Abstract
Extracellular protein–protein interactions are essential for both intercellular communication and cohesion within multicellular organisms. Approximately a fifth of human genes encode membrane-tethered or secreted proteins, but they are largely absent from recent large-scale protein interaction datasets, making current interaction networks biased and incomplete. This discrepancy is due to the unsuitability of popular high-throughput methods to detect extracellular interactions because of the biochemical intractability of membrane proteins and their interactions. For example, cell surface proteins contain insoluble hydrophobic transmembrane regions, and their extracellular interactions are often highly transient, having half-lives of less than a second. To detect transient extracellular interactions on a large scale, we developed AVEXIS (avidity-based extracellular interaction screen), a high-throughput assay that overcomes these technical issues and can detect very transient interactions (half-lives ≤ 0.1 sec) with a low false-positive rate. We used it to systematically screen for receptor–ligand pairs within the zebrafish immunoglobulin superfamily and identified novel ligands for both well-known and orphan receptors. Genes encoding receptor–ligand pairs were often clustered phylogenetically and expressed in the same or adjacent tissues, immediately implying their involvement in similar biological processes. Using AVEXIS, we have determined the first systematic low–affinity extracellular protein interaction network, supported by independent biological data. This technique will now allow large-scale extracellular protein interaction mapping in a broad range of experimental contexts.
Extracellular protein interactions such as those made between secreted and membrane-tethered proteins are important for diverse cellular behaviors, such as initiating differentiation pathways, directing migration and pathfinding routes, and mediating intricate recognition processes. Approximately a fifth of human genes encode extracellular and membrane-associated proteins, but despite their importance and abundance, they are significantly underrepresented in recent large-scale protein interaction datasets (Futschik et al. 2007). This discrepancy is remarkable since their accessibility to systemically delivered drugs makes extracellular proteins excellent therapeutic targets (Clark et al. 2003). There are several reasons for this disparity. First, cell surface proteins are biochemically difficult to manipulate; for example, their hydrophobic membrane-spanning region renders them insoluble. Second, functionally important post-translational modifications such as disulfide bonds and large hydrophilic glycans are not added in commonly used expression systems such as bacteria and cell-free systems. Finally, interactions between cell surface proteins have very fast dissociation rates and are therefore often highly transient, having half-lives of fractions of a second (van der Merwe and Barclay 1994); this makes purification protocols involving wash steps impractical. The transient nature of these interactions and necessity for posttranslational modifications makes current high-throughput protein interaction assays, such as yeast-two-hybrid and biochemical purification methods, generally unsuitable to identify this important class of interactions.
Existing methods to directly detect extracellular transient interactions often rely on experimentally increasing the overall avidity of the interaction by multimerizing a soluble recombinant protein, mirroring the way these interactions occur in vivo through arrayed proteins on apposing membranes. These methods include oriented display around microbeads (Wright et al. 2000; Letarte et al. 2005) or tags producing dimers (such as Fc-fusion proteins), trimers, and, often most potently, pentamers (Holler et al. 2000; Voulgaraki et al. 2005). No broad assessment for the suitability of any of these techniques to be used in systematic high-throughput screening has been made, since only individual interactions have been reported (Lin et al. 2003; Gonzalez et al. 2005).
To address the need for a high-throughput technique that can detect low-affinity extracellular protein interactions, we have developed a novel assay termed AVEXIS (avidity-based extracellular interaction screen) and used it to identify novel extracellular receptor–ligand pairs within the zebrafish immunoglobulin superfamily (IgSF). Independent support for identified interactions was provided by quantifying interaction strengths, performing a phylogenetic analysis, and showing that genes encoding interacting pairs were expressed in either the same or adjacent tissues.
Results
AVEXIS can specifically detect low-affinity extracellular protein interactions with a low false-positive rate
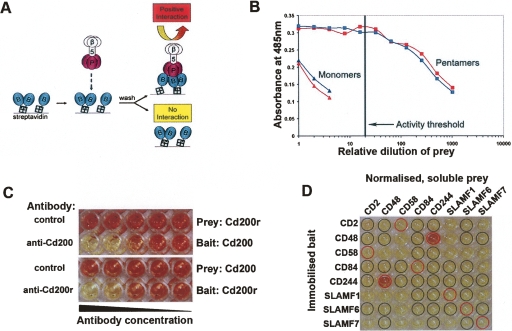
To retain the extracellular binding function while removing the insoluble transmembrane region, the entire ectodomains of cell surface proteins were produced as soluble recombinant proteins in mammalian cells. Ectodomains were expressed in two different forms: a monomeric biotinylated “bait,” which could be captured on streptavidin-coated microtiter plates, and a pentamerized “prey” tagged with β-lactamase to allow detection. The prey pentamers were produced by C-terminally tagging proteins with a coiled-coil sequence from the rat cartilage oligomeric matrix protein (Tomschy et al. 1996). These proteins were then used in the AVEXIS assay as shown (Fig. 1A). Despite a low monomeric interaction affinity (2 μM; t1/2 = 0.9 sec at 37°C) (Wright et al. 2000), interactions between rat Cd200 and its receptor could be robustly detected independently of bait–prey orientation. Increasing the avidity of prey proteins through pentamerization improved the sensitivity of detection over monomeric prey proteins by at least 250-fold (Fig. 1B). Interaction specificity was demonstrated by preventing interactions in either orientation with blocking anti-bait monoclonal antibodies (Fig. 1C). Purification of either the bait or prey was not necessary, but activities of both were measured and normalized prior to use in the assay.
