studies of the mechanism (Gratias and Bétermier 2003). However, up until now nothing was known of the efficiency of the IES excision process. The availability of ∼13× WGS sequencing data allowed identification of 1860 TA-indels, of which the majority result from errors in IES excision judging by their size distribution and terminal inverted repeat consensus. Given that a MAC contains ∼800 copies of each chromosome and that the sequence coverage is only 13×, our analyses detect only a fraction of the existing excision variability. By analyzing the frequency distribution of the number of sequence reads corresponding to each TA-indel, we estimate that there are at least 7000 loci with excision variability (see Methods). Thus, there appear to be many IESs for which excision is not 100% efficient. The situation is comparable for intron splicing from pre-mRNAs, where the cost of very efficient splicing has been postulated to be so high that inefficient splicing is tolerated (Lynch 2006; Roy and Irimia 2008). However, the undesirable effects of retained introns are greatly reduced by the mRNA surveillance mechanism (nonsense-mediated decay, or NMD) that detects and degrades mRNA molecules that contain premature termination codons (PTC), during the pioneer round of translation (Maquat 1995, 2004).
It has recently been shown that short introns, not only in Paramecium but also in plants, fungi, and animals, are under strong selective pressure to cause premature termination of mRNA translation in the case of intron retention (Jaillon et al. 2008). This study also showed that NMD is active in Paramecium and is involved in degradation of unspliced Paramecium transcripts that contain a PTC. The bias in the size and sequence of the 1860 TA-indels and 42 previously known IESs (Table 1) is highly reminiscent of the bias initially discovered in Paramecium introns. The data are consistent with selection acting on IESs that interrupt coding sequences to ensure that excision failure will result in transcripts that can be recognized by NMD. Confirmation of this hypothesis must await direct experimental validation (e.g., by measuring the rate of retention of IESs in mRNAs from NMD deficient cells). However, the use of NMD to degrade the transcripts of nonproductively rearranged genes is already well documented, in the case of immunoglobulin and T-cell receptor genes of vertebrates. PTCs often arise as a consequence of programmed V(D)J rearrangements during lymphocyte development and they efficiently trigger NMD, thus avoiding translation of truncated immunoglobulin chains or T-cell receptors that might be toxic to the cell (Li and Wilkinson 1998).
We can further conclude that unfaithful IES excision (whether it be incomplete or cryptic IES excision) is deleterious. This is based on the lower frequency of TA-indels in highly constrained genes and the counter-selection of 3 n TA-indels, unless they contain an in-frame stop codon. Interestingly, part of the variability we have observed may correspond to regulatory processes whereby the excision or not of a given IES determines a particular cellular state, which might then be maternally inherited across sexual generations. Indeed, it is well documented that the presence of an IES in the maternal MAC is sufficient to inhibit excision of that IES from the zygotic MAC, for a subset of known IESs (Duharcourt et al. 1995; Meyer and Keller 1996; Duharcourt et al. 1998).
Germline chromosomes
In contrast with the heterogeneity observed during the development of somatic MAC chromosomes, MIC chromosomes appear to remain highly stable during evolution. Indeed, comparison of paralogous chromosomes resulting from the most recent of the three WGDs revealed very few genomic rearrangements (six simple translocations, one reciprocal translocation, and 76 local inversions). Notably the ends of MAC chromosomes (which correspond to fragmentation points of MIC chromosomes) are almost perfectly conserved in location. In some cases we detected heterogeneity in the fragmentation of MAC chromosomes (e.g., Fig. 3C), but the location of the fragmentation points is conserved. An intriguing question is why the Paramecium genome has undergone so few large-scale rearrangements on the evolutionary time scale. The high coding density of the MAC-destined regions of the genome (78%) may exert a strong constraint, since chromosome breakage will usually fall within a gene and be counter-selected. It is thus possible that most large-scale rearrangements are eliminated by selection.
