Abstract
Mammalian genomes contain a high load of mobile elements among which long terminal repeat (LTR)- retrotransposons may represent up to 10% of the genomic DNA. The murine intracisternal A-type particle (IAP) sequences, the prototype of these mammalian “genetic parasites,” have an intracellular replicative life cycle and are responsible for a very large fraction of insertional mutagenesis in mice. Yet, phylogenetic analyses strongly suggest that they derive from an ancestral retrovirus that has reached the germline of a remote rodent ancestor and has been “endogenized.” A genome-wide screening of the mouse genome now has led us to identify the likely progenitor of the intracellular IAP retrotransposons. This identified “living fossil”—that we found to be present only as a single fully active copy—discloses all the characteristics of a bona fide retrovirus, with evidence for particle formation at the cell membrane, and release of virions with a mature morphology that are infectious. We show, by generating appropriate chimeras, that IAPs derive from this element via passive loss of its env gene, and gain of an endoplasmic reticulum targeting signal, resulting in its “intracellularization” and in the gain of transpositional activity. The identification within the mouse genome of the still active retroviral progenitor of the IAP endogenous mobile elements and the experimental dissection of the molecular events responsible for the shift in its life cycle provide a conclusive illustration of the process that has led, during evolution, to the generation of very successful intracellular retrotransposons from ancient retroviruses.
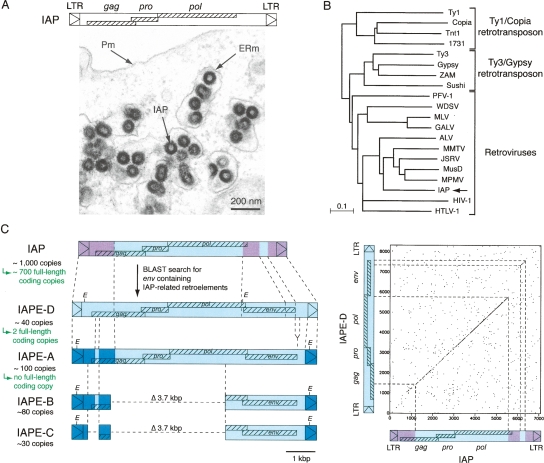
IAP sequences are among the most active murine retrotransposons, with a thousand copies dispersed throughout the genome and hundreds of copies still active for autonomous intracellular retrotransposition (for review, see Kuff and Lueders 1988; Heidmann and Heidmann 1991; Dewannieux et al. 2004). These elements have a structure of long terminal repeat (LTR)-retrotransposons with functional gag, pro, and pol genes—each required for retrotransposition—and no env gene (Mietz et al. 1987; Dewannieux et al. 2004). They produce virus-like particles (VLPs) with an immature morphology that assemble at the membrane of the endoplasmic reticulum (ER) and bud into the lumen of the cisternae (IAP, intracisternal A-type particles) (Fig. 1A), with no evidence for the release of extracellular particles. Based on the phylogenetic analysis of their pol gene, IAPs are unambiguously part of the retrovirus family (Fig. 1B; Malik and Eickbush 2001; Baillie et al. 2004) and are distinct from the bona fide LTR retrotransposons such as Ty1 in yeast or copia in Drosophila, which never possessed an env gene and are most probably more primitive retroelements. Yet, this unusual site of assembly and their strictly intracellular life cycle make IAPs atypical—albeit very successful—retroviral elements. Accordingly, it has been proposed that IAPs are endogenous retroviruses, that is, they are the genomic traces of primitive infection of the germline of a rodent ancestor by an infectious retrovirus that would have then been transmitted in a Mendelian manner while being subjected to specific genetic modifications resulting in their present-day murine retrotransposon status. Consistent with this still hypothetical scenario, previous studies have identified cDNAs with an env-like gene associated with IAP-related gag and pol sequences, originating from a still poorly characterized and polymorphic family of murine elements that were named “IAPE” (for IAP-related retroviral elements containing an Envelope gene) (Reuss and Schaller 1991). This family was estimated to contain ∼250 copies, with both full-length (IAPE-A) and deleted (IAPE-B and IAPE-C) elements (Reuss et al. 1996). Although expression of the envelope glycoprotein was detected in some cell lines (Reuss 1992), functional full-length elements have not been reported. IAPE elements were found in many laboratory inbred mouse strains and in wild mice, with a sexual dimorphism in the distribution of full-length copies in Mus musculus (Fennelly et al. 1996; Reuss et al. 1996). In order to characterize this murine family of repetitive elements and to tentatively identify copies that are still functional and that could represent the “missing link” between retroviruses and the intracellular IAP retrotransposons, we have now screened the mouse genome for such elements and have further characterized—after cloning—their biological activity. As a result, we identified a single fully active IAPE proviral copy, which proved to harbor all the characteristics of an infectious retrovirus. Furthermore, we could identify the molecular determinants that minimally allowed the—reciprocal—shift from this retroviral IAPE element into an IAP with an intracellular life cycle. This study therefore recapitulates the molecular events that have led to the endogenization of an ancestral bona fide retrovirus, with, in the present case, the rather unprecedented situation where both the progenitor element and the derived intracellularized retrotransposon still coexist in a functional state within their host.
Figure 1.
Structure of IAPs and related elements. (A, top) Genomic organization of an IAP LTR-retrotransposon, with the LTRs flanking the three ORFs homologous to the retroviral gag, pro, and pol genes. (Bottom) Electron microscopy of 293T cells transfected with an expression vector for the retrotransposition competent IAP(92L23) copy, disclosing numerous typical IAP VLPs accumulated in the cisternae of the ER. (Pm) Plasma membrane; (ERm) endoplasmic reticulum membrane. (B) Phylogeny of LTR-retroelements, based on their reverse transcriptase (RT) domain. The tree was determined by the neighbor-joining method using the seven blocks of conserved residues found in the RT domain of all retroelements and was rooted using non-LTR retrotransposons. The retroviruses listed include alpha- (ALV), beta- (MMTV, JSRV, MPMV), gamma- (MLV, GALV), delta- (HTLV-1), epsilon- (WDSV) retroviruses; lentiviruses (HIV-1); and foamyviruses (PFV-1). All sequences are readily accessible from GenBank and previous reports (e.g., Malik and Eickbush 2001). (C) Search for IAP-related retroelements containing a coding-competent env gene. BLAST search in the C57BL/6 mouse genome identifies the already known IAPE-A, -B, and -C elements, together with the yet-unreported IAPE-D subfamily. Sequences of the representative IAPE-D element and the homologous conserved domains in other IAP and IAPE elements are indicated in light blue; non homologous domains between IAP and IAPE elements are indicated in purple; non homologous domains between IAPE-D and the other IAPE subfamily members are indicated in dark blue. The limits of the conserved domains between IAPs and IAPEs were determined by DotPlot analysis (see IAP/IAPE-D dot plot analysis on the right; window size = 15, threshold = 45). For each subfamily, the number of copies in the C57BL/6 genome is indicated together with the number of full-length coding copies.
Results
Full-length, entirely coding IAP elements with an env gene in the mouse genome
The sequences of previously described IAP-related env-containing cDNAs (Reuss and Schaller 1991) were used to screen the C57BL/6 mouse genome for env-containing IAP-related elements (BLAST program, UCSC Genome Bioinformatics, genome.ucsc. edu). As previously estimated by Southern Blot analysis (Reuss and Schaller 1991), about 250 copies were identified in the mouse genome, belonging to distinct groups: the first group contains full-length elements of 8.3 kb with gag-, pro-, and pol-related sequences associated with an env gene flanked by LTRs, as well as internally deleted 4.6- and 4.2-kb elements (Fig. 1C). It is likely that these full-length and deleted elements correspond to the IAPE-A, -B, and -C elements, respectively, previously identified by Southern Blot analysis of EcoRI digests of mouse genomic DNA (Reuss et al. 1996). Of note, most IAPE-A copies are found on the Y-chromosome, leading to the previously observed sexual dimorphism (Fennelly et al. 1996; Reuss et al. 1996). The second group of elements emerging from our in silico search is more homogeneous and is composed essentially of full-length proviral elements, with approximately 40 members, dispersed throughout the autosomal chromosomes. Their LTRs are distinct from those of the previously identified elements, with no EcoRI sites (Fig. 1C). For the sake of consistency with the previously identified subfamilies and the observed differences with the full-length IAPE-A elements, we named this yet unreported subfamily “IAPE-D.”
Then, we screened each individual full-length copy wherever present, that is, within the IAPE-A and IAPE-D subfamilies, for the presence of potentially functional elements, with as a first requirement the presence of full-length coding gag, pro, and pol genes. This search disclosed only five such copies, among the approximately 140 full-length copies present in the C57BL/6 genome, four of them belonging to the IAPE-D subfamily (the IAPE-A copies proved to be much more degenerated). These copies have a canonical proviral organization, with the gag, pro, and pol genes organized so as to be translated via −1 frameshifting. Finally, only two of these copies, both belonging to the IAPE-D subfamily and that we named IAPE-D1 and -D2, contain in addition an intact env gene (as well as identical 5′- and 3′-LTRs) and are therefore likely candidates for being active, autonomous IAPE copies (see the IAPE-D1 complete sequence in Supplemental Fig. S1).