Figure 1.
AVEXIS specifically detects low-affinity extracellular interactions with a low false-positive rate. (A) Schematic representation of AVEXIS. Biotinylated bait ectodomains (blue) are captured in an orientated manner on a streptavidin-coated microtiter plate and probed with soluble pentameric β-lactamase-tagged prey ectodomains (pink). Positive binding is detected by colorimetric enzymatic turnover of nitrocefin (yellow to red). (B) Pentamerization increases the avidity of prey ectodomains. Pentameric (squares) and monomeric (triangles) Cd200 (blue) and Cd200r (red) prey constructs were normalized for β-lactamase activity, serially diluted, and tested against the appropriate bait construct. The prey activity threshold used for AVEXIS is shown, corresponding to 50 units/mL. (C) AVEXIS interaction specificity. (Top panel) Low-affinity Cd200 (bait)–Cd200r (prey) interaction can be detected in the presence of a non-binding control antibody but is specifically blocked with an anti-Cd200 blocking monoclonal antibody; the blocking effect can be titrated by decreasing antibody concentration. The interaction specificity is independent of bait–prey orientation and can also be blocked with an anti-Cd200r antibody, bottom panel. (D) AVEXIS false-positive and -negative rate evaluation. An AVEXIS screening plate with bait proteins arrayed in rows and the prey proteins normalized to threshold levels in columns; positive binding results can be seen as red wells. Expected positive and negative wells are circled in red and black, respectively.
To determine the false-positive and -negative rates of AVEXIS, a small screen was performed using eight proteins from the SLAM/CD2 subfamily of the human IgSF within which there are quantified positive hetero- and homophilic low-affinity interactions and also, importantly, published negative interactions. This set represents a “gold standard” of positive and negative interactions for assay benchmarking and validation. A prey activity threshold was selected which was able to detect the 8 μM CD244–CD48 interaction (Brown et al. 1998) in both bait–prey orientations, and all published negative interactions were confirmed (Fig. 1D). At this prey activity, the weaker 9–22 μM CD2–CD58 interaction (van der Merwe et al. 1994) and all homophilic interactions including the very weak 200 μM SLAMF1 interaction (Mavaddat et al. 2000) were not detected. All expected interactions except the SLAMF6 homophilic could be detected at a higher prey activity, although the false-positive rate consequently increased (data not shown). To eliminate false positives during screening we selected a stringent prey threshold (Fig. 1B), inevitably increasing our false-negative rate as a result. In particular, AVEXIS may not be generally suitable for detecting homophilic extracellular interactions possibly due to highly avid prey–prey association, which may then prevent prey–bait interactions. We did, however, reproducibly detect some homophilic interactions in our large screen (Fig. 2).
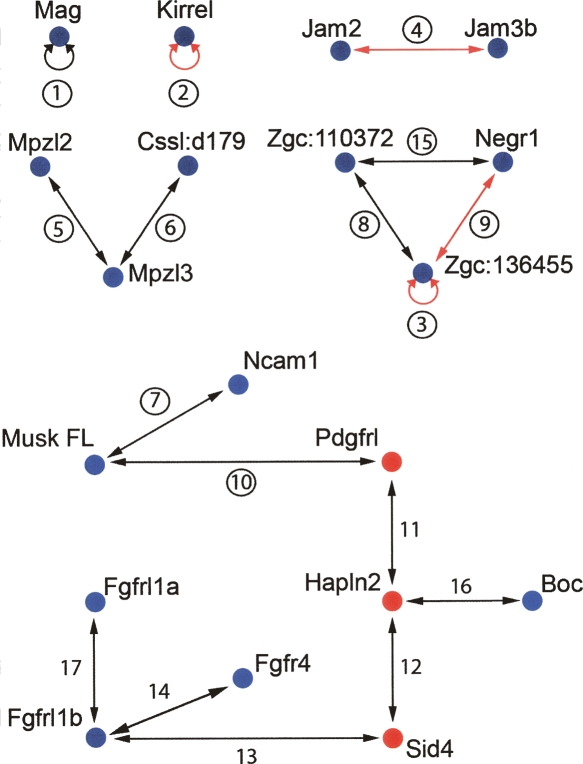
Figure 2.
Interaction network of zebrafish IgSF proteins as determined using AVEXIS. Proteins are identified by their current official nomenclature, with blue and red nodes representing cell surface and secreted proteins, respectively. Interactions are numbered for reference and circled if quantified by SPR. Entirely novel interactions are represented by black arrows and those previously detected using an orthologous mammalian protein with red arrows.