What does a MIC chromosome look like? We know from molecular, cellular, and genetic studies of Paramecium that MIC chromosomes are larger than MAC chromosomes and that they contain functional centromeres, unlike MAC chromosomes. The observation that G+C content is inversely correlated with MAC chromosome size suggests a very simple relationship between MAC and MIC chromosomes. It has been shown that recombination drives G+C content in the human genome through biased gene conversion (Meunier and Duret 2004), and there is evidence that this process is widespread in eukaryotes (Birdsell 2002). In many eukaryotes there is a strong negative correlation between chromosome size and recombination rate, because of the requirement of at least one crossing-over per chromosome arm per meiosis (Kaback et al. 1992; Kaback 1996; Lander et al. 2001; Pardo-Manuel de Villena and Sapienza 2001; International Chicken Genome Sequencing Consortium 2004; Meunier and Duret 2004). In Paramecium, the available data indicate a high frequency of meiotic recombination. Le Mouël et al. (2003) used three allelic markers along one chromosome to measure 5%–37.5% recombination frequencies for intervals of ∼100 kb. The genes ND6 (Gogendeau et al. 2005) and ND7 (Skouri and Cohen 1997), for which mutant alleles are available, behave like unlinked loci (J. Beisson and M. Rossignol, unpubl.) although separated by ∼400 kb on the same chromosome. The data are thus consistent with the hypothesis that there is also a requirement of one crossing-over per chromosome arm per meiosis in Paramecium. This model therefore predicts a higher recombination rate and hence a higher G+C content on short chromosomes. Of course, meiotic recombination occurs on MIC chromosomes and not on the fragmented MAC chromosomes. The simplest explanation for higher G+C content on short MAC chromosomes is therefore that MAC chromosomes correspond to MIC chromosome arms.
Two arguments support such a simple relationship between MAC and MIC chromosomes. First, we could not find any correlation between MAC chromosome size and G+C content in the T. thermophila somatic genome (R2 = 0.027), using the 125 scaffolds from the draft genome assembly that are capped by telomeres at both ends (Supplemental Table S3 from Eisen et al. 2006). This is consistent with our model, since MAC chromosome fragmentation results from site-specific cleavage of the germline DNA in Tetrahymena (Yao et al. 1990) and there does not appear to be any correlation between the sizes of the ∼225 MAC chromosomes and the size of the five MIC chromosomes from which they derive, judging from available assignment of telomere-capped MAC scaffolds to MIC linkage groups (Supplemental Fig. 4 from Eisen et al. 2006). A second argument is the respective number of MAC and MIC chromosomes in P. tetraurelia. Our analysis of the MAC genome assembly identifies a maximum of 160 MAC chromosomes. Jones (1956) counted ∼50 pairs of MIC chromosomes for several P. aurelia species, and this value is probably an underestimate since the cytogenetic techniques available would not have detected small chromosomes. Consequently, the ratio of MAC chromosomes to MIC chromosomes could be close to 2:1, consistent with the view that most Paramecium MAC chromosomes are stripped down MIC chromosome arms.
Methods
WGS sequencing and assembly have been detailed in Aury et al. (2006). The strain used for genome sequencing, d4-2 (Sonneborn 1974), was originally obtained by crossing strain 29 with strain 51, followed by exhaustive back crosses to strain 51, so that strain d4-2 is isogenic with strain 51 at almost all loci. The DNA that was used to construct sequencing libraries (Aury et al. 2006) comes from the same DNA preparation previously used to make plasmid libraries for complementation cloning (Keller and Cohen 2000). This DNA was extracted from purified macronuclei, obtained from a population of autogamous cells that were cultured for ∼10 vegetative cell divisions after autogamy. The DNA had no detectable mitochondrial or micronuclear DNA contamination, according to the complete absence of hybridization of 60,000 library clones (3× redundancy) with a variety of mitochondrial and micronuclear probes (Keller and Cohen 2000).
The ends of the raw sequence reads were trimmed from the position where the average quality fell below 10 (i.e., 90% chance that the base call is correct) in a window of 30 nt. In addition, individual bases with values <10 were masked. The 1,254,160 trimmed reads were then used as the database for pattern match searches for telomeric repeats, defined as at least three repeats of the hexanucleotide CCC[CA]AA with at most one mismatch. We found 15,242 reads with telomere repeats. The repeats were masked and then remapped to the assembly by alignment with megablast (McGinnis and Madden 2004), using no filter and a requirement of 98% sequence identity. To calculate sequencing depth, the whole set of trimmed reads was aligned to the assembly using megablast, with the same parameters.