A functional env gene within the IAPE-D elements
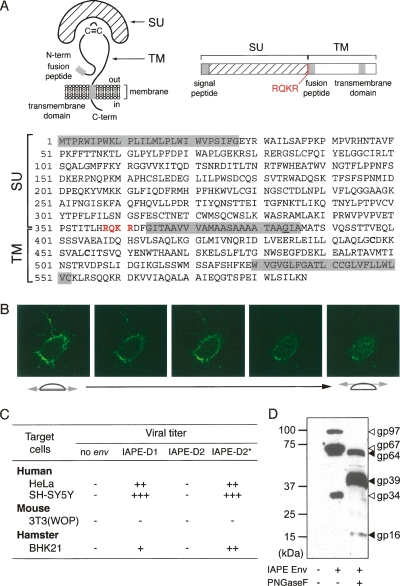
Sequence analysis of the IAPE-D1 and IAPE-D2 putative Env proteins discloses the canonical structural features of retroviral Envs with an N-terminal signal peptide (predicted using SignalP program at http://www.cbs.dtu.dk/services/SignalP), a canonical RQKR cleavage site between the SU and TM moieties, and within the latter two hydrophobic domains most probably corresponding to the fusion peptide and the transmembrane domain (Fig. 2A). These env genes were subcloned into an expression vector under the control of the strong hCMV promoter, and cells were transfected to assay for protein expression. As illustrated in Figure 2B for the IAPE-D1 Env, using a rabbit antiserum that we raised against a recombinant Env protein (corresponding to part of the SU domain), expression can be observed at the cell surface, as expected for a retroviral envelope. The functionality of the Env proteins was then assessed via a pseudotyping assay, in which retroviral particles encoded by env-defective viral cores (either from an onco- [MLV] or a lenti- [SIV] virus) can express the IAPE Env proteins at their surface and be infectious (Blaise et al. 2004). As illustrated in Figure 2C, the IAPE-D1 Env proved to be functional in this assay, the pseudotyped particles being infectious in several cell lines. Of note, IAPE-D2 Env is defective, but this appears to be only due to one point mutation, besides six others, in the hydrophobic fusion peptide. This could be demonstrated by replacing the charged glutamic acid residue (E) in IAPE-D2 Env with the neutral glycine residue (G) present at the same position in the IAPE-D1 Env (Fig. 2C, IAPE-D2*). It suggests that other env genes may not be very far from being functional, although a similar pseudotyping assay performed with 14 additional full-length coding-competent IAPE env genes was negative.
Figure 2.
Characterization of the IAPE Env glycoprotein. (A) Schematic structure and primary sequence of the IAPE-D1 Env protein. The SU and TM subunits are delineated, with the canonical furin cleavage site (R/K-X-R/K-R) between the two subunits indicated in red. The hydrophobic signal peptide, fusion peptide, and the transmembrane domain are shaded in gray. The G384 residue, mutated to E in the IAPE-D2 Env, is underlined. (B) Successive confocal images of a living cell stained for IAPE Env, demonstrating its localization at the cell surface. Human HeLa cells were grown on glass coverslips, transfected with the IAPE-D1 Env expression vector, and stained 48 h post-transfection, without permeabilization, with a rabbit anti-IAPE Env antiserum and an Alexa-Fluor 488-conjugated anti-rabbit IgG secondary antibody. (C) Infectivity assay of SIV particles pseudotyped with the IAPE-D Env proteins. Supernatants of human 293T cells cotransfected with an expression vector for the SIV core proteins, a lacZ gene-marked defective retroviral vector, and an expression vector for IAPE-D1, -D2, or -D2 E384G Env proteins (IAPE-D2*), or a control plasmid (no env) were used to infect human, murine, or hamster target cells. Viral titers, corresponding to the number of LacZ+ cell foci per milliliter of supernatant, are indicated as follows: (−) <10; (+) 10–100; (++) 100–1000; (+++) >1000. (D) Western blot analysis of SIV virions contained in the supernatant of transfected cells, pseudotyped (or not) with the IAPE-D1 Env protein. The corresponding samples from the infectivity assay in C were analyzed using a rabbit anti-IAPE Env antiserum. Supernatants were treated or not with peptide-N-glycosidase F (PNGaseF) as indicated.
To further characterize the IAPE env gene products, we performed immunoblotting experiments on the pseudotyped particles (Fig. 2D). Two high-molecular-weight bands can be specifically detected (not observed in the control without Env) with an apparent molecular mass of 97 and 67 kDa, most probably corresponding to the full-length Env precursor and the processed SU subunit, respectively, as classically observed for bona fide retroviral Envs. These proteins are glycosylated: in vitro treatment with peptide-N-glycosidase F, which removes all N-linked carbohydrates, results in a shift in the mobility of the Env precursor and the SU subunit to molecular weights of 64 and 39 kDa, respectively, consistent with the theoretical values (62 and 38 kDa). Consistent with this assignment, a mutant that we constructed for the SU-TM cleavage site (with the canonical RQKR sequence mutated to AQAR) lacks the 39-kDa band (data not shown). An additional low-molecular-weight glycosylated Env-specific band (34 kDa shifted to 16 kDa after deglycosylation) can also be observed, which is also incorporated into the viral particles and most probably correspond to a noncanonical SU cleavage product.
The IAPE-D1 provirus makes extracellular viral particles
Preliminary transient transfection experiments using an IAPE-D LTR-driven reporter gene revealed very low promoter activity in a series of cell lines that are classically used in ex vivo cell culture assays, including the human HeLa and murine 3T3 cells (data not shown). To characterize the functional properties of the IAPE-D1 provirus, the promoter U3 domain of the IAPE-D1 5′-LTR was therefore replaced by the strong hCMV promoter, in such a way as to preserve the natural retroviral start site and transcript structure (see Fig. 3A). Heterologous human cells (HeLa and 293T) were transfected with this expression vector, and cells were analyzed for both viral transcripts and proteins.
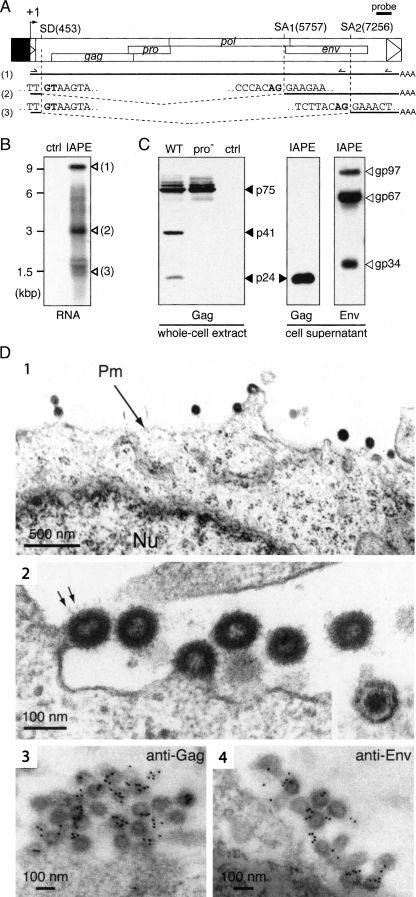
Figure 3.
Structure of the IAPE-D1 provirus and characterization of its gene products. (A) Genomic organization of the IAPE-D1 provirus, cloned under the control of the CMV promoter (in black) and structure of the corresponding viral transcripts. (SD) Splice donor site; (SA) splice acceptor site. The splice sites of the IAPE subgenomic transcripts (2 and 3) were determined by sequencing the RT-PCR products obtained using total RNAs and the two sets of primers indicated. (B) Northern blot analysis of total RNA extracted from 293T cells transfected with pCMV-β (ctrl) or the IAPE-D1 expression vector (IAPE), using a probe (shown in A) complementary to a 3′ domain of the IAPE genome. (C) Western blot analysis of IAPE-D1 proteins. Whole-cell extracts and virion-containing supernatants of human 293T cells transfected with IAPE-D1 (WT), a protease-deficient mutant (pro−), or a control plasmid (ctrl) were analyzed by Western blotting, using rabbit anti-IAPE Gag or anti-IAPE Env antisera, as indicated. (D, 1–4) Electron microscopy of cells transfected with IAPE-D1 and of the released viral particles. (1) Representative low-magnification image of transfected 293T cells, with particles budding at the plasma membrane. The nucleus (Nu) and plasma membrane (Pm) are indicated. (2) High-magnification view of budding and extracellular particles. Prominent spikes, corresponding to the Env protein, are indicated (arrows); (inset) view of an extracellular mature particle, with a condensed central core. (3–4) Immunogold labeling of IAPE particles in transfected 293T cells using the anti-IAPE Gag (3) or anti-IAPE Env (4) rabbit antiserum and a secondary antibody linked to gold beads, observed by electron microscopy. Gold beads are preferentially associated with viral particles (439 ± 165 gold beads/μm2 associated with VLPs vs. 5.6 ± 3.0 and 2.8 ± 2.6 gold beads/μm2 associated with cytoplasm and VLP-free extracellular medium for the anti-IAPE Gag antiserum and 302 ± 119 gold beads/μm2 associated with VLPs vs. 18.1 ± 9.1 and 4.2 ± 2.4 gold beads/μm2 associated with cytoplasm and VLP-free extracellular medium for the anti-IAPE Env antiserum).