A large systematic protein interaction screen identifies novel extracellular receptor–ligand pairs
To identify novel extracellular receptor–ligand pairs, a protein library containing the entire ectodomains of 110 proteins, mainly from the zebrafish IgSF, was expressed in mammalian cells as both bait and prey molecules, and their activities normalized (Table S1). This protein family was chosen since they are known to form receptor–ligand pairs between themselves (Barclay 2003), yet many have no documented extracellular binding partner or function. The zebrafish vertebrate model was selected to facilitate rapid determination of gene expression patterns during early vertebrate development. All proteins were tested against each other for interactions in a systematic unbiased matrix fashion. In total, 9751 interaction tests were performed; 7292 of these were screened in a reciprocal two-way fashion and the rest in a one-way orientation, representing a total of 6105 unique possible interactions tested. The protein pairs that interacted were re-expressed as both baits and preys and were all systematically rescreened in both orientations. Interactions were only considered positive if they were independent of the bait–prey orientation. Using these criteria, 17 interactions were identified between 19 proteins (16 cell surface, three secreted), representing the first systematic extracellular interaction network (Fig. 2). Only 0.28% of all unique interactions tested were positive, putting a stringent upper limit on a false-positive rate. All interactions detected by AVEXIS are detailed in Table S2, including some lower-confidence interactions not meeting these stringent criteria; for example, interactions that were dependent on the bait–prey orientation.
Within our interaction network we found that approximately half of the proteins had multiple binding partners, suggesting competition between potential ligands in tissues where they are co-expressed. The paucity of interaction data for zebrafish proteins made the identification of entirely novel interactions and the assessment of a false-negative rate very difficult. For proteins where a clear mammalian ortholog could be identified (protein sequence identity > 40%, see Table S1) we were able to detect within our network some interactions previously reported in mammals (Fig. 2).
Novel interactions identified in the screen included proteins with no known extracellular ligand (orphan receptors). This included a subnetwork involving the Mpzl2 protein, a gene expressed during mouse thymus development (Guttinger et al. 1998). We also identified new ligands for well-characterized proteins. For example, we found that neural cell adhesion molecule 1 (Ncam1) interacted with the muscle-specific kinase receptor (Musk). Interestingly, there are functional data that suggest that both proteins are necessary for correct neuromuscular junctions (NMJ) formation and maintenance (DeChiara et al. 1996; Polo-Parada et al. 2001) (see Discussion). We believe that this is the first example of a membrane-tethered receptor–ligand pair that could mediate the initial contact between motor neuron axons and muscles to form the NMJ. We also demonstrated interactions between Fgfr4 and Fgfrl1b, which in turn interacted with Fgfrl1a. To our knowledge, this is the first report of inter-receptor signaling within this important family of signaling molecules. Knockdown of either fgfrl1a or fgfrl1b in zebrafish again results in very similar phenotypes (Hall et al. 2006) (see Discussion).
AVEXIS can detect interactions with a half-life ≤ 0.1 sec
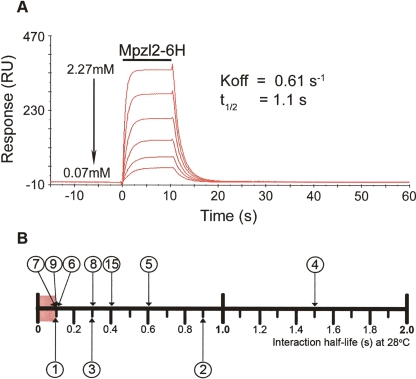
To confirm interactions and determine the affinity detection threshold of AVEXIS, we used SPR to measure off-rate constants (koff) which have the advantage of being independent of active protein concentration estimates. These comparative quantitative data are also important to address functional questions, such as ligand binding competition, when multiple potential ligands are simultaneously displayed on the same cell surface. We injected serial dilutions of purified monomeric extracellular ligands over appropriate biotinylated baits and controls immobilized on a streptavidin-coated sensor chip. A typical experiment is shown in Figure 3A, and the traces for all interactions are shown in Figure S1. The measured off-rate constants were converted to half-lives (t1/2 = ln(2)/koff) and are shown on the scale in Figure 3B. Eleven out of the 17 identified interactions were tested by SPR, and all were verified (Table S2; Fig. S1), confirming the assay’s low false-positive rate. Four of these interactions were very transient having half-lives at or below the limit of SPR sensitivity (t1/2 ≤ 0.1 sec), demonstrating that AVEXIS can detect extremely low-affinity interactions. Nine of the 11 dissociation curves fitted a simple first-order 1:1 dissociation model, but other interactions had more complex behaviors, perhaps due to multisite binding models that have been reported for other extracellular interactions (Aricescu et al. 2006). Nine of the lower-confidence interactions were also tested, and all but one gave positive results (Table S2).
Figure 3.
