Abstract
MicroRNAs (miRNAs) are emerging as important, albeit poorly characterized, regulators of biological processes. Key to further elucidation of their roles is the generation of more complete lists of their numbers and expression changes in different cell states. Here, we report a new method for surveying the expression of small RNAs, including microRNAs, using Illumina sequencing technology. We also present a set of methods for annotating sequences deriving from known miRNAs, identifying variability in mature miRNA sequences, and identifying sequences belonging to previously unidentified miRNA genes. Application of this approach to RNA from human embryonic stem cells obtained before and after their differentiation into embryoid bodies revealed the sequences and expression levels of 334 known plus 104 novel miRNA genes. One hundred seventy-one known and 23 novel microRNA sequences exhibited significant expression differences between these two developmental states. Owing to the increased number of sequence reads, these libraries represent the deepest miRNA sampling to date, spanning nearly six orders of magnitude of expression. The predicted targets of those miRNAs enriched in either sample shared common features. Included among the high-ranked predicted gene targets are those implicated in differentiation, cell cycle control, programmed cell death, and transcriptional regulation.
MicroRNAs (miRNAs) are short RNA molecules, 19–25 nucleotides (nt) in length, that derive from stable fold-back substructures of larger transcripts (Cai et al. 2004). In most cases, primary miRNA transcripts (pri-miRNAs) are cleaved by a complex of Drosha (currently known as RNASEN) and its cofactor DGCR8, producing one or more pre-miRNA hairpins, each with a 2-nt 3′ overhang (Lund et al. 2004). Nuclear export of these precursors is mediated by Exportin 5 (Lund et al. 2004), after which they are released as short double-stranded RNA duplexes following a second cleavage by Dicer1 (Lund et al. 2004). Based on the thermodynamic stability of each end of this duplex, one of the strands is thought to be preferentially incorporated into the RNA-induced silencing complex (RISC), producing a biologically active miRNA and an inactive miRNA* (O'Toole et al. 2006).
Once assembled within the RISC protein complex, miRNAs can elicit down-regulation of target genes by blocking their translation, inducing EIF2C2 (formerly AGO2) mediated degradation, or potentially inducing deadenylation (Aravin and Tuschl 2005; Giraldez et al. 2006). In animals, miRNA–target interactions occur through semicomplementary base pairing, usually within the 3′ untranslated region (UTR) of the target transcript. The relaxed complementarity between miRNAs and their target sites poses many problems in computational target prediction; hence, this is an active field of study with numerous emerging approaches that rely on differing methodologies and assumptions. Common target prediction algorithms assess complementarity of miRNAs to their targets (John et al. 2004), many enforcing strong pairing within the seed region of the miRNA (positions 2–8) (Lewis et al. 2005; Grimson et al. 2007). As they were discovered only relatively recently, the study of miRNAs is a young and rapidly changing field in which undiscovered genes remain and the sequence modifications and activity of mature miRNAs are not well understood.
Before the effect of miRNAs on gene regulation can be globally studied, a robust method for profiling the expression level of each miRNA in a sample is required. The current commercially available high-throughput methodologies rely on primers or probes designed to detect each of the current reference miRNA sequences residing in miRBase, which acts as the central repository for known miRNAs (Griffiths-Jones 2006). These systems, which are often available in array form, allow concurrent profiling of many miRNAs (Zhao et al. 2006). These approaches feature good reproducibility and facilitate clustering of samples by similar miRNA expression profiles (Davison et al. 2006; Porkka et al. 2007). However, probe-based methodologies are generally restricted to the detection and profiling of only the known miRNA sequences previously identified by sequencing or homology searches.
Sequencing-based applications for identifying and profiling miRNAs have been hindered by laborious cloning techniques and the expense of capillary DNA sequencing (Pfeffer et al. 2005; Cummins et al. 2006). Nevertheless, direct small RNA sequencing has several advantages over hybridization-based methodologies. Discovery of novel miRNAs need not rely on querying candidate regions of the genome but rather can be achieved by direct observation and validation of the folding potential of flanking genomic sequence (Berezikov et al. 2006a; Cummins et al. 2006). Direct sequencing also offers the potential to detect variation in mature miRNA length, as well as enzymatic modification of miRNAs such as RNA editing (Kawahara et al. 2007) and 3′ nucleotide additions (Aravin and Tuschl 2005; Landgraf et al. 2007). In contrast with capillary sequencing, recently available “next-generation” sequencing technologies offer inexpensive increases in throughput, thereby providing a more complete view of the miRNA transcriptome. With the added depth of sequencing now possible, we have an opportunity to identify low-abundance miRNAs or those exhibiting modest expression differences between samples, which may not be detected by hybridization-based methods. Next-generation miRNA profiling has already been realized in a few organisms (Rajagopalan et al. 2006; Fahlgren et al. 2007; Kasschau et al. 2007; Yao et al. 2007) using the massively parallel signature sequencing (MPSS) methodology (Nakano et al. 2006) and more recently the Roche/454 platform (Margulies et al. 2005). The recently released Illumina sequencing platform provides approximately two orders of magnitude greater depth than current competing technologies (Berezikov et al. 2006b), yielding up to several million sequences from a single flow cell lane (http://www.solexa.com).
We sought to use this new technology to extensively profile and identify changes in miRNA expression that occur within a previously characterized model system. Pluripotent human embryonic stem cells (hESCs) can be cultured under nonadherent conditions that induce them to differentiate into cells belonging to all three germ layers and form cell aggregates termed embryoid bodies (EBs) (Itskovitz-Eldor et al. 2000; Bhattacharya et al. 2004). Samples of undifferentiated hESCs and differentiated cells from EBs were chosen for miRNA profiling, first because the pluripotency of ESCs is known to require the presence of miRNAs (Bernstein et al. 2003; Song and Tuan 2006; Wang et al. 2007) and second because specific changes in miRNA expression are thought to accompany differentiation (Chen et al. 2007). The hESC messenger RNA transcriptome has been extensively studied by ourselves and others (Sato et al. 2003; Abeyta et al. 2004; Bhattacharya et al. 2004,2005; Boyer et al. 2005; Hirst et al. 2007), but to date, very little is known about the specific miRNAs that may play roles in their pluripotency or differentiation (Suh et al. 2004).