As illustrated in Figure 3B, Northern blot analysis of the RNA extracted from the transfected cells, using a probe complementary to the 3′UTR IAPE genome (base pairs 7766–8105), discloses a transcript of ∼9 kb, most probably corresponding to the full-length viral transcript, together with two shorter transcripts (but no transcript in cells transfected with a control plasmid). These were further characterized by RT-PCR using appropriate primers (schematized in Fig. 3A) and sequencing of the corresponding products, which identified one donor splice site located in the 5′UTR of the IAPE genome and two distinct acceptor splice sites: The first one generates a subgenomic env-coding mRNA, as classically observed for retroviruses, whereas the second generates a shorter mRNA of unknown function (with coding capacity for at least a severely truncated Env protein, from amino acids 498–587).
Viral protein and particle formation was then assessed by Western blot analysis of both whole-cell extracts and cell supernatants. As illustrated in Figure 3C, using an anti-Gag rabbit antiserum (initially raised against the major homology region [MHR] domain of the IAP Gag Capsid [CA] moiety, which shares 82% homology with the MHR domain of IAPE) (Dewannieux et al. 2004), IAPE-D1 generates several Gag-associated fragments (not observed in cells transfected with a control plasmid): high-molecular-weight proteins of 75 kDa, most probably corresponding to uncleaved Gag polyprotein precursors, and lower-molecular-weight fragments (41 and 24 kDa), corresponding to Gag cleavage products, with the 24-kDa protein most probably corresponding to the CA protein. Consistently, only high-molecular-weight fragments are observed in cells transfected with an IAPE protease mutant (Fig. 3C; see structure in Fig. 4C). Finally, Western blot analysis of the supernatant of the IAPE-D1 transfected cells shows both the processed Gag p24 protein and the Env products (with a pattern similar to that in Fig. 2D), consistent with the release of viral particles by the IAPE-D1-transfected cells.
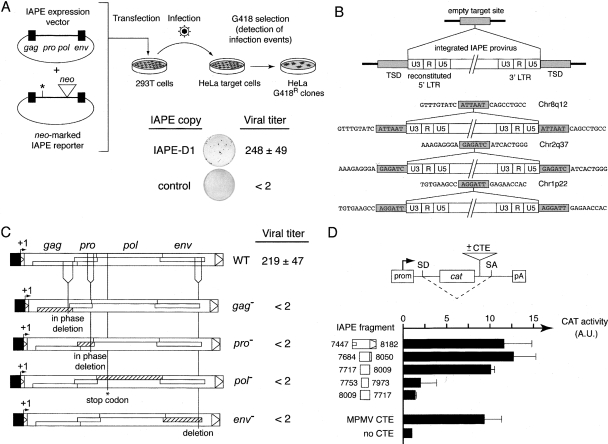
Figure 4.
Infectivity and functional characterization of IAPE-D1 and mutant derivatives. (A) Rationale of the infectivity assay and viral titer of IAPE-D1. 293T cells were cotransfected with an expression vector for the wild type (see Fig. 3A) or mutant IAPE-D1 derivatives (or pCMV-β as a control) and a neo-marked defective IAPE reporter. Supernatants from the transfected cells were collected 48 h post-transfection and used to infect HeLa target cells. After a 3-d growth period, infection events were detected upon G418 selection of the cells, and viral titers were quantified by counting the number of G418R clones per milliliter of supernatant (mean values ± SD, n = 3). (B) Structure of de novo integrated IAPE proviruses. The complete characterization of integrated IAPE elements and their insertion sites was performed using individual HeLa clones obtained after infection and G418 selection. A provirus insertion and the corresponding empty, pre-insertion site are schematized at the top, with the sequences of three characterized IAPE de novo insertions shown below. Target-site duplications of 6 bp (TSD, light gray) are found in all cases, associated with reconstituted 5′-LTRs. (C) IAPE genes required for infection. Infectivity assays were performed with either the wild-type IAPE-D1 copy or the same element rendered defective for gag, pro (via in-frame deletions, from nucleotides 1909–2313 and from nucleotides 2876–3139, respectively), pol (via introduction of a stop codon at nucleotide 3802), or env (via an out of frame deletion from nucleotide 7194–7453). The number of G418R clones obtained per milliliter of supernatant for each construct is indicated. (D) Rationale of the assay and evidence for CTE activity in IAPE-D. The reporter vector contains a cat gene flanked by splice donor and acceptor sites (SD and SA) placed under the control of a simian virus 40 promoter (prom) and polyadenylation signal (pA), and sequences to be tested for CTE activity are inserted as indicated. The presence of a CTE should promote the export of unspliced RNAs from the nuclei of cells transfected with the reporter plasmid, leading to a detectable chloramphenicol acetyltransferase (CAT) activity, whereas the absence of a CTE-like activity should lead to the export of spliced RNA and no CAT activity. 293T cells were transiently transfected with the reporter vector containing the indicated IAPE fragments, placed in the forward or reverse orientation (numbers indicate the nucleotide positions of the 5′- and 3′-ends of each fragment in the IAPE sequence). Forty-eight hours post-transfection, cells were lysed, and CAT activity was determined as described in Methods. The mean ratio of CAT activity between the reporter vector containing an IAPE sequence (or a control MPMV CTE) versus the empty vector (no CTE) was calculated from two to four independent experiments (A.U., arbitrary units; error bars indicate standard deviation).
Retroviral particles encoded by IAPE-D1 were finally characterized by electronic microscopy. As illustrated in Figure 3D, particles budding at the plasma membrane can be observed in the transfected cells (not observed with a control vector). Viral cores seem to assemble directly at the plasma membrane, as classically observed for gamma- or delta-retroviruses, with no pre-assembled particles detected in the cytoplasm. Two morphologies can be distinguished among the released particles, either with a hollow interior surrounded by a dense ring of stain corresponding to an immature morphology, or with a condensed central core. The latter most probably corresponds to mature particles in which the viral protease has cleaved the polyprotein precursors. Indeed, the protease-deficient IAPE mutant, unable to cleave the Gag polyprotein precursor (Fig. 3C), only produces viral particles with an immature morphology (data not shown). Of note, the morphology of the IAPE immature particles is very similar to that of the IAP particles (see Dewannieux et al. 2004 and Fig. 1A).
To definitively demonstrate that the observed particles are, indeed, the product of the IAPE-D1 genes, the transfected cells were analyzed by electron microscopy after immunogold labeling using the antisera directed against either IAPE Gag or Env. As illustrated in Figure 3D, the particles were specifically labeled with both antisera, as expected.
The IAPE-D1 encoded retroviral particles are infectious
To assess whether IAPE-D1 encodes a functional retrovirus, we conducted infection assays using a neo-marked element that allows detection of infection events after selection of the infected target cells with G418. In order not to disrupt the open-reading frames of the IAPE genome, we tentatively inserted the neo gene into the 3′UTR of the element. Yet, this insertion proved to be deleterious due to the presence of a non-coding regulatory sequence in this domain (see below). The infection assay was therefore conducted in trans with neo inserted directly into the IAPE-D1 env gene, and the resulting inactivated marked element (further rendered non-coding for the Gag-Pro-Pol polyprotein by a stop codon and an internal deletion) being complemented in trans by the unmarked native IAPE-D1 element. 293T cells were cotransfected with IAPE-D1 (or a control plasmid) and the neo-marked reporter, and the supernatant of the transfected cells was then harvested 48 h post-transfection and used to infect HeLa cells. As illustrated in Figure 4A, infection events could be detected upon G418 selection of the target cells, which yielded viral titers >100 CFU/mL, with no G418R clones detected with the control plasmid. To definitely demonstrate retroviral integration, the structure of the IAPE-D1 copies and their integration sites in G418R HeLa clones were determined, using a reverse PCR strategy as in Heidmann and Heidmann (1991) and Dewannieux et al. (2004). As illustrated in Figure 4B for three such infection events, full-length integrated proviral copies were observed, with target site duplications of 6 bp (as naturally occurring for IAPE copies in the mouse genome) and complete LTRs (with, e.g., a reconstituted U3 domain in the 5′ LTR in place of the CMV promoter, as expected for a canonical retroviral cycle).