Validation and quantification of interactions by surface plasmon resonance. (A) Reference-subtracted surface plasmon resonance traces of soluble Mpzl2-Cd4-6H protein interacting with surface-immobilized Mpzl3-Cd4-bio. The binding curves of six twofold serial dilutions (as shown) of the purified protein were injected over 1016RU of immobilized Mpzl3 as indicated by the bar. The wash-out phase of the binding was fitted to a 1:1 dissociation model, and the off-rate constant (koff) was determined and a half-life calculated (see Methods). (B) Relative monomeric half-lives of interactions quantified by SPR as measured at 28°C. Interactions are numbered as in Fig. 2. Heterophilic interactions are placed above the scale, and homophilic interactions below. The red box represents the limits of SPR sensitivity, suggesting that several interactions have half-lives ≤ 0.1 sec.
Extracellular receptor–ligand pairs are enriched for paralogs
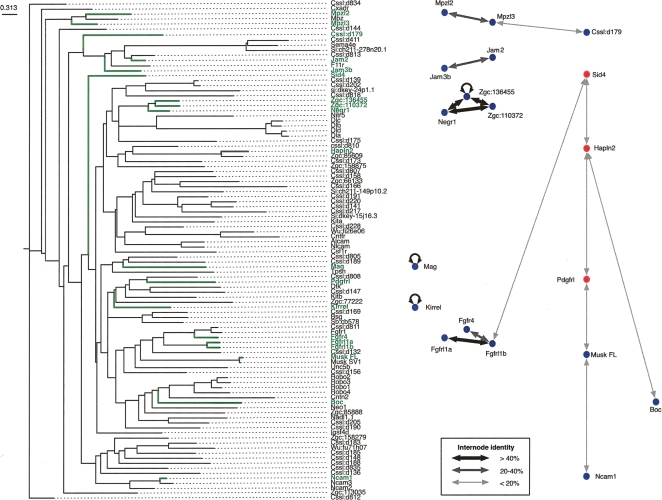
Large-scale proteomics experiments in yeast, which mainly detect stable cytoplasmic complexes, are enriched for interactions between paralogous proteins (Ispolatov et al. 2005; Pereira-Leal et al. 2007). This property, often coupled with close genetic linkage, has also been observed for heterophilic IgSF receptor–ligand pairs (Wright et al. 2000; Sidorenko and Clark 2003) and is thought to have evolved from the genomic segmental duplication of a gene encoding a protein capable of homophilic association (Williams and Barclay 1988). To determine if this was a property of our network, the relationships between all non-redundant proteins within the library were quantified using percentage sequence identities from optimal local alignments and are shown as a phylogenetic tree. The projection of the extracellular interaction network onto the tree revealed that, in 50% of all heterophilic pairs, the interacting proteins were clustered on the nearest or next-nearest branch (Fig. 4). On average, sequence identity between interacting pairs was 32.4%, whereas networks made by replacing the nodes with other proteins randomly selected from within the library, shared only 23.2% ± 1.0% (SD), a statistically significant difference (P = 1.78 × 10−20; n = 1000 replacements). Similarly, interacting proteins shared only 22.8% ± 0.7% (P = 4.29 × 10−39; n = 1000 rewirings) sequence identity in networks that were randomly rewired by edge swapping but with a fixed degree distribution. Further evidence of this possible evolutionary mechanism came from the two closest human homologs for the zebrafish Mpzl2 and Mpzl3 proteins, MPZL2 and MPZL3, which are closely linked and have identical gene structures on chromosome 11q24. This analysis suggested that networks involving transient extracellular interactions evolved in the same manner as those already described for stable cytoplasmic interactions and are more likely to occur between paralogous proteins.
Figure 4.
Interactions between paralogous proteins are enriched within the extracellular interaction network. A phylogenetic tree showing the relationships of the non-redundant protein ectodomains used within the screen is shown. Names and branches representing the interacting proteins from the network are highlighted in green. The line widths in the interaction network reflect the sequence identities between interacting proteins as described in the key. Homophilic interactions are shown for completeness.