Results
Sequencing and annotation of small RNAs
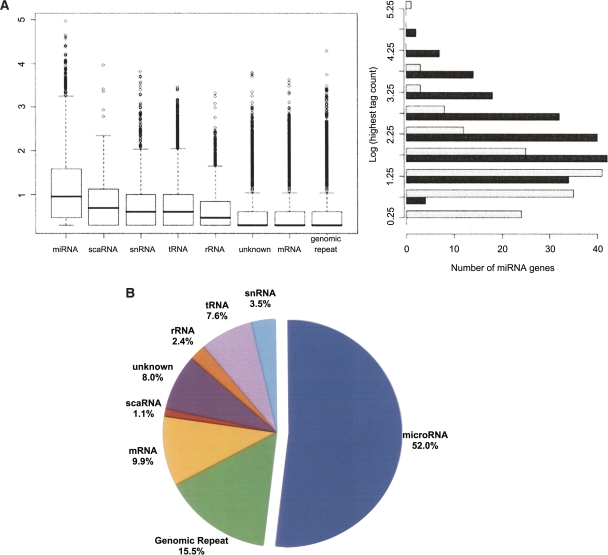
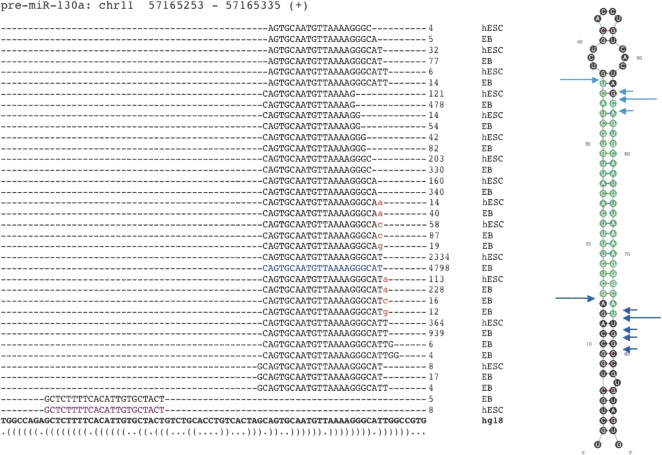
Sequencing of small RNA libraries yielded 6,147,718 and 6,014,187 37-nt unfiltered sequence reads from hESCs and EBs, respectively. After removal of reads containing ambiguous base calls, 5,261,520 (hESC) and 5,192,421 (EB) unique sequences remained with counts varying between 1 and 38,390. These reads were mapped to the genome by forcing perfect alignments beginning at the first nucleotide and retaining the longest region of each read that could be aligned to the reference genome, along with all alignment positions. After mapping, a total of 766,199 (hESC) and 724,091 (EB) unique error-free trimmed small RNA sequences were represented by 4,351,479 and 3,886,865 reads. Genomic positions of all non-singleton small RNA sequences can be viewed in our genome browser (http://microrna.bcgsc.ca/cgi-bin/gbrowse/hg18). Any sequence observed more than three times was considered a reliable representation of a small RNA molecule. These sequences were annotated as one of the known classes of small RNA genes or degradation fragments of larger noncoding RNAs (see Methods). Briefly, sequences were annotated based on their overlap with publicly available genome annotations including miRNAs, rRNAs, tRNAs, other small RNAs, and genomic repeats. Sequences deriving from 334 distinct miRNA genes were identified. The miRNAs were the most abundant class of small RNAs on average, but spanned the entire range of expression, with sequence counts up to ∼120,000 (Fig. 1A). The PIWI-associated RNAs (piRNAs), previously thought to exist only in mammalian germline cells, were also observed at relatively low levels. Searching the mapped sequences from the hESC and EB libraries against known piRNAs identified 460 and 378 unique sequences (including singletons) with counts of 9012 and 4606, respectively (median count = 2). These sequences correspond to 118 and 94 distinct known piRNAs (Supplemental Table 1). The sampling depth provided by one lane of the Illumina sequencing apparatus extends the dynamic range of miRNA expression within a cell, previously thought to span only ∼3 orders of magnitude (Berezikov et al. 2006a).
Figure 1.
Distributions of sequence counts for the different classes of small RNAs. (A) The box plot (left) shows the relative expression levels of sequences in each of the eight major classes of small RNAs (log10 transformation) in the hESC small RNA library. miRNAs were the most highly expressed class (mean sequence count = 253, median sequence count = 9). The most abundant miRNA in this library was miR-103, which had 91,398 instances of the most common isomiR in this library and nearly 120,000 in the matched library (EB). The highest log-transformed count between the two libraries (right) for all miRNAs identified as differentially expressed (black) is roughly normal (mean = 2.32, median = 2.17), representing a tag count of 1743 and 151, respectively. The miRNAs detected in at least one of the libraries but not significantly differentially expressed are shown in light gray for comparison. There is a slight enrichment of miRNAs with lower absolute expression in this group (mean = 1.35, median = 1.24), suggesting miRNAs with higher absolute expression levels are more likely to be identified as differentially expressed. (B) Total counts for the eight classes in the box plot are summarized. They are represented as a fraction of the total sequences that had at least one perfect alignment to the human reference genome (1,631,559 total).
Variability in microRNA processing
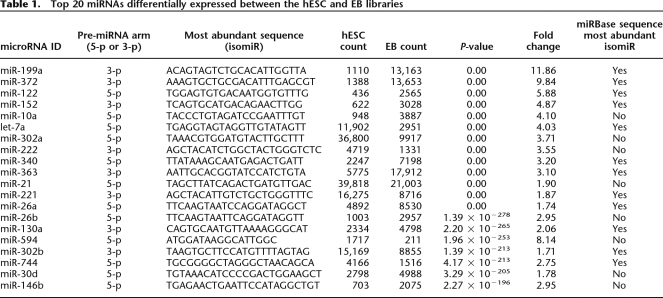
In both libraries, miRNAs frequently exhibited variation from their “reference” sequences, producing multiple mature variants that we hereafter refer to as isomiRs. In many cases, the miRNA* sequence and its isomiRs were also observed in our libraries. The existence of isomiRs (e.g., Fig. 2) is commonly reported in miRNA cloning studies but generally dismissed with either the sequence matching the miRBase record or the most frequently observed isomiR chosen as a reference sequence (Cummins et al. 2006; Ruby et al. 2006; Landgraf et al. 2007). It appears that much of the isomiR variability can be explained by variability in either Dicer1 or Drosha cleavage positions within the pre-miRNA hairpin (Fig. 2). All isomiRs of this class including their sequences and abundances are summarized in Supplemental Table 2. Notably, our data show that choosing a different isomiR sequence for measuring miRNA expression level can affect the ability to detect differential miRNA expression. Based on our analysis, the read count for the most abundant isomiR, rather than the miRBase reference sequence, provides the most robust approach for comparing miRNA expression between libraries (Supplemental Table 3; Supplemental Methods). In 107 cases, this most abundant sequence did not correspond exactly to the current miRBase reference sequence (Supplemental Methods). This suggests either that the relative abundance of isomiRs may vary across tissues or that the original submission of this miRNA to miRBase was incorrect. Following the most recent update to miRBase, far more of the most abundant sequences in our libraries corresponded with the updated miRBase reference, pointing toward the latter explanation.
Figure 2.
The repertoire of isomiRs and 3′ modifications of hsa-miR-130a. A diverse variety of isomiRs were observed for many of the known and novel miRNAs. Sequences representing the miR* were also commonly observed (highlighted in purple). The reference miRNA sequence from miRBase was, in many cases, not the most frequently observed isomiR (shown in blue). In this example, the most abundant miR* did not correspond to the current miRBase entry, although within the predicted pre-miRNA structure it derives from the correct 2-nt offset position relative to the reference miRNA (far right). Although variation at the 3′ end is generally much more common than at the 5′ end, target preference of the 5′ isomiRs may differ. Sequences with evidence of 3′ additions of nucleotides (red) were common, with certain miRNAs more heavily modified than others (summarized in Supplemental Table 4). The predicted structure of the pre-miRNA is represented at the bottom in dot-bracket notation, and as a graphic (far right). Experimentally inferred Drosha and Dicer1 cleavage positions are indicated in dark and light blue arrows, respectively. Large arrows represent cleavage sites for the most abundant isomiR whereas small arrows indicate those for the other isomiRs.