All known Paramecium Surface Proteins were extracted from GenBank and mapped to the assembly using BLASTX (Altschul et al. 1997) default parameters, an E-value cutoff of 10−40, and validation by visual inspection of the alignments.
The internal comparison of the 188 chromosome-sized scaffolds was carried out using MUMmer version 3.05 (Delcher et al. 2002). The nucmer script from the MUMmer package was used with default parameters. The segments of nucleotide similarity between scaffolds were grouped into blocks. Only blocks consisting of at least three segments and covering at least 5% of one of the two scaffolds were retained for transitive clustering. In order to draw the 73 resulting clusters, all of the data were put into a Bio∷DB∷GFF database using scripts from GBrowse software (Stein et al. 2002). Instead of displaying the data in a browser, a Perl script was written to draw the clusters using scaled vector graphics. A synteny browser was implemented using SynBrowse software (Pan et al. 2005). The drawings and the synteny browser are available at ParameciumDB (Arnaiz et al. 2007), http://paramecium.cgm.cnrs-gif.fr/tool/synteny.
The drawings of scaffolds related by the recent WGD allowed us to refine the classification of the scaffolds into (1) complete MAC chromosomes with telomeric regions at either end, (2) incomplete MAC chromosomes, and (3) MAC chromosomes that belong to clusters that reveal large-scale rearrangements since the recent WGD.
To assess the level of heterogeneity generated by the process of excision of IESs or repeated sequences, we compared all sequence reads (trimmed for quality) to the genome assembly with megablast. All cases where the megablast alignments showed an insertion or deletion (indel) in the sequence read compared to the assembly were realigned with the more accurate SIM alignment software (Huang and Miller 1991). We then retained high quality alignments (i.e., covering at least 85% of the length of the sequence read with >95% identity) containing indels of at least 10 bp. Note that in some cases there can be more than one optimal sequence alignment. For example the two following alignments have exactly the same score:
In such cases, when possible, we retained the optimal alignment where the indel is flanked by a T in 5′ and an A in 3′ (called TA-indels).
Among the 2169 indels detected in high-quality alignments, we identified 1860 TA-indels (86%). To determine the number of TA-indels expected under the null hypothesis that indels are randomly distributed across the genome we performed simulations. We randomly sampled 1000 sequences (1 kb long) in the assembly, and introduced a deletion (60 bp) at a random position. We then compared these 1000 sequences to the assembly, using the same protocol as described previously. In this simulation we found only 6% of TA-indels. The TA-indels can be downloaded from ParameciumDB, http://paramecium.cgm.cnrs-gif.fr/download/fasta/TAindels/TA_indels.fa.
To estimate the number of loci that display excision variability, we developed a model for TA-indel distribution. Given that the MAC contains ∼800 copies of each chromosome, and that the average coverage of the whole-genome shotgun sequencing is 13×, the TA-indels we detected represent only a fraction of the loci for which there is some excision variability in the MAC. If there is some excision variability at a given locus, the probability of detecting this variability depends on the number of sequence reads covering that locus (x) and on the frequency of the variant excision forms among the 800 chromosomes. Let f be the frequency of the major form. The probability of detecting only the major form is f x and the probability of detecting only the minor form is (1 − f)x. Thus, the probability of detecting at least one of the excision variants (p) is:
Among the 1860 detected TA-indels, 1485 were represented by a single sequence read, 216 by two reads, and 159 by three or more reads. This indicates that, in most TA-indels, the frequency of the minor form is <1/13 (i.e., f is close to 1).