Genes encoding interacting proteins are often expressed in the same or neighboring tissues
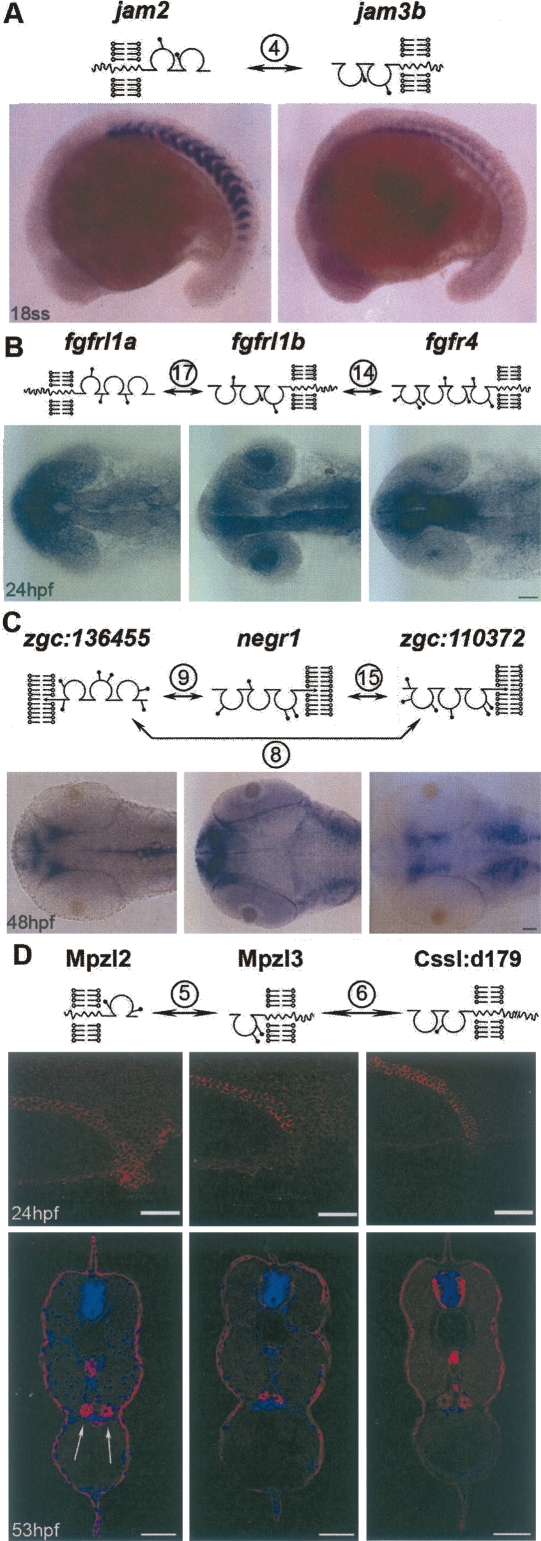
To mediate adhesion or initiate signaling, membrane proteins are likely to be expressed on the surface of the same or neighboring cells at the same stage of development. To determine which receptor–ligand pairs are co-expressed and also to provide clues for the in vivo function of identified interactions, the transcriptional expression patterns for all genes encoding interacting proteins were determined by whole-mount in situ hybridization at several stages during the first 48 h of zebrafish development. Of the 19 genes in our interaction network, 18 were expressed during the first 24 h and all by 48 h, suggesting that these genes have roles during the earliest stages of vertebrate development. IgSF-domain–containing proteins are known to have diverse functions (Rougon and Hobert 2003), and accordingly, the genes were transcribed in a broad variety of tissues. The gene expression patterns for the cell surface proteins could be split into two classes: those expressed in neighboring tissues and presumably involved in intercellular signaling and those that are expressed in the same tissue and are likely to have adhesive roles, although the two functions are not mutually exclusive (Fig. 5). For example, jam2 was detected in the mesenchyme and epithelium of somites just after their formation, while its binding partner, jam3b, showed abutting expression at the posterior somite boundaries (Fig. 5A), hinting at a novel role for this interaction during segmentation. The expression patterns of fgfr4, fgfrl1a, and fgfrl1b are known to be highly dynamic (Thisse et al. 1995; Hall et al. 2006). Our expression data showed they indeed had the potential to interact at several stages during early development but perhaps most notably within the forming telencephalon and anterior medial part of the lens at 24 hpf (Fig. 5B) where they showed overlapping expression patterns. The three interacting GPI-linked proteins belonging to the IgLON family (Reed et al. 2004) were expressed in complementary territories in the zebrafish head, suggesting roles in patterning the embryonic brain (Fig. 5C). Finally, all three proteins in the network involving Mpzl2 were expressed in the same tissue: the cuboidal epithelium of the pronephric ducts. Antisera raised against these proteins demonstrated that they were all localized at the cell surfaces of the pronephric duct epithelium and the epidermis (Fig. 5D): These interactions may therefore perform a role in cellular adhesion.
Figure 5.
Expression patterns of genes encoding interacting proteins during early vertebrate development. Schematic diagrams representing the domain architecture and location of potential N-linked glycosylation sites are shown above the corresponding expression patterns. Whole-mount in situ expression patterns of zebrafish jam2 and jam3b at 18 somite stage (A), fgfrl1a, fgfrl1b, and fgfr4 at 24 hpf (B), and zgc:136455, negr1, and zgc:110372 at 48 hpf (C) showing complementary expression patterns. (D) (Upper panels) Lateral views of whole-mount embryos with anterior to the left showing Mpzl2, Mpzl3, and Cssl:d179 protein expression as detected by immunohistochemistry in the pronephric ducts at 24 hpf. (Lower panels) Immunohistochemistry of transverse 53-hpf zebrafish sections counterstained for nuclei with DAPI (blue) showing co-expression in the paired pronephric ducts (arrows) and epidermis (red). Scale bars = 50 μm.