Enzymatic modification of microRNAs
Although some reads were detected that may represent the result of pre-miRNA editing by adenosine deaminases (observed as A to G transitions) or cytidine deaminases (producing C to U transitions), examples of these were infrequent and few were significantly above the background level of other apparent sequencing errors. The more prevalent type of modification noted among the miRNAs were single-nucleotide 3′ extensions, which were observed in multiple isomiRs from nearly all observed miRNA genes (316 miRNA genes). These modifications produce an isomiR that matches the genome at every position except the terminal nucleotide. The nucleotide most commonly added was adenine (1130 distinct examples), followed by uridine (1008 examples), cytosine (733 examples), then guanine (508 examples).
The prevalence of modifications differed among the miRNAs, with some miRNAs having more reads representing modified than unmodified forms. End modifications were not limited to the most common isomiR, nor did they show significant differences between the two libraries, suggesting that they may not bear direct significance in hESCs, but rather may reflect a general cellular process of miRNA modification. Figure 2 shows an example of 3′ modification, and Supplemental Table 4 summarizes the extensions that were most prevalent in the two libraries. For some of the miRNAs showing the highest ratios of modified/unmodified isomiRs, the same modification was previously observed across multiple human and mouse tissues (Landgraf et al. 2007). For example, hsa-miR-326 was found with a terminal adenine addition in 66% of the corresponding reads in both of our libraries, and multiple members of the family hsa-miR-30 showed uridine additions in more than half of their reads. Both of these specific modifications were noted in the previous study (Landgraf et al. 2007) where the same modifications were also observed in the orthologous mouse miRNAs. Library preparation involves a ligation reaction mediated by T4 ligase in the presence of ATP (see Methods), which might explain the addition of adenosines. However, to our knowledge this cannot explain the addition of other nucleotides nor the apparent preference for 3′ extension of some miRNAs over others.
MicroRNAs differentially expressed between hESCs and EBs
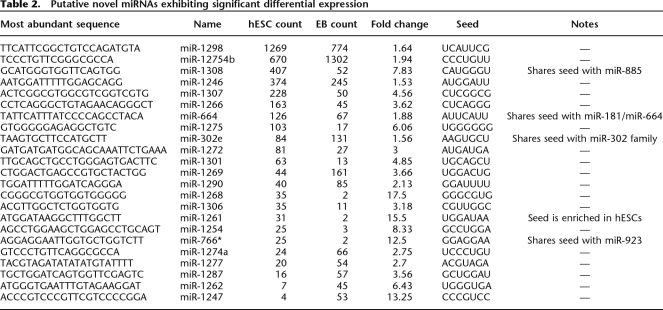
We tested for differential expression between the hESC and EB libraries using all sequences, regardless of their annotation; hence, each isomiR was independently tested for differential expression. The total number of differentially expressed sequences corresponding to the miRBase reference sequence (119 total from 100 miRNA genes) was less than the total number of miRNAs identified as differentially expressed using the most abundant isomiR for each pre-miRNA arm (171 total from 145 miRNA genes) (Table 1; Supplemental Table 5). All 100 of the miRNA genes identified as differentially expressed, based only on the sequences corresponding perfectly with those in miRBase, were among the 145 identified by the most abundant sequence in our data. Employing the count of the most abundant sequence for the calculation of differential expression also allows identification of the supposed miR* sequences that are differentially expressed between hESCs and EBs but are currently absent from miRBase (22 total). Taken together, this reinforces the concept that the most abundant, rather than the reference miRNA sequence, is the most useful for identifying differentially expressed miRNAs.
Table 1.
Top 20 miRNAs differentially expressed between the hESC and EB libraries
Novel microRNA genes
We sought to identify novel miRNA genes among the unclassified sequences in our libraries. After annotation, 40,699 of the small RNA sequences in the hESC library and 40,598 in the EB library (ignoring singletons) remained unclassified because they derived from unannotated regions of the human genome. As they were of generally low expression and often similar in sequence to known genomic repeats, we assumed many of these were either randomly derived fragments of various RNAs as well as unidentified piRNAs or regulatory siRNAs (Berezikov et al. 2006b; Watanabe et al. 2006). To identify candidate novel miRNAs among these, we employed both in-house and publicly available algorithms (see Methods). First, small hot-spot regions of the genome with alignments to more than one small RNA in our library were identified, followed by folding flanking genomic sequence with RNALfold. In parallel, we classified these folded structures using a publicly available miRNA classifier (MiPred) and an in-house classifier based on a support vector machine (SVM). Once good candidates were identified, we compared both their mature sequences and the predicted pre-miRNA sequences to those of all currently known miRNAs to aid in classifying them into families. The total set of novel miRNA candidates, included in Supplemental Table 6, comprises 83 unique miRNA sequences from up to 104 distinct genes. Compared with other miRNAs in our data, these miRNAs exhibited modest expression (mean count = 45), perhaps indicating that most of them may not perform significant functions in these cell types. However, 23 exhibited significant differential expression between the two libraries and are thus potentially biologically important in the cells in which they were present (Table 2).
Table 2.
Putative novel miRNAs exhibiting significant differential expression
Targets of differentially expressed microRNAs
Strong base pairing between the seed region of a miRNA and the UTR of its target mRNA is important for its activity; hence, many target prediction algorithms enforce strong seed complementarity and evolutionary conservation in the complementary region of potential targets (Grimson et al. 2007). As such, the repertoire of predicted targets of miRNAs with identical seeds often overlaps considerably. We sought to determine whether miRNAs sharing identical seeds demonstrated coexpression. We identified 1009 distinct seed sequences when considering all unique isomiRs from both libraries. Many of these represented the noncanonical seeds of isomiRs arising from variation at their 5′ end. One hundred fourteen of these seeds were significantly over-represented in the hESC library, while 106 are over-represented in the EB library (Fisher Exact Test, alpha = 0.05/1009) (Table 3; Supplemental Table 7).
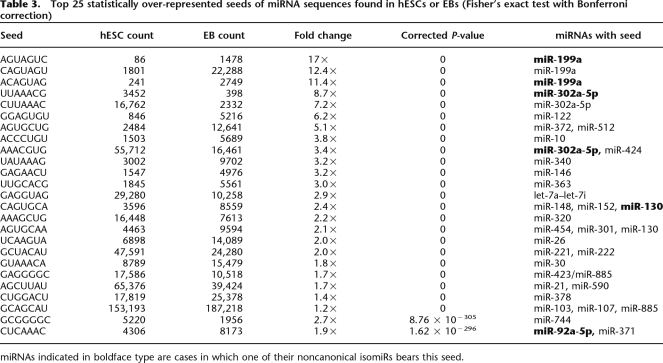
Table 3.