To characterize the role of the IAPE genes in the generation of infection events, we inactivated each of them independently, by in-frame deletions for gag and pro so as to preserve translation of the downstream genes, and by introducing a premature stop codon in pol or a deletion in env (Fig. 4C). We checked that the introduced mutations had an effect neither on the level of transcription of the mutants nor on the level of Gag expression (data not shown). As illustrated in the figure, none of the four mutants was infectious in the assay, indicating that the four genes are necessary for the IAPE-D1 retroviral cycle.
Finally, the deleterious effect of the neo insertion into the IAPE-D1 3′UTR (see above) led us to identify a Constitutive Transport Element (CTE) within this domain, as found in many bona fide retroviruses. Such sequences promote nuclear export of unspliced, full-length viral transcripts to be packaged within the viral particles, which would otherwise be subjected to nuclear splicing (for review, see Hammarskjold 2001). This was demonstrated by a specific assay described in Figure 4D, that uses a previously devised indicator vector (pDM128/PL) (Hope et al. 1990; Fridell et al. 1993) into which we inserted the IAPE-D1 3′UTR or deletion derivatives thereof, or the MPMV CTE as a positive control. As illustrated in the figure, a 300-bp domain with a strong CTE activity could be delineated, inactive when inserted in the backward orientation, as expected for such sequences known to act at the RNA level.
In conclusion, the IAPE-D1 contains all the genes—as well as the post-transcriptional regulatory sequences—that are normally present in bona fide retroviruses.
The IAP/IAPE relationship: Molecular determinants of the intracellularization process
The identification of a family of endogenous elements among which one element proved to be an infectious provirus (IAPE-D1) raises the question of the phylogenetic relationship between IAPE and the closely related IAP intracellular retrotransposons. Both classes of elements disclose strong sequence similarities (Fig. 1C), consistent with the hypothesis that the IAP retrotransposon simply derives from the IAPE endogenous retrovirus. Yet, these elements have completely different life cycles: IAPEs make viral particles, which are assembled at the plasma membrane and are released into the supernatant as infectious viral particles, whereas IAPs make intracellular particles, which assemble at the membrane of the ER into the lumen of which they bud, with no particles released outside the cells. Close comparison of their sequences disclose strong similarities in the pro and pol genes (see DotPlot in Fig. 1C) with major divergences essentially located in the env region—highly degenerated for IAPs, with only an ∼180-bp fragment of the IAPE env still recognizable in the IAP sequence—and most importantly at the 5′-end of the gag gene that contains unrelated matrix (MA) domains, followed, in contrast, by closely related capsid (CA) and nucleocapsid domains (NC).
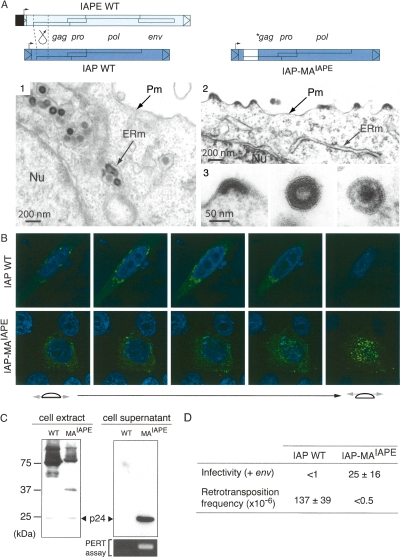
To determine whether the molecular determinants of IAP and IAPE particle assembly could be carried by the divergent N-terminal Gag domains, the strategy that we used was to construct chimeras between the IAPE-D1 and a previously characterized functional IAP copy (Dewannieux et al. 2004). We first attempted to switch the site of IAP assembly from the ER to the plasma membrane by replacing its MA domain with that of IAPE-D1, resulting in the IAP-MAIAPE chimera (see structure in Fig. 5A). As illustrated in the figure, electron microscopy analysis of cells transfected with the wild-type IAP and the IAP-MAIAPE chimera shows that the replacement of the IAP MA domain has a dramatic effect: The modified IAPs are now budding at the plasma membrane instead of the ER membrane, and particles are released into the extracellular space. The morphology of the released particles is closely related to that observed for IAPE-D1 (but without Env) with both immature and mature particles, the latter never being observed for the wild-type IAP in the ER.
Figure 5.
Recovery of an extracellular life cycle for an IAP chimera. (A) Structure of the wild-type and chimeric IAPs and morphology of associated VLPs. The IAP-MAIAPE element was constructed as illustrated by replacing the IAP Gag N terminus with that of IAPE. (1–3) Electron microscopy of 293T cells transfected with either wild-type or chimeric IAPs. (1) Representative low-magnification image of wild-type IAP particles accumulated in the cisternae of the ER. No particle can be observed at the cell membrane or in the extracellular space. (2) Low-magnification image of cells transfected with the chimeric IAP, disclosing particles budding at the cell membrane. No particle can be observed in the ER. (3) High-magnification images of chimeric IAP-associated particles. Images represent (from left to right) budding, free immature, and free mature particles. (B) Immunofluorescence confocal analysis of human HeLa cells transfected with wild-type or chimeric IAPs, fixed, permeabilized, and stained with the anti-Gag antibody (in green). Nuclei are stained with TO-PRO-3 iodide (in blue). (C) Western Blot analysis of whole-cell lysates or cell supernatants from 293T cells transfected with either the wild-type (WT) or the chimeric IAP (MAIAPE). (Bottom right) Detection of RT activity in the corresponding cell supernatant, using the PERT assay. (D) Functional characterization of wild-type and chimeric IAPs. For the intracellular retrotransposition assay, HeLa cells were cotransfected with the wild-type or chimeric IAP, and a neoTNF-marked defective IAP reporter in which a (backward) neomycin resistance gene—with the neo ORF interrupted by a (forward) intron—becomes functional only after splicing out of the intron, upon achievement of a complete replicative cycle. Retrotransposition events are detected upon G418 selection of the transfected cells, and frequencies are expressed as numbers of G418R clones per transfected cells in selection (mean values ± SD, n = 4); see Dewannieux et al. (2004) and Supplemental Figure S2 for a detailed description of the assay. For the infectivity assay, 293T cells were cotransfected with the same IAP plasmids as above, but with in addition the IAPE-D1 Env expression vector. Supernatants from the transfected cells were collected and used to infect naive HeLa cells, as in Figure 4A, and the viral titers were determined upon G418 selection of the target cells (mean values ± SD, n = 3).
This remarkable change in the site of VLP assembly was confirmed by whole-cell confocal immunofluorescence analysis with anti-Gag antibodies, which shows a peri-nuclear localization of the wild-type IAPs and a plasma-membrane-associated localization of the chimeric IAP element particles (Fig. 5B). Western blot analysis of the cells transfected with the wild-type IAP and IAP-MAIAPE reveals the cleaved p24 Gag protein in the cell supernatant for the IAP chimera, not observed with the wild-type IAP and consistent with the switch in IAP targeting (Fig. 5C). Noteworthily, the supernatant of the cells transfected with IAP-MAIAPE further discloses an RT activity detectable by the sensitive PERT assay (Pyra et al. 1994), thus, strongly suggesting that the released particles are close to be functional retroviruses. We finally tested whether the chimeric IAP released particles could be rendered infectious, simply by complementation for the absence of an env gene within the IAP genome. To do so, cells were cotransfected with IAP-MAIAPE (or wild-type IAP as a control) and an expression vector for the IAPE-D1 Env, together with the neo-marked IAP reporter described in Dewannieux et al. (2004) to assay for infectivity of the cell supernatants, using HeLa target cells and G418 selection as in Figure 4A. G418R clones were obtained with IAP-MAIAPE but not with the wild-type IAP (Fig. 5D), and no resistant clones were obtained in the absence of the IAPE env gene (data not shown). This indicates that the IAP-MAIAPE chimera makes functional particles that are able to carry out a retroviral infection cycle, provided that they are pseudotyped by an Env glycoprotein. Of note, the lack of G418R clones using the supernatant of cells transfected with the wild-type IAP and the IAPE-D1 Env (Fig. 5D) strongly suggests that the sole expression of IAPE Env is not sufficient to re-address an IAP particle to the plasma membrane for its release (consistent with the absence of effect of IAPE Env expression on the site of IAP assembly as observed by electronic microscopy) (data not shown). Finally, the shift in the site of particle assembly observed with the IAP-MAIAPE chimera and the release of the viral particles outside the cells resulted in a complete loss of retrotransposition activity (Fig. 5D), as measured by an assay as in Dewannieux et al. (2004) using a neoTNF-marked IAP element (for a description of the neoTNF indicator gene for retrotransposition and of the rationale of the assay, see Supplemental Fig. S2).
Conclusively, the N terminus of the IAP/IAPE Gag protein is the sole determinant of the site of VLP assembly, with the IAPE MA domain (that we have shown to contain the canonical sequences responsible for myristylation and targeting of the Gag proteins to the plasma membrane) (see Supplemental Fig. S3) allowing export of the chimeric IAP VLPs outside of the cells, and the addition in trans of the IAPE env gene finally “reconstituting” an infectious retrovirus most probably very close to the IAP/IAPE retroviral progenitor.