Discussion
We have described the development of AVEXIS, an assay that overcomes the technical difficulties associated with detecting extracellular protein interactions and can be used in both small and larger—including genome-wide—interaction screens. The assay has several features that will allow it to be widely useful. First, it is not system-specific and can therefore be applied to screen for interactions between extracellular proteins from most metazoans. In our study, for example, we have used both human and zebrafish proteins. Second, we have demonstrated that AVEXIS is suitable for systematic large-scale screens and have provided a full screening assessment showing that it has a very low false-positive rate. Thirdly, we have shown that it can detect extremely low-affinity interactions, certainly at or below half-lives of a tenth of a second. Finally, we have provided independent biological support for several of the interactions identified in our screen that suggest they occur in vivo. Taken together, AVEXIS is therefore a versatile technique to detect and study low-affinity extracellular interactions in a high-throughput format.
The number of identified protein–protein interactions has increased rapidly with the use of high-throughput interaction assays, with the yeast-two-hybrid (Y2H) and biochemical purification “TAP-tagging” techniques being especially successful. Due to the heterogeneity of protein–protein interactions and the practical limitations of large-scale screening, each assay must inevitably accept a certain rate of false positives and negatives. For example, the Y2H and TAP-tagging methods are unsuitable to detect transient extracellular interactions because they do not add essential post-translational modifications (Y2H) or the stringent wash steps preclude detection of weak interactions. These limitations are well appreciated and have led to the development of other scalable assays to detect specialized classes of protein interactions unsuited to these methods; for example, the split-ubiquitin system to detect interactions between membrane proteins embedded within the same membrane (Stagljar et al. 1998). In developing AVEXIS, we sought to eliminate false positives while recognizing that high-throughput screening assays, due to in vitro limitations such as non-native protein glycosylation, presentation, and surrounding milieu, would inevitably lead to false negatives. The relative lack of zebrafish protein interaction data made assessing a false-negative rate difficult, although by using the closest mammalian ortholog it was clear that the main class of interactions not detected by AVEXIS was homophilic, as expected.
By using AVEXIS, we have described the first low-affinity extracellular protein interaction network. We have shown that many cell surface and secreted proteins have more than one binding partner. Given that we have screened only around a fifth of the total zebrafish IgSF family and a fraction of the total extracellular proteome, it is likely that extracellular interaction networks will be highly connected, as described for intracellular networks. Also similar to other protein interaction networks, we observed an enrichment of paralogous pairs between interacting partners, which suggests gene duplication as a common mechanism for the evolution of novel interactions.
Within the zebrafish IgSF, we have identified interactions between previously uncharacterized proteins. For example, we have discovered a subnetwork of three proteins: Mpzl2, Mpzl3, and Cssl:d179, which are all localized to the cell surface of the developing pronephric duct epithelium and epidermis, suggesting a role in cellular adhesion. The interaction strength between Mpzl3 and Cssl:d179 is one of the weakest that we measured (t1/2 ≤ 0.1 sec) and yet both proteins have a striking co-localization: even highly transient interactions could therefore have a physiological role. Both Mpzl2 and Mpzl3 share sequence similarity and domain architecture with myelin protein P0. This single-IgSF-domain-containing protein capable of homophilic interactions is involved in the tight packing of apposing membranes within compacted myelin sheaths to facilitate saltatory conduction in nerves (Quarles 2002). Proteins in the Mpzl2 network may similarly be involved in the tight packing of the epithelial membranes, contributing to the osmotic seal ensuring that both the blood filtrate and external freshwater are separated from the body tissues.
Using AVEXIS, we have also discovered novel interactions for well-studied proteins. We have demonstrated that the full-length muscle-specific kinase (Musk FL) isoform (Zhang et al. 2004) interacts with neural cell adhesion molecule, Ncam1. Mice lacking either Musk or Ncam proteins both have defects in the formation of functional NMJs (DeChiara et al. 1996; Polo-Parada et al. 2001). We have found that expression of these genes during zebrafish development is also consistent with a role in NMJ formation: ncam1 is expressed in the neural tube and musk FL in the formed somites, the precursors of the axial muscle (data not shown). This interaction could therefore mediate the initial contact between motor axons and muscle to form the NMJ and/or be involved in NMJ maintenance.
Consistent with the Fgfrl1a–Fgfr11b interaction identified in our screen, a functional study of both genes in zebrafish points toward their involvement in the same biological process: knocking down either gene resulted in the same striking phenotype, a loss of the ceratobranchial arches in the gill cartilage (Hall et al. 2006). While the authors interpreted this as the non-redundant requirement for Fgf-mediated signaling through both receptors, our data allow another mechanistic interpretation: the direct interaction of Fgfrl1a and Fgfrl1b at the cell surface could initiate inter-receptor signaling, leading to correct ceratobranchial arch formation. Taken together, the interaction and distribution data, along with examples of functional studies from the literature, provide strong biological support to suggest that the AVEXIS assay is able to identify interactions which are physiologically relevant in vivo.