Top 25 statistically over-represented seeds of miRNA sequences found in hESCs or EBs (Fisher’s exact test with Bonferroni correction)
miRNAs indicated in boldface type are cases in which one of their noncanonical isomiRs bears this seed.
As expected, we found that many of the seeds with the largest changes in relative levels between the two libraries corresponded with the differentially expressed miRNAs in Table 1. Notably, in some cases, we observed pairs of miRNAs that shared a common seed yet exhibited inverse expression changes. A clear example of this from our data is the inverse abundances of hsa-miR-302a (hESC-enriched) and hsa-miR-372 (EB-enriched), which share the seed sequence AAGUGCU. This behavior may mask the effect of differential expression of some miRNAs in each library. Hence, focusing on the net change in seed levels, rather than distinct miRNAs, may be important in this context. Some miRNAs appear in this table more than once, suggesting that more than one isomiR from a single miRNA gene can contribute to net changes in miRNAs with a given seed during differentiation, and these isomiRs could potentially regulate different sets of transcripts.
The cooperative action of multiple miRNAs can be multiplicative and, in some cases, synergistic (Grimson et al. 2007); hence, transcripts with more predicted target sites for coexpressed miRNAs should be most drastically affected by those miRNAs. In an attempt to highlight potentially significant targets of differentially expressed miRNAs, we identified genes with predicted target sites for multiple hESC-enriched miRNAs or EB-enriched miRNAs using TargetScan. Measures were taken to compensate for UTR length, miRNAs with identical target sites, and a general preponderance of target sites in some genes (see Methods). A total of 591 likely cooperative targets of EB-enriched and 461 targets of hESC-enriched miRNAs were identified by this approach. As these genes are likely under redundant post-transcriptional regulation by multiple miRNAs, they could comprise genes of central importance to the maintenance of these cells. Surprisingly, these two gene lists showed a significant overlap of 64 genes (P < 0.0001, permutation test), suggesting that some of the miRNA-regulated genes in hESCs may also be regulated in EBs by a different set of miRNAs.
The genes that have been highlighted herein as likely targets of the differentially expressed miRNAs would be expected to be significant to hESC biology. This was supported by examining the significant Gene Ontology (GO) “biological process” classifications that are over-represented among these genes. This analysis revealed that many of the genes in both groups have been previously associated with differentiating stem cells and included those involved in differentiation, development, and regulation of transcription (Skottman et al. 2005). Interestingly, some of the biological processes were enriched only in the predicted targets of hESC-enriched or EB-enriched miRNAs. For example, genes involved in programmed cell death were enriched among the predicted targets of hESC-enriched miRNAs while those involved in cell proliferation were enriched among the predicted targets of EB-enriched miRNAs (Figure 3).
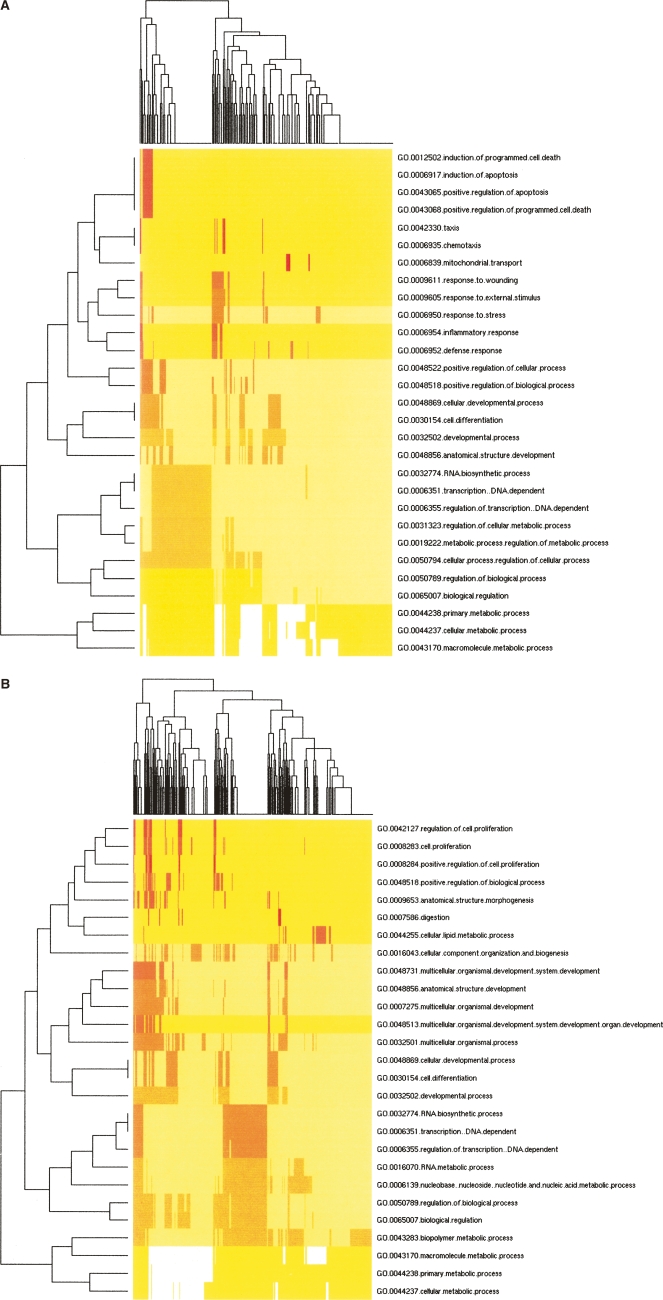
Figure 3.
Clustering of over-represented Gene Ontology (GO) classes in predicted targets of differential microRNAs. Shown are heat map representations of GO (Ashburner et al. 2000) terms over-represented among predicted cooperative targets (Y-axis) of hESC-enriched miRNAs (A) and EB-enriched miRNAs (B). All genes with statistically over-represented GO annotations were included (P < 0.01, X-axis) as identified by GoStat (Beissbarth and Speed 2004). GO terms common to both sets of genes were those involved in transcriptional regulation, differentiation, and development. Those GO terms unique to hESC-enriched miRNA targets were associated with programmed cell death, response to stress, and cell motility while those unique to EB-enriched miRNA targets describe various aspects of cell proliferation regulation as well as nucleotide and nucleic acid metabolism.
Discussion
These data highlight the potential of a new massively parallel sequencing strategy to profile miRNA expression in differentiating hESCs. Between the two libraries, we identified 171 differentially expressed known and 23 novel miRNAs corresponding with 168 distinct miRNA genes. This is a striking advance in contrast with a recent array-based study of murine ESCs, which revealed a much smaller number of miRNAs differentially expressed between ESCs and EBs (Chen et al. 2007). The latter study identified only 23 candidate miRNAs as either ESC-specific or down-regulated during differentiation, as well as 10 miRNAs that appear to be enriched in EBs. It is encouraging that, among the 27 listed mouse miRNAs with identifiable human orthologs, 14 exhibited expression patterns consistent with the hESC results presented here, while many of the others exhibited insignificant changes in expression. Further, of the additional differential miRNAs reported here (Table 1), many were originally discovered in human or murine ESCs (Houbaviy et al. 2003; Suh et al. 2004).