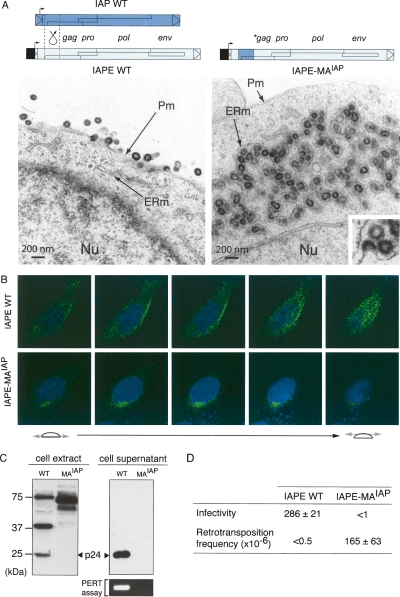
To tentatively recapitulate the molecular events that have led to the “intracellularization” of the IAP retrotransposon, starting from an infectious retrovirus, we reciprocally modified the IAPE gag gene and constructed the IAPE-MAIAP chimera in which the IAPE MA domain was replaced by that of IAP (see structure in Fig. 6A). Electron microscopy analyses of cells transfected with the wild-type IAPE and the IAPE-MAIAP chimera clearly show that all IAPE VLPs, initially budding at the plasma membrane, are now budding at the ER and accumulate into the cisternae, as observed for the IAPs. This modification in VLP targeting was further confirmed by whole-cell confocal immunofluorescence analysis with the anti-Gag antibodies, which discloses a plasma-membrane-associated localization for the wild-type IAPE and a peri-nuclear localization for the IAPE-MAIAP chimera (Fig. 6B). As further illustrated in Figure 6C, Western blot analyses of the transfected cells revealed that Gag proteins were still efficiently produced by the IAPE-MAIAP chimera, but with a very low level of protease-dependent cleavage (a property also shared, incidentally, by the wild-type IAPs and not incompatible with high retrotransposition activity and release of an active Pol from the Gag-Pro-Pol precursor) (Dewannieux et al. 2004). Neither Gag proteins nor RT activity could be detected in the cell supernatant, consistent with the shift of IAPE-MAIAP chimera Gag targeting.
Figure 6.
Structural and functional IAPE to IAP transition. (A) Structure of the wild-type and chimeric IAPEs and morphology of associated VLPs. The IAPE-MAIAP element was constructed as illustrated by replacing the IAPE Gag N terminus with that of IAP. Representative low-magnification images of 293T cells transfected with the wild-type IAPE disclose particles budding at the cell membrane (left). No particle can be observed at the level of the ER. Low-magnification images of cells transfected with the chimeric IAPE (right) disclose particles accumulated in the cisternae of the ER, with a high-magnification image of a particle budding into a cisternae (bottom right inset). No particle can be observed at the level of the cell membrane. (B) Immunofluorescence confocal analysis of human HeLa cells transfected with wild-type or chimeric IAPEs, fixed, permeabilized, and stained with the anti-Gag antibody (green). Nuclei are stained with TO-PRO-3 iodide (blue). (C) Western blot analysis of whole-cell lysates or cell supernatants from 293T cells transfected with either the wild-type (WT) or the chimeric IAPE (MAIAP). (Bottom right) Detection of RT activity in the corresponding cell supernatant, using the PERT assay. (D) Functional characterization of wild-type and chimeric IAPE. Infectivity was assayed as in Figure 4 either with the wild-type or the chimeric IAPE (mean values ± SD, n = 3). For the retrotransposition assay, HeLa cells were transfected with the wild-type or chimeric IAPE element in which the neoTNF indicator gene was inserted into the env gene, thus rendered defective (see structures in Supplemental Fig. S2). Retrotransposition frequencies were determined upon G418 selection of the transfected cells, as in Figure 5D (mean values ± SD, n = 3).
We finally asked whether this “intracellularized” IAPE would be able to retrotranspose, as a bona fide IAP element. The env gene in both the wild-type IAPE and the IAPE-MAIAP chimera was inactivated by deleting a 260-bp fragment within the gene (same deletion as in Fig. 3C) to prevent any possible reinfection event, and a retrotransposition assay was performed using constructs in which the neoTNF reporter gene was inserted in place of the deleted fragment. As illustrated in Figure 6D, G418R clones were obtained with neoTNF-marked IAPE-MAIAP, indicating that this element is autonomous for retrotransposition, but none were obtained with the neoTNF-marked IAPE copy. Conversely, an infectivity assay carried out using the neo-marked IAPE reporter cotransfected with the wild-type IAPE or the IAPE-MAIAP chimera, as described in Figure 3A, demonstrated a complete loss of infectivity for the latter, as expected from its now strictly intracellular localization. Conclusively, these data show that the sole modification of the N-terminal domain of the IAPE Gag protein results in both a shift in the site of IAPE VLP assembly, from the plasma membrane to the ER—as observed for bona fide IAP elements—and in the acquisition of retrotransposition activity. This transition most probably recapitulates the molecular events—together with the loss of a functional env gene—that have been associated with the emergence of a strictly intracellular retrotransposon from an infectious retrovirus.
Discussion
The present investigation has led to the characterization of the IAPE family of murine endogenous retroviruses and the identification of a fully active member of this family disclosing all the characteristics of a bona fide retrovirus. The identified element possesses all four gag, pro, pol, and env genes, and mutations introduced into each of them have shown that they are all required for a complete retroviral cycle. It also possesses non-coding sequences involved in RNA processing and export, with an identified Constitutive Transport Element within its 3′UTR and splice sites for the generation of subgenomic env transcripts. Electron microscopy analysis of the associated viral particles shows that IAPE behaves like a type-C retrovirus with particle formation and budding at the cell membrane and release of particles with an immature morphology, which then acquire a condensed central core, as commonly observed for gamma-retroviruses (Nermut and Hockley 1996; Vogt 1997). This morphological maturation is associated with Gag protein cleavage as shown by Western blotting using specific anti-Gag antibodies, and is further IAPE protease-dependent, with only immature particles—and uncleaved Gag—observed with a protease mutant that we constructed. Finally, the released particles are infectious and generate insertions into target cells with the canonical features of proviral integrations, full-length proviruses being flanked by target site duplications of a fixed length, 6 bp, that is, the same as observed for the resident genomic IAPE copies already present in the mouse genome. The identified IAPE provirus is therefore likely to be the prototypic element of the IAPE family, and its identified life cycle provides compelling evidence that amplification of this family occurred via a reinfection process, as already strongly suggested for a family of possibly still active human endogenous retroviruses, namely, the HERV-K(HML2) elements (Belshaw et al. 2004; Dewannieux et al. 2006). In this process, it is likely that packaging of IAPE transcripts with internal deletions and/or altered structures within infectious particles might have been responsible for the generation in trans of the presently observed IAPE subfamilies and especially of the previously described IAPE-B and -C subtypes (Reuss et al. 1996). Of note, one cannot exclude that the identified IAPE functional Env protein is further involved in the horizontal transfer of other endogenous retroviruses, with the present demonstration of its efficient pseudotyping of heterologous infectious retroviruses (Fig. 2C). Finally, the identification of a functional copy from the IAPE family of retroelements indicates that this family should still be active, and it therefore adds to the already characterized—and not so numerous—families of active endogenous retroviruses in mice, with the MLV and the MMTV elements (for review, see Boeke and Stoye 1997; Maksakova et al. 2006). Yet, despite the extensive screening that was performed, the IAPE-D1 copy was the only one found to be entirely coding and functional for infectious viral particle production, although we additionally identified four other proviral copies with entirely coding gag-pro-pol genes—but a defective env—that could possibly be active provided that a functional env gene is supplied in trans. Such a limited number of still active copies is not an unusual feature among retroelements, with, for instance, one of the most active family of mouse retroelements responsible for numerous insertional mutations in mice, that is, the MusD/ETn retrotransposons that contain only three active copies among the hundreds of such elements in the mouse genome (Ribet et al. 2004). Yet, one cannot exclude that the IAPE family of elements is on its way to extinction, and in this respect, it is also noteworthy that among the characterized de novo insertions of LTR-retroelements in the mouse genome, none corresponds to IAPE insertions but rather to MusD/ETn and IAP insertions (for review, see Ostertag and Kazazian 2001; Maksakova et al. 2006). In this respect, it is likely that IAPs are much more efficient insertional mutagens than IAPEs, and one of the important outcomes of the present investigation is precisely to have shown that IAPs can quite simply derive from IAPEs, via a limited number of molecular changes that we have identified and that have resulted in the transition from a bona fide infectious retrovirus to a very “successful” intracellular retrotransposon.