Protein–protein interactions, although they are likely to have evolved using similar processes, are mechanistically very diverse, varying in both affinity and surrounding subcellular environment. It therefore seems unlikely that any one particular technique will be suitable to detect all classes of protein–protein interaction; rather, techniques tailored to detect particular classes will be necessary. The use of AVEXIS will now make the identification of low-affinity extracellular interactions possible on a large scale and contribute to the defining of a complete and accurate protein interaction network. AVEXIS is not organism specific and can therefore be used in many contexts, including extracellular host–pathogen interaction mapping to identify pathogen receptors. Indeed, the interaction between pathogen proteins and their host cell surface receptors is often the initiating event during infections. Since extracellular proteins are readily accessible to systemically delivered drugs, AVEXIS can therefore be used to identify novel therapeutic opportunities to target both genetic and infectious diseases.
Methods
Sources of zebrafish genes and ectodomain protein expression library
A comprehensive list of all genes used in this study is provided in Table S1. Zebrafish genes encoding immunoglobulin superfamily (IgSF) domains were obtained from three sources: (1) “known” genes already deposited in gene databases (32 genes) and amplified by RT-PCR from mixed-stage zebrafish cDNA; (2) zebrafish IMAGE clones (85 genes) that were predicted to encode IgSF domains and a complete signal peptide were identified from EST databases by TBLASTN searches using all human and mouse IgSF-domain-encoding protein sequences; and (3) novel genes identified from the zebrafish genome sequence (32 genes). The complete extracellular region and signal peptide were obtained by both 5′ and 3′ RACE procedures. Bait and prey ectodomain expression clones were made for 125 genes; 110 (88%) produced sufficient protein (>0.1 μg/mL). All clones were fully sequenced.
Mammalian expression system
All proteins were produced by transient transfection using a HEK293E expression system as described in (Durocher et al. 2002), except that cells were grown in Freestyle medium (Invitrogen) supplemented with 1% fetal calf serum. Transfections were left for 6 d before removing cells and cell debris by centrifugation at 3220g for 5 min. Supernatants were then filtered (0.2-μm filter) and stored at 4°C until use.
Bait protein expression
The bait protein expression vector was constructed from a derivative of pTT3 (Durocher et al. 2002) that contains an expanded multiple cloning site. This vector provided in-frame fusion of the ectodomains to rat Cd4 domains 3+4 (Cd4d3+4) (Brown and Barclay 1994) and a 17-amino acid biotinylation peptide (Schatz 1993; Brown et al. 1998). Bait proteins were enzymatically biotinylated during synthesis by co-transfecting an expression plasmid containing a modified E. coli biotin ligase, BirA, together with the bait expression construct in a 1:9 ratio. The BirA coding sequence was fused downstream of the rat Cd4 5′ UTR and signal peptide to direct expression to the secretory pathway essentially as described by Parrott and Barry (2001). When the culture medium was supplemented with 100 μM D-biotin, at least 95% of the target protein was biotinylated (data not shown). Supernatants were dialyzed extensively against PBS to remove any excess D-biotin; no purification was necessary.
Prey protein constructs
The prey protein expression vector was also derived from pTT3 and provided in-frame fusion of the ectodomains to Cd4d3+4, the pentamerization domain of rat cartilaginous oligomeric matrix protein (COMP) as described (Tomschy et al. 1996), and the ampicillin resistance protein beta-lactamase. A protein linker sequence (-LDRNLPPLAPLGP-) was designed according to principles described in George and Heringa (2002) and inserted between the COMP and beta-lactamase domains using complementary oligonucleotides.
Bait and prey protein normalization
Despite standardized procedures, we observed a wide range in the level at which each ectodomain protein was secreted, that correlated between bait and prey proteins and spanned at least four orders of magnitude. Expression levels of both were normalized by dilution or concentration prior to AVEXIS screening.
Biotinylated bait protein concentrations were determined by ELISA using streptavidin-coated plates (Nunc). Protein activities were measured with a mouse anti-rat Cd4 antibody (OX68, Serotec) followed by incubation with an anti-mouse alkaline phosphatase secondary and detected with p-nitrophenyl phosphate. Biotinylated protein concentrations were normalized by either concentrating using spin concentrators (Vivascience, 10 k MWCO) or diluting with conditioned tissue culture supernatant or PBS containing 0.2% BSA; 50 μL of a 10–100 nM biotinylated protein concentration was found to be sufficient to saturate the available streptavidin on the plates.
Prey protein solutions used for interaction screening were normalized using the beta-lactamase enzymatic activity. One unit of activity was defined as the amount of prey construct required to turnover 1 nmol of nitrocefin (Calbiochem) in 1 min at room temperature. Twenty microliters of serial dilutions of prey supernatants were incubated with 60 μL of 250 μg/mL nitrocefin at room temperature for 20 min, and nitrocefin turnover was quantitated by measuring the absorbance at 485 nm. Prey proteins were normalized by concentrating with 20 k MWCO spin concentrators or diluting in the same manner as the bait proteins to within an order of magnitude, with a minimum threshold of 50 units/mL.