Under the assumption that both f and x are constant across the genome, it is possible to estimate f from the frequency distribution of the number of sequence reads corresponding to the minor sequence variant. The expected proportion of TA-indels represented by a single sequence read is:
The expected proportion of TA-indels represented by two sequence reads is:
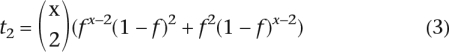 |
Given that f is close to 1 these two expressions can be approximated by:
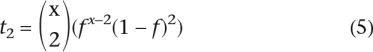 |
Thus, the ratio of the number of TA-indels represented by one or two sequence reads is given by:
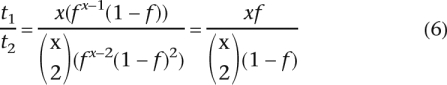 |
Hence:
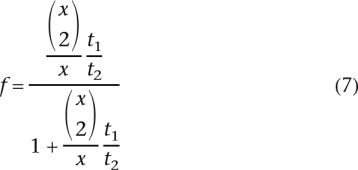 |
Given the observed number of TA-indels with one or two sequence reads, and the sequence coverage (x = 13), this model would predict that f = 97.6% (NB: when analyzed separately, residual and low-frequency TA-indels give similar estimates of f: respectively 97.4% and 97.8%). Given Equation 1, the probability of detecting a TA-indel given this value of f is 27%. Thus, we can estimate that there are ∼7000 loci with excision variability, each showing on average 20 unfaithful excisions among the 800 chromosomes. Note that this is a rough and probably minimal estimate because in reality x varies along chromosomes (see Fig. 2) and it is also likely that f varies among IESs. Furthermore, we have excluded TA-indels inferior in size to 10 nt.
Sequence data for the T. thermophila scaffolds were obtained from The Institute for Genomic Research ftp site (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/t_thermophila/Assemblies_and_Sequences).
Custom Perl scripts written with the Bioperl library (Stajich et al. 2002) and/or the EMBOSS package (Rice et al. 2000) were used for most analyses. Statistical calculations and graphics were made using R (a language and environment for statistical computing: http://www.R-project.org) unless otherwise indicated.
Acknowledgments
We thank Marie Sémon for suggesting a model for TA-indel distribution. This work was supported by the CNRS, the Consortium National de Recherche en Génomique, the ACI IMPBio contract 2004#14 to L.S., the ANR contract NT05-2_1522 to J.C., the Ligue National Contre le Cancer (grant RS06/75-32 to MB), and the Federation pour la Recherche Médicale (grant INE20061108404 to M.B.).
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074534.107.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar L., Dubrana K., Dubrana K. Epigenetic control of chromosome breakage at the 5′ end of Paramecium tetraurelia gene A. Eukaryot. Cell. 2004;3:1136–1146. doi: 10.1128/EC.3.5.1136-1146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Cain S., Cohen J., Sperling L., Cain S., Cohen J., Sperling L., Cohen J., Sperling L., Sperling L. ParameciumDB: A community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J.M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Duret L., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Noel B., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Jubin C., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Porcel B.M., Segurens B., Daubin V., Anthouard V., Aiach N., Segurens B., Daubin V., Anthouard V., Aiach N., Daubin V., Anthouard V., Aiach N., Anthouard V., Aiach N., Aiach N., et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Baird S.E., Klobutcher L.A., Klobutcher L.A. Differential DNA amplification and copy number control in the hypotrichous ciliate Euplotes crassus. J. Protozool. 1991;38:136–140. doi: 10.1111/j.1550-7408.1991.tb06033.x. [DOI] [PubMed] [Google Scholar]
- Baroin A., Prat A., Caron F., Prat A., Caron F., Caron F. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 1987;15:1717–1728. doi: 10.1093/nar/15.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétermier M. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res. Microbiol. 2004;155:399–408. doi: 10.1016/j.resmic.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Bétermier M., Duharcourt S., Seitz H., Meyer E., Duharcourt S., Seitz H., Meyer E., Seitz H., Meyer E., Meyer E. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol. 2000;20:1553–1561. doi: 10.1128/mcb.20.5.1553-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell J.A. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K.H., Hokamp K., Wolfe K.H., Wolfe K.H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam K.R., Seoighe C., Sharp P.M., Wolfe K.H., Seoighe C., Sharp P.M., Wolfe K.H., Sharp P.M., Wolfe K.H., Wolfe K.H. G+C content variation along and among Saccharomyces cerevisiae chromosomes. Mol. Biol. Evol. 1999;16:666–675. doi: 10.1093/oxfordjournals.molbev.a026149. [DOI] [PubMed] [Google Scholar]