Actually, a second important issue of the present investigation is the demonstration that IAPs should be the descendants of the IAPEs, and the identification of the molecular determinants that most probably resulted in the transformation of a classical infectious retrovirus, with an extracellular life cycle, into a strictly intracellular retrotransposon. Indeed, the chimera that we generated from the identified active IAP and IAPE copies clearly demonstrates transformation of the intracisternal IAP particles into an infectious IAPE-like retrovirus, simply by replacing its Gag N-terminal domain with that of IAPE (and adding the IAPE Env in trans), whereas the infectious IAPE was transformed by reciprocal exchange into an IAP-like intracisternal particle with retrotransposition activity. For both chimeras, the specific sites of particle assembly, the extent of Gag processing, the structure of the particles, and their functional properties were “shifted” precisely to that found in the IAP and IAPE “native” counterparts.
In fact, it is a well-documented issue that the N-terminal domain of Gag retroviral proteins is an essential determinant of the virus life cycle and site of assembly, with a series of critical signals identified for these roles. Among them, the N terminus of most retrovirus Gag proteins possesses a myristylation signal (Rhee and Hunter 1987; Saad et al. 2006) that is responsible for the targeting of Gag (either already assembled in a cytoplasmic particle for type B/D retroviruses, or as single molecules that will assemble at the cell membrane for type C retroviruses). We actually identified such a signal within the functional IAPE copy that turned out to be essential for plasma membrane targeting of IAPE Gag and particle formation. Noteworthily, no myristylation signal could be identified in the IAP Gag protein. Yet, previous studies have shown that the IAP Gag N-terminal end is a signal peptide- (SP-) like sequence that is recognized by the signal recognition particle (SRP) translation machinery and targets the IAP Gag to the ER membrane (Fehrmann et al. 2003). In addition, its replacement by a plasma-membrane-targeting signal (from the Src protein) was shown to result in a readdressing of the IAP particles and their release from the cell (Welker et al. 1997).
The chimera experiments then strongly suggest that replacement of the IAPE Gag N-terminal domain has been the founding event in the generation of the IAP retrotransposon. In addition, this “intracellularization” has been associated with the loss of the env gene (with “traces” still visible in the IAP sequence) (see Fig. 1C), most probably due to the loss of any selective pressure for the maintenance of this gene, rendered useless in the now strictly intracellular life cycle of the newly generated element. Consistently, we have also shown that expression of an IAPE Env protein has no effect on the IAP life cycle, as expected, with no IAP particles being addressed to the plasma cell membrane by this sole addition (Fig. 5D; data not shown). Accordingly, the present investigation allows a recapitulation of the rather simple molecular events that have been associated with a major change in the retrovirus life style, still resulting in viable structures albeit with completely distinct modes of amplification. The IAPE elements have retained from their ancestral progenitor its infectivity, that is, their ability to effect cell-to-cell transfer and consequently to propagate their genome, whereas the IAP sequences are only capable of self-propagation within their host cell genome. Yet, the latter process is very efficient, most probably as a consequence of the sequestration of the particles within the cell, without “dilution” in the extracellular medium, a feature that makes them prone to direct reintegration—after reverse transcription—of their genome. Such a “physical” constraint, as similarly observed for classical bona fide retrotransposons such as the yeast Ty1 and Ty3 retrotransposons (for review, see Sandmeyer and Menees 1996) or for the MusD/ETn elements (Ribet et al. 2004, 2007), is most probably the key feature that allowed these elements to very efficiently transpose and consequently maintain within the host genomes, despite genetic drift, the active copies that can be found today. One important feature of the present investigation is that in the case of the intracellularized IAP retrotransposon, its infectious progenitor is still present and coexists with its “progeny,” in the form of the still active—although possibly close to extinction—IAPE family of elements.
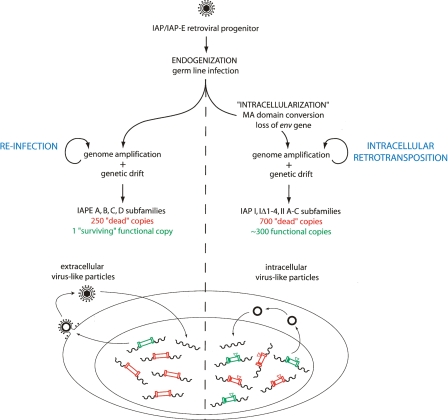
In summary, a model can be proposed (see the scheme in Fig. 7 and its accompanying legend) that summarizes the data and accounts for the generation of the IAP retrotransposon from an infectious retroviral progenitor. The model includes “intracellularization” of the endogenized retrovirus progenitor, via alteration of its canonical plasma membrane targeting signal and loss of its env gene rendered useless by the intracellular life cycle, which results in IAP elements with high transpositional efficacy and their conservation in the mouse genome. In parallel, evolution of the infectious IAPE progenitor has resulted in the “survival” of a single fully active copy—among a family of defective elements—that most probably represents a “living fossil” of the ancestral founder retrovirus.
Figure 7.
A model for the “intracellularization” of infectious retroviruses. A bona fide infectious retrovirus—for example, the IAP/IAPE progenitor—is “endogenized” upon infection of the germline of a remote ancestor and Mendelian transmission to the following generations, thus resulting in a so-called Endogenous RetroVirus (ERV) (de Parseval and Heidmann 2005; Bannert and Kurth 2006). This ERV may retain the characteristics of retroviruses, that is, produce infectious extracellular particles with a functional envelope protein, which are prone to horizontal transmission and can also amplify in their proper host genome by reinfection of the germline (the IAPEs). “Intracellularization” is expected to correspond to a further adaptation, in which the produced virus-like particles are no more able to exit or to re-enter the cell. For IAPs this has been achieved via the conversion of the plasma membrane targeting function of the Gag MA domain with the acquisition of a signal for Gag-targeting to the ER membrane, and the loss of the env gene. The resulting strictly intracellular life cycle resembles that of primitive Ty-like LTR-retrotransposons (although both classes of elements should be evolutionarily distinct) and is associated—at least in the case of the IAP element—with high-efficiency retrotransposition. For both types of elements, that is, the infectious IAPEs and the strictly intracellular IAPs, defective elements resulting from genetic drift (e.g., point mutation, deletion, recombination, reverse transcription error, DNA-editing) can be complemented in trans by active copies, resulting in the present-day multiple-copy IAP and IAPE subfamilies, with a single “surviving” functional IAPE element, but ∼300 full-length autonomous IAP copies still present due to their high “multicopying” activity.
Methods
Plasmids
All primers used are listed in Supplemental Table S1.
Each coding-competent IAPE env gene was PCR-amplified from the corresponding BAC DNA (BACPAC resources) by using the proofreading PfuTurbo Hotstart DNA polymerase (Stratagene) and appropriate primers. Each PCR product was then cloned in place of the VSV-G gene into the phCMV-VSV-G vector (GenBank accession no. AJ318514), opened by XhoI. The furin cleavage site mutant of the IAPE-D1 envelope glycoprotein was generated by PCR by mutating the canonical RQKR motif into AQAR using the QuickChange II XL Site-Directed Mutagenesis kit (Stratagene).
The R-U5 domain of the identified IAPE copies (AC123738, pos: 161181–152862 for IAPE-D1; AC131339, pos: 143356–135028 for IAPE-D2) was PCR-amplified from the corresponding BAC DNA and cloned 3′ to the transcription start site of the CMV promoter contained in a SacI–SalI fragment derived from pCMV-β (Clontech; pCMV-R-U5 plasmids). IAPE expression vectors were obtained by inserting SacII–SacII internal fragments of each IAPE copy, excised from BAC DNA, into the corresponding pCMV-R-U5 plasmid, opened by SacII. The defective neo-marked IAPE reporters were obtained by inserting the blunt-ended HindIII–XhoI fragment from pSVneo* (Esnault et al. 2002) into the IAPE expression vectors, between the two SpeI sites of the env gene (nucleotides 7186 and 7446), and by further introducing a premature stop codon into the gag gene at the MluI site (nucleotide 804) and a 3819-bp internal deletion (from nucleotides 1507–5325). The neoTNF-marked IAPE elements were obtained by inserting the blunt-ended HindIII–XhoI fragment from pSVneoTNF (Esnault et al. 2002) into the IAPE expression vectors, between the two SpeI sites of the env gene (nucleotides 7186 and 7446). The functional IAP copy (IAP-92L23) and the neoTNF-marked IAP reporter (with a premature stop codon introduced into the gag gene) are described in Dewannieux et al. (2004).
The IAP-MAIAPE chimera was obtained by replacing the 608–1180-nt fragment of the IAP copy (corresponding to the 5′ part of the gag gene) by the 683–1408-nt fragment of IAPE-D1 (the resulting IAP Gag product thus containing the first 242 N-terminal residues of IAPE Gag). The IAPE-MAIAP chimera was symmetrically obtained by replacing the 674–1417-nt fragment of the IAPE copy by the 473–1189-nt fragment of IAP-92L23 (the resulting IAPE Gag product thus containing the first 194 N-terminal residues of IAP Gag).