Some of these previous studies generalized these miRNAs as hESC-specific in their expression. However, compared with EBs, our method detected only two miRNAs that appeared exclusively in hESCs (miR-486 and miR-187); this is likely a consequence of the increased sampling depth of the method used here. On the other hand, we found 14 miRNAs in EBs (all with counts of at least 10) that were not observed in hESCs (Supplemental Table 5). This is consistent with the postulate that the number of expressed miRNAs increases during differentiation (Strauss et al. 2006) and further supports the importance of miRNAs during ESC differentiation (Wang et al. 2007). However, because EBs likely represent a more diverse population of cells, the total numbers of expressed miRNAs in any given cell type may not necessarily increase. Thus, the miRNAs highlighted by this method include those previously implicated in pluripotency and differentiation, but the added sampling depth demonstrates that most of these miRNAs are not uniquely expressed by undifferentiated hESCs. This result may suggest that the processes involved in differentiation that are under miRNA-mediated regulation are dictated by the repertoire and relative miRNA levels, rather than the number of distinct miRNAs being expressed.
With few exceptions (Berezikov et al. 2006b), recent large-scale cloning efforts have provided minimal yields of new miRNA genes. This may be due to the dominance of the highly expressed miRNAs in small RNA libraries as well as difficulties associated with library normalization (Shivdasani 2006; Landgraf et al. 2007). Here, we present 104 novel miRNA genes that have passed multiple levels of annotation criteria including expression andfolding verification. This gene list was obtained by combining two separate miRNA classification tools relying on different assumptions and input data. MiPred relies on a Random Forest (RF) algorithm to classify miRNAs based on structural properties (Jiang et al. 2007). Our custom classifier, which uses an SVM, assigns a classification score based on a separate set of structural descriptors as well as data specific to this sequencing method. As a result, this classifier has provided the best classification accuracy of any machine learning-based miRNA method (see Methods).
At least 23 of the novel miRNAs identified in this study show evidence of regulated expression during hESC differentiation (Table 2), suggesting that they may also have roles in maintaining the pluripotent status of hESCs or their ability to self-renew. In support of this, five of these miRNA sequences were found to share a common seed sequence with at least one of the differentially expressed known miRNAs (Table 2). Many of the remaining novel miRNAs with overall low abundance and insignificant expression differences may later prove unimportant in the context of hESC biology. It is encouraging, however, that many of the known miRNAs reside in the same range of expression as these novel miRNAs (Fig. 1A, black bars), suggesting that many miRNAs may exhibit low-level expression in hESCs detectable only by deep sequencing. Like many of the known miRNAs present in these libraries, these low-abundance novel miRNAs may be observed in higher quantities among other tissues where they are functionally important.
In addition to novel miRNA genes, this study has aided in identifying a diverse population of variants of known miRNAs, collectively termed isomiRs. The functional implications of the widespread 3′ modifications (Supplemental Table 4) and RNA editing are unclear in the context of hESCs as neither process appeared to vary significantly between the two libraries. It is important to note that the diverse 3′ nucleotide additions observed here reiterate previous observations of this nature (Landgraf et al. 2007). By comparing some of the most prevalent 3′ additions in our libraries with those from diverse human and mouse tissues, it is evident that the nature of the added nucleotide is nonrandom and evolutionarily conserved.
Some of the remaining noncanonical isomiRs, which appear to derive from variability in Dicer1 and Drosha cleavage sites, were quite abundant. By including all isomiRs and grouping those that share an identical seed sequence, we revealed 222 of seeds that were represented in significantly different quantities these two libraries (Table 3; Supplemental Table 7). As the noncanonical isomiRs share most of their sequence with the most highly expressed isomiRs, it is possible that these variants share a common set of targets with the canonical miRNAs. Those isomiRs resulting from variation at the 5′ end may be of particular interest as they bear a different seed sequence than the reference miRNA, thus indicative of their potential to target different transcripts. However, whether any of these noncanonical variants associate within RISC remains to be experimentally determined. If they are found to associate within RISC, the presence of isomiRs may have implications in future annotation of miRNAs and the development of new target prediction algorithms. A recent update to miRBase resulted in better agreement between the most abundant sequences in our libraries, but some of the most prevalent isomiRs in our libraries still do not perfectly correspond to the miRBase entry. This suggests that the most abundant sequences have yet to be reliably determined for all miRNAs. If this is the case, further large-scale efforts such as this should result in the truly most common isomiR replacing the erroneous sequences residing in miRBase.
There was a large overlap of predicted target genes for the most prevalent miRNAs enriched in either hESCs or EBs. Since a large proportion of these genes were predicted by TargetScan to be targets of numerous miRNAs, genes were weighted based on their total number of predicted target sites. By taking only the genes with weights above a certain threshold, we derived two relatively small groups of genes that are strong candidates for cooperative targeting by hESC-enriched or EB-enriched miRNAs. This enabled resolution of key gene classes in each group (Fig. 3), but it does not assert that these are true in vivo targets, nor does it provide a comprehensive list of the targets of these miRNAs. Further directed validation and gene expression profiling will be necessary to elucidate the true in-vivo targets. These lists do, however, provide the foundation for an alternate view of gene regulation in hESCs. Previously, focus has been placed on changes to mRNA levels during differentiation. By shifting focus to the genes targeted by miRNAs in this context, it is plausible that genes previously unlinked to differentiation and pluripotency may be discovered.
Adaptation of a common adapter ligation method to a novel sequencing platform has allowed us to generate a view of the small RNA component of differentiating hESCs at an unprecedented depth. Following a combination of novel miRNA annotation and discovery techniques, we have revealed the largest list to date of miRNA sequences. This list includes a diverse population of miRNA variants, termed isomiRs, with variation at both the 5′ and 3′ ends. We also present numerous novel human miRNAs, some of which may be important in the control of hESC pluripotency or differentiation. Notably, many of miRNAs exhibiting the most significant differences between hESCs and EBs appear to regulate genes involved in transcriptional regulation, differentiation and development.
Methods
Small RNA library preparation
Undifferentiated H9 hESCs (Hirst et al. 2007) were cultured on Matrigel (BD Biosciences) coated dishes in maintenance medium consisting of Dulbecco’s Modified Eagle Medium (DMEM)/F12 containing 20% Knockout Serum Replacer (Invitrogen), 0.1 mM β-mercaptoethanol, 0.1 mM nonessential amino acids, 1 mM glutamine, and 4 ng/mL FGF2 (R&D Systems) and conditioned by mitotically inactivated mouse embryonic fibroblasts (Xue et al. 2005). For EB differentiation, hESCs were harvested via 0.05% trypsin (Invitrogen) supplemented with 0.5 mM CaCl2, and the resultant cell aggregates were cultured in nonadherent dishes (BD Biosciences) for up to 30 d in maintenance medium lacking FGF2 (medium changes performed as necessary) (Itskovitz-Eldor et al. 2000; Dvash et al. 2004). At appropriate time points, RNA was extracted into TRIzol, and aliquots of total RNA from day 0 (hESC) and day 15 (EB) were subjected to miRNA library construction as follows.