- Caron F. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J. Mol. Biol. 1992;225:661–678. doi: 10.1016/0022-2836(92)90393-x. [DOI] [PubMed] [Google Scholar]
- Coleman A.W. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 2005;52:68–77. doi: 10.1111/j.1550-7408.2005.3327r.x. [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Phillippy A., Carlton J., Salzberg S.L., Phillippy A., Carlton J., Salzberg S.L., Carlton J., Salzberg S.L., Salzberg S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donhoff T., Klein A., Klein A. Timing of differential amplification of macronucleus-destined sequences during macronuclear development in the hypotrichous ciliate Euplotes crassus. Chromosoma. 1996;105:172–179. doi: 10.1007/BF02509498. [DOI] [PubMed] [Google Scholar]
- Dubrana K., Amar L., Amar L. Programmed DNA under-amplification in Paramecium primaurelia. Chromosoma. 2000;109:460–466. doi: 10.1007/s004120000108. [DOI] [PubMed] [Google Scholar]
- Duharcourt S., Butler A., Meyer E., Butler A., Meyer E., Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes & Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duharcourt S., Keller A.M., Meyer E., Keller A.M., Meyer E., Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol. Cell. Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., Badger J.H., Ren Q., Amedeo P., Jones K.M., Ren Q., Amedeo P., Jones K.M., Amedeo P., Jones K.M., Jones K.M., et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L.M., Forney J.D., Forney J.D. Mendelian and non-Mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol. Cell. Biol. 1984;4:1583–1590. doi: 10.1128/mcb.4.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney J.D., Blackburn E.H., Blackburn E.H. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol. Cell. Biol. 1988;8:251–258. doi: 10.1128/mcb.8.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frels J.S., Tebeau C.M., Doktor S.Z., Jahn C.L., Tebeau C.M., Doktor S.Z., Jahn C.L., Doktor S.Z., Jahn C.L., Jahn C.L. Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol. Biol. Cell. 1996;7:755–768. doi: 10.1091/mbc.7.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Piganeau G., Mouchiroud D., Duret L., Piganeau G., Mouchiroud D., Duret L., Mouchiroud D., Duret L., Duret L. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogendeau D., Keller A.M., Yanagi A., Cohen J., Koll F., Keller A.M., Yanagi A., Cohen J., Koll F., Yanagi A., Cohen J., Koll F., Cohen J., Koll F., Koll F. Nd6p, a novel protein with RCC1-like domains involved in exocytosis in Paramecium tetraurelia. Eukaryot. Cell. 2005;4:2129–2139. doi: 10.1128/EC.4.12.2129-2139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratias A., Bétermier M., Bétermier M. Developmentally programmed excision of internal DNA sequences in Paramecium aurelia. Biochimie. 2001;83:1009–1022. doi: 10.1016/s0300-9084(01)01349-9. [DOI] [PubMed] [Google Scholar]
- Gratias A., Bétermier M., Bétermier M. Processing of double-strand breaks is involved in the precise excision of paramecium internal eliminated sequences. Mol. Cell. Biol. 2003;23:7152–7162. doi: 10.1128/MCB.23.20.7152-7162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes W.J., Ling K.Y., Preston R.R., Saimi Y., Kung C., Ling K.Y., Preston R.R., Saimi Y., Kung C., Preston R.R., Saimi Y., Kung C., Saimi Y., Kung C., Kung C. The cloning and molecular analysis of pawn-B in Paramecium tetraurelia. Genetics. 2000;155:1105–1117. doi: 10.1093/genetics/155.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Miller W., Miller W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Bouhouche K., Gout J.F., Aury J.M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Bouhouche K., Gout J.F., Aury J.M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Gout J.F., Aury J.M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Aury J.M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Nowacki M., Serrano V., Porcel B.M., Ségurens B., Serrano V., Porcel B.M., Ségurens B., Porcel B.M., Ségurens B., Ségurens B., et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–363. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- Jones K.W. 1956. Nuclear differentiation in Paramecium. Ph.D. thesis, University of Wales, Aberystwyth. [Google Scholar]
- Jones J.M., Gellert M., Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Kaback D.B. Chromosome-size dependent control of meiotic recombination in humans. Nat. Genet. 1996;13:20–21. doi: 10.1038/ng0596-20. [DOI] [PubMed] [Google Scholar]