The pDM128/PL plasmid (Hope et al. 1990; Fridell et al. 1993) contains the cat gene and a polylinker flanked by a donor and an acceptor splice site. pDM128/MPMV-CTE contains the full-length CTE of MPMV inserted into the polylinker BglII site present in pDM128/PL (Bogerd et al. 1998). The IAPE 3′UTR and deletion derivatives were PCR-amplified and inserted into pDM128/PL between the BglII and KpnI sites of the polylinker.
Cell culture, transposition, and infectivity assay
Human HeLa and 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (VWR), 100 μg/mL streptomycin, and 100 U/mL penicillin. The day prior to transfection, cells were seeded at 5.5 × 105 or 7.5 × 105 cells per 60-mm dish for HeLa and 293T cells, respectively. Transfection of HeLa cells was performed with 3 μg of DNA using Lipofectamine (Invitrogen). Transfection of 293T cells was performed with 4.5 μg of DNA using the MBS transfection kit (Stratagene).
To assay for retrotransposition, transfected HeLa cells were expanded for 6 d, seeded at 5 × 105 cells per 100-mm dish and allowed to settle for 24 h before adding G418 (560 μg/mL; GIBCO BRL). After a 15-d selection, G418R foci were fixed, stained, and counted or individually picked and expanded.
To assay for infection, target HeLa cells were seeded in six-well plates at 1.2 × 105 cells per well the day prior to infection. Supernatants from transfected 293T cells were harvested 48 h post-transfection, filtered through 0.45-μm-pore-size PVDF membranes, supplemented with Polybrene (4 μg/mL), and transferred to target HeLa cells. To increase sensitivity, target cells were subjected to spinoculation at 1200g for 2.5 h at 25°C. After removal of the supernatants, cells were incubated in regular medium for 72 h at 37°C. HeLa cells were then split into 100-mm dishes (5 × 105 cells per dish), allowed to settle for 24 h, and subjected to G418 selection. G418R foci were fixed, stained, and counted or individually picked and expanded.
Reverse transcriptase activity assay
Supernatants of 293T cells were harvested 48 h post-transfection, centrifuged for 5 min at 1500 rpm, filtered through a 0.45-μm-pore-sized PVDF membrane, and used for a Product Enhanced Reverse Transcriptase (PERT) assay (Pyra et al. 1994). Briefly, the reverse transcriptase (RT) contained in 2.5 μL of supernatant was used to reverse-transcribe 0.3 μg of MS2 phage RNA, beforehand annealed to 16 pmol of RT-1 primer (5′-CACAGGTCAAA CCTCCTAGGAATG-3′). cDNA synthesis was then assayed using 1/50 of the RT reaction as a template for a 25-cycle PCR reaction conducted with RT-1 and RT-2 (5′-TCCTGCTCAACTTCCTGTC GAG-3′) primers, using Tth polymerase (Promega) and leading to the amplification of a 112-bp fragment.
Northern blot and RT-PCR
Total RNA was extracted from 293T transfected cells using a Nucleospin RNA II kit (Macherey Nagel). For Northern blot analysis, 10 μg of total RNA was fractionated on 1.0% (w/v) agarose gels in 1× MOPS (pH 7.0), 0.66 M formaldehyde buffer. RNAs were transferred to a positively charged nylon filter (Hybond N; Amersham) in 0.15 M NH4Ac buffer. Filters were pre-hybridized for 30 min, hybridized for 18 h at 65°C in Church solution (7% SDS, 1 mM EDTA at pH 8, 0.5 M Na2HPO4 at pH 7), and washed twice for 15 min at 65°C in 2× SSC, 0.1% SDS. The probe used was a 339-bp AvrII–PflmI fragment encompassing the IAPE 3′UTR and a part of 3′-LTR, 32P-labeled using a random priming kit (Amersham).
One microgram of total RNA of 293T transfected cells was reverse-transcribed in a 20-μL sample reaction using 50 U of Moloney murine leukemia virus RT (Applied Biosystems), 20 U of RNasin (Applied Biosystems), 1 mM dA/T/C/G (Amersham), 5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 10 mM KCl, and 15 pmol of primer IAPE-U3-R (5′-GCACGTAGATTGCAGCCCTTT-3′). PCR was carried out using IAPE-U3-R, IAPE-ENV-R (5′-CCTTT TAATTGGGCACTGAG-3′), and IAPE-U5-F (5′-ACGGGGATA AGCTCGGGATA-3′) primers and Expand Long template PCR system (Roche). Amplification products were then sequenced to identify splicing events.
IAPE antisera and Western blot analysis
IAPE Gag was detected using a previously described rabbit antiserum directed against the CA domain of the IAP MIA14 Gag protein (Dewannieux et al. 2004). To generate an anti-IAPE Env rabbit antiserum, a 737-bp fragment (nucleotides 2406–3142), corresponding to part of the SU moiety of the IAPE Env, had been PCR-amplified from the IAPE copy AC107817 (pos 125299–129927), and cloned into the pET19b vector (Novagen) opened by NdeI–BlpI, to allow prokaryotic expression as a histidine-tagged protein. The recombinant protein was then purified on a HiTrap column (Amersham) and used for rabbit immunization.
For analysis of IAP and IAPE proteins in cell extracts, transfected 293T cells were lysed 48 h post-transfection in Laemmli buffer, and whole-cell lysates were separated by SDS-PAGE using gels containing 12% polyacrylamide (37.5:1 ratio of acrylamide to N,N′-methylenebisacrylamide). Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and incubated with rabbit antisera against IAPE proteins.
For analysis of IAP and IAPE proteins in cell supernatants, the culture medium of transfected 293T cells was harvested 48 h post-infection, and the particulate material was concentrated by ultracentrifugation (1.5 h, 38,000 rpm) onto a 20% sucrose cushion and recovered in PBS. Virions were either treated by peptide-N-glycosidase F (PNGase-F; NEB Biolabs) or directly lysed in Laemmli buffer before analysis by SDS-PAGE and immunoblotting.
Characterization of IAPE de novo insertions sites
Integration sites were analyzed by inverse PCR using individual G418R HeLa clones obtained in the infectivity assay. Five micrograms of genomic DNA from each clone was digested by a mix of DraI, SacI, and RsrII, which should restrict the de novo integrated proviruses in the neo gene and in the 3′-flanking sequence at an unknown distance from the integration site. Restriction fragments were blunt-ended and self-ligated at a high dilution (500 μL) using 3200 U of T4 DNA Ligase (Biolabs). The circularized DNAs containing the neo gene, the IAPE 3′-LTR, and the 3′-flanking sequence were amplified by two nested PCRs using sets of divergent primers. Amplification products were cloned and sequenced thus yielding the 3′ target site sequence. Primers were then designed within the putative 5′-flanking region (according to the human genome sequence database at http://www.ncbi.nlm.nih.govsite), and the 5′-flanking region was finally PCR-amplified and sequenced. Primers are listed in Supplemental Table S1.
CAT activity assay
293T cells transfected with pDM128/PL or its derivatives containing IAPE sequences were lysed 48 h post-transfection in 0.25 M Tris-HCl (pH 7.0) by three freeze–thaw cycles. Cell extracts were incubated for 1 h at 37°C in the presence of 0.625 μCi/mL D-threo-[dichloroacetyl-1,2-14C]chloramphenicol (60 mCi/mmol; Perkin Elmer Life Sciences) and 1.25 mM N-acetyl-coenzyme A (Roche Diagnostics). Chloramphenicol and its derivatives were extracted with ethylacetate. Acetylated and unacetylated forms of chloramphenicol were separated by thin layer chromatography on TLC silica plates (Macherey-Nagel) using chloroform-methanol (19:1). The chloramphenicol acetyltransferase (CAT) activity was measured by PhosphorImager quantitation using a FLA-3000 scanner.
Immunofluorescence microscopy
Transfected HeLa cells were grown on 14-mm glass coverslips. Forty-eight hours post-transfection, cells for Gag detection were fixed in 1× PBS–4% paraformaldehyde at room temperature and permeabilized in PBS–0.1% SDS. After blocking in PBS–3% BSA, cells were immunostained with the anti-IAPE Gag polyclonal rabbit serum. After subsequent washes in PBS, cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) as secondary antibody. Cell nuclei were stained with TO-PRO-3 iodide (Molecular Probes). For Env detection and cell surface staining, incubation of the transfected cells with the anti-Env polyclonal rabbit serum was performed on living cells without prior fixation and permeabilization. Cells were then fixed in 1× PBS–4% paraformaldehyde and incubated with the secondary antibody as above. Fluorescence patterns were visualized with a Zeiss LSM 510 laser-scanning confocal microscope.