For each library, 10 μg of DNase I (DNA-free kit; Ambion) treated total RNA was size fractionated on a 15% tris-borate-EDTA (TBE) urea polyacrylamide gel and a 15–30 base pair fraction was excised. RNA was eluted from the polyacrylamide gel slice in 600 μL of 0.3 M NaCl overnight at 4°C. The resulting gel slurry was passed through a Spin-X filter column (Corning) and precipitated in two 300-μL aliquots by the addition of 800 μL of ethanol and 3 μL of mussel glycogen (5 mg/mL; Invitrogen). After washing with 75% ethanol, the pellets were allowed to air dry at 25°C and pooled in diethylpyrocarbonate (DEPC) water. The 5′ RNA adapter (5′-GUUCAGAGUUCUACAGUCCGAC GAUC-3′) was ligated to the RNA pool with T4 RNA ligase (Ambion) in the presence of RNase Out (Invitrogen) overnight at 25°C. The ligation reaction was stopped by the addition of 2× formamide loading dye. The ligated RNA was size fractionated on a 15% TBE urea polyacrylamide gel, and a 40–60 base pair fraction was excised. RNA was eluted from the polyacrylamide gel slice in 600 μL of 0.3 M NaCl overnight at 4°C. The RNA was eluted from the gel and precipitated as described above followed by resuspension in DEPC-treated water.
The 3′ RNA adapter (5′-pUCGUAUGCCGUCUUCUGC UUGidT-3′; p, phosphate; idT, inverted deoxythymidine) was subsequently ligated to the precipitated RNA with T4 RNA ligase (Ambion) in the presence of RNase Out (Invitrogen) overnight at 25°C. The ligation reaction was stopped by the addition of 10 μL of 2× formamide loading dye. Ligated RNA was size fractionated on a 10% TBE urea polyacrylamide gel, and the 60–100 base pair fraction was excised. The RNA was eluted from the polyacrylamide gel and precipitated from the gel as described above and resuspended in 5.0 μL of DEPC water. The RNA was converted to single-stranded cDNA using Superscript II reverse transcriptase (Invitrogen) and Illumina’s small RNA RT-Primer (5′-CAAGCAGAAGACGGCATACGA-3′) following the manufacturer’s instructions. The resulting cDNA was PCR-amplified with Hotstart Phusion DNA Polymerase (NEB) in 15 cycles using Illumina’s small RNA primer set (5′-CAAGCAGAAGACG GCATACGA-3′; 5′-AATGATACGGCGACCACCGA-3′).
PCR products were purified on a 12% TBE urea polyacrylamide gel and eluted into elution buffer (5:1, LoTE: 7.5 M ammonium acetate) overnight at 4°C. The resulting gel slurry was passed through a Spin-X filter (Corning) and precipitated by the addition of 1100 μL of ethanol, 133 μL of 7.5 M ammonium acetate, and 3 μL of mussel glycogen (20 mg/mL; Invitrogen). After washing with 75% ethanol, the pellet was allowed to air dry at 25°C and dissolved in EB buffer (Qiagen) by incubation at 4°C for 10 min. The purified PCR products were quantified on the Agilent DNA 1000 chip and diluted to 10 nM for sequencing on the Illumina 1G.
Small RNA genome mapping and quantification
Virtually no reads aligned to the genome after position 28, so we trimmed all reads at 30 nt to reduce the number of unique sequences. We counted the occurrences of each unique sequence read and used only the unique sequences for further analysis. We aligned each sequence to the human reference genome (NCBI build 36.1) using Mega BLAST (version 2.2.11). We filtered these alignments and retained only those that included the first nucleotide of the read and were devoid of insertions, deletions, and mismatches. For every read, the longest alignment was determined, and this subsequence, as well as the positions for every alignment of this length, was stored in a database (to a maximum of 100 alignments). The sequence following the aligned region was checked for presence of the 3′ linker sequence. The counts for all reads containing the same aligned subsequence were summed to provide a metric of the total frequency of that small RNA molecule in the original RNA sample. The presence and length of intervening sequences between the alignment and the linker was recorded to enable identification and separate quantification of small RNAs with 3′ additions.
Differential expression detection
All unique small RNA sequences were compared between the two libraries (hESC and EB) for differential expression using the Bayesian method developed by Audic and Claverie (1997). This approach was developed for analysis of digital gene expression profiles and accounts for the sampling variability of tags with low counts. Sequences were deemed significantly differentially expressed if the P-value given by this method was <0.001 and there was at least a 1.5-fold change in sequence counts between the two libraries. Unless stated otherwise, the most frequently observed isomiR was used as the diagnostic sequence for comparison of miRNA expression between libraries.
Small RNA annotation
Each small RNA sequence was annotated if its full sequence contained one or more nucleotides of recognizable 3′ linker sequence, had at most 25 perfect full-length alignments to the human genome, and was detected at least twice. Genomic positions of each sequence were compared with genome annotations obtained from the UCSC Genome Browser download page (http://hgdownload.cse.ucsc.edu/downloads.html). The data used for annotation included the Ensembl genes, RepeatMasker, and sno/miRNA tables. The positions of all miRNA genes were also downloaded from miRBase and used for this positional annotation (http://microrna.sanger.ac.uk/sequences/ftp.shtml). Sequences overlapping annotations from these tables were classified into one of the following classes, listed in the priority used: miRNA, tRNA, rRNA, scaRNA, CDBox, scRNA, snoRNA, snRNA, srpRNA, genomic repeat, or known transcript (exonic or intronic). All mapped sequences were also searched against the currently known human piRNAs (Aravin et al. 2007) using sequences retrieved from piRNABank (Lakshmi and Agrawal 2007). As piRNAs are considerably longer than miRNAs, we used the first 18 nt of the piRNAs to search for perfect matches among our sequences. Sequences that did not overlap any of these annotations (in any one of their genomic positions) or that had more than 25 perfect alignments to the genome were automatically classified as “unknown.” The unknown sequences with 25 or fewer perfect alignments to the genome were used, along with those corresponding to introns and genomic repeats, for novel miRNA prediction. Sequences identified as miRNAs were named based on the specific miRNA gene they overlapped as well as the arm (either 5′ or 3′) with respect to the pre-miRNA. Where miRNA naming was ambiguous due to identical sequences shared by related miRNA genes, sequences were named arbitrarily by one of their possible parent miRNA genes. For those miRNA genes producing identical mature sequences (e.g., hsa-miR-9-1), the trailing number was dropped from the name. MiRBase (release 10.0) reference sequences were used for the comparison of the common isomiRs to the reference miRNA sequences.