- Kaback D.B., Guacci V., Barber D., Mahon J.W., Guacci V., Barber D., Mahon J.W., Barber D., Mahon J.W., Mahon J.W. Chromosome size-dependent control of meiotic recombination. Science. 1992;256:228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- Keller A.M., Cohen J., Cohen J. An indexed genomic library for Paramecium complementation cloning. J. Eukaryot. Microbiol. 2000;47:1–6. doi: 10.1111/j.1550-7408.2000.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Klobutcher L.A., Herrick G., Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: Resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M., Mayer K., Forney J.D., Mayer K., Forney J.D., Forney J.D. Developmentally regulated excision of a 28-base-pair sequence from the Paramecium genome requires flanking DNA. Mol. Cell. Biol. 2000;20:8390–8396. doi: 10.1128/mcb.20.22.8390-8396.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terza A., Miceli C., Luporini P., Miceli C., Luporini P., Luporini P. Differential amplification of pheromone genes of the ciliate Euplotes raikovi. Dev. Genet. 1995;17:272–279. doi: 10.1002/dvg.1020170312. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Le Mouël A., Butler A., Caron F., Meyer E., Butler A., Caron F., Meyer E., Caron F., Meyer E., Meyer E. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell. 2003;2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wilkinson M.F., Wilkinson M.F. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Mol. Biol. Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Mayer K.M., Mikami K., Forney J.D., Mikami K., Forney J.D., Forney J.D. A mutation in Paramecium tetraurelia reveals functional and structural features of developmentally excised DNA elements. Genetics. 1998;148:139–149. doi: 10.1093/genetics/148.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S., Madden T.L., Madden T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J., Duret L., Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Meyer E. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes & Dev. 1992;6:211–222. doi: 10.1101/gad.6.2.211. [DOI] [PubMed] [Google Scholar]
- Meyer E., Chalker D., Chalker D. Epigenetics of ciliates. In: Allis D., et al., editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 127–150. [Google Scholar]
- Meyer E., Keller A.M., Keller A.M. A Mendelian mutation affecting mating-type determination also affects developmental genomic rearrangements in Paramecium tetraurelia. Genetics. 1996;143:191–202. doi: 10.1093/genetics/143.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Garber M., Gentles A.J., Goodstadt L., Heger A., Gentles A.J., Goodstadt L., Heger A., Goodstadt L., Heger A., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A., Gorovsky M.A. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M., Zagorski-Ostoja W., Meyer E., Zagorski-Ostoja W., Meyer E., Meyer E. Nowa1p and Nowa2p: Novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 2005;15:1616–1628. doi: 10.1016/j.cub.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Nowacki M., Vijayan V., Zhou Y., Schotanus K., Doak T.G., Landweber L.F., Vijayan V., Zhou Y., Schotanus K., Doak T.G., Landweber L.F., Zhou Y., Schotanus K., Doak T.G., Landweber L.F., Schotanus K., Doak T.G., Landweber L.F., Doak T.G., Landweber L.F., Landweber L.F. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Stein L., Brendel V., Stein L., Brendel V., Brendel V. SynBrowse: A synteny browser for comparative sequence analysis. Bioinformatics. 2005;21:3461–3468. doi: 10.1093/bioinformatics/bti555. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C., Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome. 2001;12:318–322. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Phan H.L., Forney J., Blackburn E.H., Forney J., Blackburn E.H., Blackburn E.H. Analysis of Paramecium macronuclear DNA using pulsed field gel electrophoresis. J. Protozool. 1989;36:402–408. doi: 10.1111/j.1550-7408.1989.tb05535.x. [DOI] [PubMed] [Google Scholar]
- Prescott D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roy S.W., Irimia M., Irimia M. Intron mis-splicing; no alternative? Genome Biol. 2008;9:208. doi: 10.1186/gb-2008-9-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouri F., Cohen J., Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: Identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn T. In: Paramecium aurelia. Handbook of genetics. King R., editor. Plenum Press; New York: 1974. pp. 469–594. [Google Scholar]
- Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Fuellen G., Gilbert J.G., Korf I., Lapp H., Gilbert J.G., Korf I., Lapp H., Korf I., Lapp H., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Stajich J.E., Harris T.W., Arva A., Harris T.W., Arva A., Arva A., et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yao M.C., Yao C.H., Monks B., Yao C.H., Monks B., Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]