Electron microscopy
For ultrastructural studies, transfected cells were fixed in phosphate buffer (pH 7.2), 1.6% glutaraldehyde, for 1 h, and post-fixed in 0.1 M cacodylate buffer, 1% osmium tetroxyde, for 2 h. After being rinsed for 5 min in water and 15 min in 0.1 M cacodylate buffer, cells were transferred to 0.2 M cacodylate buffer for 30 min. Cells were washed in 30% methanol for 10 min, stained in 2% uranyl acetate in 0.1 M cacodylate buffer–30% methanol for 1 h, and washed in 30% methanol. Cells were then dehydrated through a graded ethanol series and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Zeiss 902 microscope at 80 kV.
For immuno-electron microscopy experiments, transfected cells were fixed for 1 h at 4°C with either 4% formaldehyde or 1.6% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3). Cell pellets were dehydrated in increasing concentrations of methanol and embedded in Lowicryl K4M at low temperature. Polymerization was carried out for 5 d at −30°C under long-wavelength UV light. Ultrathin sections of Lowicryl-embedded material were used for labeling with the rabbit antisera specific for IAPE Gag or Env proteins, followed by incubation with a goat anti-rabbit IgG conjugated to gold particles, 10 nm in diameter. Sections were contrasted with uranyl acetate.
Acknowledgments
We thank Bryan R. Cullen for pDM128 plasmids, A. Jalil for assistance with confocal microscopy, E. Pichard for technical assistance, and C. Lavialle for critical reading of the manuscript. This work was supported by the CNRS, by a grant from the Ligue Nationale contre Le Cancer (Equipe Labellisée), and by a fellowship from the Association pour la Recherche sur le Cancer to D.R.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073486.107.
References
- Baillie G.J., van de Lagemaat L.N., Baust C., Mager D.L., van de Lagemaat L.N., Baust C., Mager D.L., Baust C., Mager D.L., Mager D.L. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 2004;78:5784–5798. doi: 10.1128/JVI.78.11.5784-5798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N., Kurth R., Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Belshaw R., Pereira V., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Pereira V., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Katzourakis A., Talbot G., Paces J., Burt A., Tristem M., Talbot G., Paces J., Burt A., Tristem M., Paces J., Burt A., Tristem M., Burt A., Tristem M., Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S., Ruggieri A., Dewannieux M., Cosset F.L., Heidmann T., Ruggieri A., Dewannieux M., Cosset F.L., Heidmann T., Dewannieux M., Cosset F.L., Heidmann T., Cosset F.L., Heidmann T., Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J. Virol. 2004;78:1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Stoye J.P., Stoye J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J.M., et al., editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 343–435. [PubMed] [Google Scholar]
- Bogerd H.P., Echarri A., Ross T.M., Cullen B.R., Echarri A., Ross T.M., Cullen B.R., Ross T.M., Cullen B.R., Cullen B.R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval N., Heidmann T., Heidmann T. Human endogenous retroviruses: From infectious elements to human genes. Cytogenet. Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Dupressoir A., Harper F., Pierron G., Heidmann T., Dupressoir A., Harper F., Pierron G., Heidmann T., Harper F., Pierron G., Heidmann T., Pierron G., Heidmann T., Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- Dewannieux M., Harper F., Richaud A., Letzelter C., Ribet D., Pierron G., Heidmann T., Harper F., Richaud A., Letzelter C., Ribet D., Pierron G., Heidmann T., Richaud A., Letzelter C., Ribet D., Pierron G., Heidmann T., Letzelter C., Ribet D., Pierron G., Heidmann T., Ribet D., Pierron G., Heidmann T., Pierron G., Heidmann T., Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Casella J.F., Heidmann T., Casella J.F., Heidmann T., Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann F., Jung M., Zimmermann R., Krausslich H.G., Jung M., Zimmermann R., Krausslich H.G., Zimmermann R., Krausslich H.G., Krausslich H.G. Transport of the intracisternal A-type particle Gag polyprotein to the endoplasmic reticulum is mediated by the signal recognition particle. J. Virol. 2003;77:6293–6304. doi: 10.1128/JVI.77.11.6293-6304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennelly J., Harper K., Laval S., Wright E., Plumb M., Harper K., Laval S., Wright E., Plumb M., Laval S., Wright E., Plumb M., Wright E., Plumb M., Plumb M. Co-amplification to tail-to-tail copies of MuRVY and IAPE retroviral genomes on the Mus musculus Y chromosome. Mamm. Genome. 1996;7:31–36. doi: 10.1007/s003359900008. [DOI] [PubMed] [Google Scholar]
- Fridell R.A., Partin K.M., Carpenter S., Cullen B.R., Partin K.M., Carpenter S., Cullen B.R., Carpenter S., Cullen B.R., Cullen B.R. Identification of the activation domain of equine infectious anemia virus rev. J. Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold M.L. Constitutive transport element-mediated nuclear export. Curr. Top. Microbiol. Immunol. 2001;259:77–93. doi: 10.1007/978-3-642-56597-7_4. [DOI] [PubMed] [Google Scholar]
- Heidmann O., Heidmann T., Heidmann T. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell. 1991;64:159–170. doi: 10.1016/0092-8674(91)90217-m. [DOI] [PubMed] [Google Scholar]
- Hope T.J., Huang X.J., McDonald D., Parslow T.G., Huang X.J., McDonald D., Parslow T.G., McDonald D., Parslow T.G., Parslow T.G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: Mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuff E.L., Lueders K.K., Lueders K.K. The intracisternal A-particle gene family: Structure and functional aspects. Adv. Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- Maksakova I.A., Romanish M.T., Gagnier L., Dunn C.A., van de Lagemaat L.N., Mager D.L., Romanish M.T., Gagnier L., Dunn C.A., van de Lagemaat L.N., Mager D.L., Gagnier L., Dunn C.A., van de Lagemaat L.N., Mager D.L., Dunn C.A., van de Lagemaat L.N., Mager D.L., van de Lagemaat L.N., Mager D.L., Mager D.L. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H., Eickbush T.H. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- Mietz J.A., Grossman Z., Lueders K.K., Kuff E.L., Grossman Z., Lueders K.K., Kuff E.L., Lueders K.K., Kuff E.L., Kuff E.L. Nucleotide sequence of a complete mouse intracisternal A-particle genome: Relationship to known aspects of particle assembly and function. J. Virol. 1987;61:3020–3029. doi: 10.1128/jvi.61.10.3020-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermut M.V., Hockley D.J., Hockley D.J. Comparative morphology and structural classification of retroviruses. Curr. Top. Microbiol. Immunol. 1996;214:1–24. doi: 10.1007/978-3-642-80145-7_1. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H.J., Kazazian H.H.J. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Pyra H., Boni J., Schupbach J., Boni J., Schupbach J., Schupbach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc. Natl. Acad. Sci. 1994;91:1544–1548. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss F.U. Expression of intracisternal A-particle-related retroviral element-encoded envelope proteins detected in cell lines. J. Virol. 1992;66:1915–1923. doi: 10.1128/jvi.66.4.1915-1923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss F.U., Schaller H.C., Schaller H.C. cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J. Virol. 1991;65:5702–5709. doi: 10.1128/jvi.65.11.5702-5709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss F.U., Frankel W.N., Moriwaki K., Shiroishi T., Coffin J.M., Frankel W.N., Moriwaki K., Shiroishi T., Coffin J.M., Moriwaki K., Shiroishi T., Coffin J.M., Shiroishi T., Coffin J.M., Coffin J.M. Genetics of intracisternal-A-particle-related envelope-encoding proviral elements in mice. J. Virol. 1996;70:6450–6454. doi: 10.1128/jvi.70.9.6450-6454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.S., Hunter E., Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J. Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., Dewannieux M., Heidmann T., Dewannieux M., Heidmann T., Heidmann T. An active murine transposon family pair: Retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., Harper F., Dewannieux M., Pierron G., Heidmann T., Harper F., Dewannieux M., Pierron G., Heidmann T., Dewannieux M., Pierron G., Heidmann T., Pierron G., Heidmann T., Heidmann T. Murine MusD retrotransposon: Structure and molecular evolution of an “intracellularized” retrovirus. J. Virol. 2007;81:1888–1898. doi: 10.1128/JVI.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F., Tai J., Kim A., Ghanam R.H., Summers M.F., Kim A., Ghanam R.H., Summers M.F., Ghanam R.H., Summers M.F., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S.B., Menees T.M., Menees T.M. Morphogenesis at the retrotransposon-retrovirus interface: gypsy and copia families in yeast and Drosophila. Curr. Top. Microbiol. Immunol. 1996;214:261–296. doi: 10.1007/978-3-642-80145-7_9. [DOI] [PubMed] [Google Scholar]
- Vogt V.M. Retroviral virions and genome. In: Coffin J.M., et al., editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 27–69. [PubMed] [Google Scholar]
- Welker R., Janetzko A., Kräusslich H.G., Janetzko A., Kräusslich H.G., Kräusslich H.G. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J. Virol. 1997;71:5209–5217. doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]