Novel microRNA prediction
Candidate miRNA gene loci were identified by finding distinct small RNA sequences lacking annotations that shared partially overlapping genomic positions on the same strand (termed “hotspots”). Three hundred nucleotides of genomic sequence flanking each seed was extracted, reverse-complemented where appropriate, and folded using RNALfold (Hofacker 2003), which identifies locally stable substructures within a query RNA sequence. The largest unbranched fold-back substructure was identified from each structure, and redundant structures were then removed. Structures were also removed if the seed region (where the original small RNA sequences derived) spanned the loop. The remaining structures and their sequences comprised a set of candidate miRNA genes with expressed sequence and more than one candidate isomiR. The putative pre-miRNA sequences were then enriched for likely real pre-miRNA hairpins using two machine learning approaches. The previously published method, termed MiPred, relies on an RF algorithm (Jiang et al. 2007) and uses a combination of structural and thermodynamic parameters. The second approach was devised specifically for its application in this study and employs a SVM classifier and mostly uses parameters not used by the RF method.
The SVM functionality was provided in the e1071 package for R. The parameters used by this classifier include the relative proportion of each nucleotide and the number of each type of base pair in the optimal pre-miRNA folded structure. As well, some parameters describing the folded structure are common to other SVM approaches including minimum folding energy index (MFEI), adjusted minimum folding energy (AMFE) (Zhang et al. 2006), normalized pairing propensity (Ng Kwang Loong and Mishra 2007), and loop length. The parameters that are unique to this type of sequencing data are descriptors of the seed itself and its positioning within the pre-miRNA structure. Specifically, these include seed length, positioning with respect to the loop, as well as the ratio of paired to unpaired nucleotides within the seed. This classifier was trained using the folded flanking sequence of the miRNAs present in the two libraries for positive examples and the folded flanking sequences of small RNA sequences classified as either tRNA, rRNA, snRNA, or snoRNA for negative examples. After training, the classifier showed a sensitivity of 0.973 and specificity of 0.988, which represents the upper level of discrimination of pre-miRNAs reported for any machine learning method to date.
The intersection of the positive predictions of the SVM and RF methods was used as an initial set of reliable novel miRNA predictions. This was supplemented with miRNAs showing either significant differential expression between the two libraries, significant sequence similarity to known miRNAs, or overlap with an EvoFold prediction for an evolutionarily conserved hairpin (Pedersen et al. 2006).
Cooperative miRNA target prediction
TargetScan (release 4.0) predicted targets for known miRNAs were downloaded from the download page (http://www.targetscan.org/). Only miRNAs with counts of at least 100 in either hESC or EB were included in target analyses. Genes with target sites for at least two coexpressed miRNAs (either hESC-enriched or EB-enriched) were identified as potential cooperative targets. To compensate for potential bias, genes with numerous predicted miRNA target sites were given a lower rank than those with few predicted target sites. The rank score of a gene was calculated by dividing the number of target sites for coexpressed miRNAs by the total number of target sites for that gene. We used a cutoff of 0.15 (rank) to produce the two sets of high-ranked candidate cooperative targets of hESC-enriched and EB-enriched miRNAs (Fig. 3).
GO analysis
GoStat (Beissbarth and Speed 2004) was employed for identification of significantly enriched “biological process” GO (Ashburner et al. 2000) terms in the two lists of likely targets of hESC-enriched and EB-enriched miRNAs (P < 0.01). Custom software was used to extract genes and from the GoStat output. Genes and GO terms were clustered using hierarchical clustering in R. Heat maps were created in R using the heatmap function (default parameters).
Acknowledgments
We thank the staff at Illumina, Inc. for technical assistance. We also thank Rhonda Oshanek of the Genome Sciences Centre for her contribution in editing the draft of this manuscript. This project was funded, in part, by the Canadian Stem Cell Network with cofunding from Stem Cell Technologies, Inc as well as the BC Cancer Foundation. M.A.M. is a Michael Smith Foundation for Health Research Senior Scholar. R.D.M. receives stipends from the Michael Smith Foundation for Health Research and the Canadian Institutes for Health Research. M.O.C. is a recipient of a StemCell Technologies-sponsored Canadian Institutes of Health Research (CIHR) Industrial Postdoctoral Fellowship. F.K. is supported by the Deutsche Forschungsgemeinschaft Germany (grant no. Ku 2288/1-1).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7179508.
References
- Abeyta M.J., Clark A.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Clark A.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Rodriguez R.T., Bodnar M.S., Pera R.A., Firpo M.T., Bodnar M.S., Pera R.A., Firpo M.T., Pera R.A., Firpo M.T., Firpo M.T. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum. Mol. Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Aravin A., Tuschl T., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J., Girard A., Fejes-Toth K., Hannon G.J., Fejes-Toth K., Hannon G.J., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S., Claverie J.M., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P., Speed T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. Approaches to microRNA discovery. Nat. Genet. 2006a;38:S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Thuemmler F., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., van Laake L.W., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Kondova I., Bontrop R., Cuppen E., Plasterk R.H., Bontrop R., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006b;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Elledge S.J., Anderson K.V., Hannon G.J., Anderson K.V., Hannon G.J., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B., Miura T., Brandenberger R., Mejido J., Luo Y., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Miura T., Brandenberger R., Mejido J., Luo Y., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Brandenberger R., Mejido J., Luo Y., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Mejido J., Luo Y., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Luo Y., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Yang A.X., Joshi B.H., Ginis I., Thies R.S., Amit M., Joshi B.H., Ginis I., Thies R.S., Amit M., Ginis I., Thies R.S., Amit M., Thies R.S., Amit M., Amit M., et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B., Cai J., Luo Y., Miura T., Mejido J., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Cai J., Luo Y., Miura T., Mejido J., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Luo Y., Miura T., Mejido J., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Miura T., Mejido J., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Mejido J., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Brimble S.N., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Zeng X., Schulz T.C., Rao M.S., Puri R.K., Schulz T.C., Rao M.S., Puri R.K., Rao M.S., Puri R.K., Puri R.K. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev. Biol. 2005;5:22. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R., Hagedorn C.H., Cullen B.R., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ridzon D., Lee C.T., Blake J., Sun Y., Strauss W.M., Ridzon D., Lee C.T., Blake J., Sun Y., Strauss W.M., Lee C.T., Blake J., Sun Y., Strauss W.M., Blake J., Sun Y., Strauss W.M., Sun Y., Strauss W.M., Strauss W.M. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm. Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz A.L., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., He Y., Leary R.J., Pagliarini R., Diaz A.L., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Leary R.J., Pagliarini R., Diaz A.L., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Pagliarini R., Diaz A.L., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Diaz A.L., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Bentwich Z., Szafranska A.E., Labourier E., Szafranska A.E., Labourier E., Labourier E., et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison T.S., Johnson C.D., Andruss B.F., Johnson C.D., Andruss B.F., Andruss B.F. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- Dvash T., Mayshar Y., Darr H., McElhaney M., Barker D., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Mayshar Y., Darr H., McElhaney M., Barker D., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Darr H., McElhaney M., Barker D., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., McElhaney M., Barker D., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Barker D., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Yanuka O., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Kotkow K.J., Rubin L.L., Benvenisty N., Eiges R., Rubin L.L., Benvenisty N., Eiges R., Benvenisty N., Eiges R., Eiges R. Temporal gene expression during differentiation of human embryonic stem cells and embryoid bodies. Hum. Reprod. 2004;19:2875–2883. doi: 10.1093/humrep/deh529. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Law T.F., Grant S.R., Dangl J.L., Grant S.R., Dangl J.L., Dangl J.L., et al. High-throughput sequencing of ArabidopsismicroRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Garrett-Engele P., Lim L.P., Bartel D.P., Lim L.P., Bartel D.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M., Delaney A., Rogers S.A., Schnerch A., Persaud D.R., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., Delaney A., Rogers S.A., Schnerch A., Persaud D.R., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., Rogers S.A., Schnerch A., Persaud D.R., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., Schnerch A., Persaud D.R., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., Persaud D.R., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., O'Connor M.D., Zeng T., Moksa M., Fichter K., Mah D., Zeng T., Moksa M., Fichter K., Mah D., Moksa M., Fichter K., Mah D., Fichter K., Mah D., Mah D., et al. LongSAGE profiling of nine human embryonic stem cell lines. Genome Biol. 2007;8:R113. doi: 10.1186/gb-2007-8-6-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A., Murray M.F., Sharp P.A., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N., Yanuka O., Amit M., Soreq H., Benvenisty N., Amit M., Soreq H., Benvenisty N., Soreq H., Benvenisty N., Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Wu H., Wang W., Ma W., Sun X., Lu Z., Wu H., Wang W., Ma W., Sun X., Lu Z., Wang W., Ma W., Sun X., Lu Z., Ma W., Sun X., Lu Z., Sun X., Lu Z., Lu Z. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Aravin A., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C., Cumbie J.S., Givan S.A., Carrington J.C., Givan S.A., Carrington J.C., Carrington J.C. Genome-wide profiling and analysis of ArabidopsissiRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Hatzigeorgiou A.G., Nishikura K., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi S., Agrawal S., Agrawal S. piRNABank: A web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm696. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rice A., Kamphorst A.O., Landthaler M., Kamphorst A.O., Landthaler M., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U., Guttinger S., Calado A., Dahlberg J.E., Kutay U., Calado A., Dahlberg J.E., Kutay U., Dahlberg J.E., Kutay U., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Nobuta K., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C., Nobuta K., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C., Tej S.S., Skogen J.W., Meyers B.C., Skogen J.W., Meyers B.C., Meyers B.C. Plant MPSS databases: Signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Kwang Loong S., Mishra S.K., Mishra S.K. Unique folding of precursor microRNAs: Quantitative evidence and implications for de novo identification. RNA. 2007;13:170–187. doi: 10.1261/rna.223807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole A.S., Miller S., Haines N., Zink M.C., Serra M.J., Miller S., Haines N., Zink M.C., Serra M.J., Haines N., Zink M.C., Serra M.J., Zink M.C., Serra M.J., Serra M.J. Comprehensive thermodynamic analysis of 3′ double-nucleotide overhangs neighboring Watson-Crick terminal base pairs. Nucleic Acids Res. 2006;34:3338–3344. doi: 10.1093/nar/gkl428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lander E.S., Kent J., Miller W., Haussler D., Kent J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., van Dyk L.F., Ho C.K., Shuman S., Chien M., Ho C.K., Shuman S., Chien M., Shuman S., Chien M., Chien M., et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Porkka K.P., Pfeiffer M.J., Waltering K.K., Vessella R.L., Tammela T.L., Visakorpi T., Pfeiffer M.J., Waltering K.K., Vessella R.L., Tammela T.L., Visakorpi T., Waltering K.K., Vessella R.L., Tammela T.L., Visakorpi T., Vessella R.L., Tammela T.L., Visakorpi T., Tammela T.L., Visakorpi T., Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P., Vaucheret H., Trejo J., Bartel D.P., Trejo J., Bartel D.P., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P., Lee W., Nusbaum C., Ge H., Bartel D.P., Nusbaum C., Ge H., Bartel D.P., Ge H., Bartel D.P., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sato N., Sanjuan I.M., Heke M., Uchida M., Naef F., Brivanlou A.H., Sanjuan I.M., Heke M., Uchida M., Naef F., Brivanlou A.H., Heke M., Uchida M., Naef F., Brivanlou A.H., Uchida M., Naef F., Brivanlou A.H., Naef F., Brivanlou A.H., Brivanlou A.H. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottman H., Mikkola M., Lundin K., Olsson C., Stromberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Mikkola M., Lundin K., Olsson C., Stromberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Lundin K., Olsson C., Stromberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Olsson C., Stromberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Stromberg A.M., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Tuuri T., Otonkoski T., Hovatta O., Lahesmaa R., Otonkoski T., Hovatta O., Lahesmaa R., Hovatta O., Lahesmaa R., Lahesmaa R. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- Song L., Tuan R.S., Tuan R.S. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res. C Embryo Today. 2006;78:140–149. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- Strauss W.M., Chen C., Lee C.T., Ridzon D., Chen C., Lee C.T., Ridzon D., Lee C.T., Ridzon D., Ridzon D. Nonrestrictive developmental regulation of microRNA gene expression. Mamm. Genome. 2006;17:833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., Chung H.M., Yoon H.S., Moon S.Y., Yoon H.S., Moon S.Y., Moon S.Y., et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R., Medvid R., Melton C., Jaenisch R., Blelloch R., Melton C., Jaenisch R., Blelloch R., Jaenisch R., Blelloch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Mise K., Okuno T., Sasaki H., Minami N., Imai H., Okuno T., Sasaki H., Minami N., Imai H., Sasaki H., Minami N., Imai H., Minami N., Imai H., Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Li F., He T., Liu G.P., Li Y., Zhang X., Li F., He T., Liu G.P., Li Y., Zhang X., He T., Liu G.P., Li Y., Zhang X., Liu G.P., Li Y., Zhang X., Li Y., Zhang X., Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Guo G., Ni Z., Sunkar R., Du J., Zhu J.K., Sun Q., Guo G., Ni Z., Sunkar R., Du J., Zhu J.K., Sun Q., Ni Z., Sunkar R., Du J., Zhu J.K., Sun Q., Sunkar R., Du J., Zhu J.K., Sun Q., Du J., Zhu J.K., Sun Q., Zhu J.K., Sun Q., Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X.P., Cox S.B., Cobb G.P., Anderson T.A., Pan X.P., Cox S.B., Cobb G.P., Anderson T.A., Cox S.B., Cobb G.P., Anderson T.A., Cobb G.P., Anderson T.A., Anderson T.A. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. 2006;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.J., Hua Y.J., Sun D.G., Meng X.X., Xiao H.S., Ma X., Hua Y.J., Sun D.G., Meng X.X., Xiao H.S., Ma X., Sun D.G., Meng X.X., Xiao H.S., Ma X., Meng X.X., Xiao H.S., Ma X., Xiao H.S., Ma X., Ma X. Genome-wide microRNA profiling in human fetal nervous tissues by oligonucleotide microarray. Childs Nerv. Syst. 2006;22:1419–1425. doi: 10.1007/s00381-006-0173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]