AVEXIS screening
In total, 110 extracellular protein fragments were used in the screen which included four Deltas (A, B, C, and D) and four semaphorins (Sema4E, SemaD, and two forms of SemaC). Eleven pairs of proteins within the 102 IgSF proteins had some level of redundancy including splice variants, allelic variants, and ectodomains differing at the truncation position; one pair of ectodomain fragments had identical protein sequences.
Arrayed biotinylated bait proteins were captured on streptavidin-coated plates as described above by incubating 50 μL of normalized bait proteins for 1 h at room temperature. Plates were washed three times with PBS, and each plate was probed with 50 μL/well of a normalized beta-lactamase-tagged prey protein and incubated at room temperature for 1 h. Plates were washed twice with PBS containing 0.05% Tween-20 and then twice with PBS alone before adding 50 μL of 150 μg/mL nitrocefin dissolved according to the manufacturer’s instructions. Plates were incubated at room temperature for 3 to 4 h and then overnight at 4°C. Interactions were identified by taking absorbance readings at 485 nm and positive interactions called as having a plate Z-score of >6; that is, the positive wells were >6 standard deviations above the plate mean. Relying on absorbance readings alone, however, resulted in a significant number of technical false positives, since wells with high absorbance readings sometimes did not show visible substrate turnover; this was attributed to unavoidable imperfections/scratches on the microtiter plates or precipitated nitrocefin. Technical false positives were eliminated by photographing the screening plates and verifying substrate turnover in true-positive wells.
A total of 110 proteins were used for the interaction screening, representing a total of (1102 + 110)/2 = 6105 unique interactions screened. The majority of the library was tested in both bait–prey orientations (7292 interactions), although a proportion (2459, including all homophilic interactions) was only screened in one bait–prey orientation, making a total of 9751 tested interactions. Proteins which showed positive hits were then re-expressed and tested in a second, independent “validation” screen using both bait–prey orientations in the same matrix-style fashion. Interactions were classified from A to J according to the criteria listed in Table S2. One prey construct, Cssl:d805, showed positive interactions with >50% of bait proteins and was therefore excluded from further analysis.
AVEXIS validation
Initial parameters for the AVEXIS assay were defined using the rat Cd200–Cd200r interaction, and blocking monoclonal antibodies, OX2 and OX102 (Wright et al. 2000), which were kind gifts from Professor Neil Barclay (Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom). The appropriate antibody dilution, 50 μL/well, was added as an extra step to the AVEXIS assay just after bait immobilization. Using this assay, a threshold prey concentration of 50 units/mL (see Fig. 1) was selected, which reproducibly gave high signal–noise ratios.
The full-length coding regions of the eight members of the human CD2 and SLAM family members used for false-positive and -negative rate determination were provided by the Sanger Institute cORF cloning group. These cDNAs were cloned from either existing MGC clones or amplified from mixed-tissue human cDNA essentially as described (Collins et al. 2004). Within this set of proteins are published negative interactions (Martin et al. 2001; Kumaresan et al. 2002; Falco et al. 2004; Flaig et al. 2004; Romero et al. 2005), outlined with black circles in Figure 1D.
Protein purification and gel filtration for SPR studies
The ectodomains of proteins selected for quantitation using surface plasmon resonance were subcloned into an expression construct containing the rat Cd4d3+4 followed by a six-histidine tag for purification using standard procedures. To exclude protein aggregates that are known to influence kinetic binding measurements (van der Merwe and Barclay 1996), four 0.5-mL fractions containing the highest concentration of purified protein were fractionated by gel filtration in HBS-EP buffer immediately prior to Biacore analysis on a 125-mL Superose 6 column. The concentrations of purified proteins were calculated by measuring the absorbance at 280 nm and using a computationally predicted extinction coefficient as described elsewhere (Gill and von Hippel 1989).
Kinetic analysis of interactions using SPR
Kinetic analysis was performed on a Biacore T100 SPR machine using HBS-EP as a running buffer at 28°C (zebrafish physiological temperature), and the data were collected at a frequency of 10 Hz. Biotinylated proteins were immobilized at the indicated levels (Fig. S1) on a streptavidin-coated SA biosensor chip (Biacore), and monomeric, purified proteins were injected in increasing concentrations at the maximum flow rate of 100 μL/min to minimize rebinding effects. Dissociation curves were fitted to a first-order 1:1 dissociation model using the T100 BIAevaluation software, and when the data fitted the model well (χ2 values within 1 RU2 over the initial 90% of the dissociation) an off-rate constant and half-life were calculated. For rapidly dissociating interactions, only a lower limit for the off-rate constant could be determined (≥6/sec) corresponding to a t1/2 ≤ 0.1 sec. To determine relative interaction strengths, first-order off-rate constants are more suitable than equilibrium measurements because they are independent of active protein concentration measurements, which are difficult to determine, particularly for low-affinity and homophilic interactions.
Phylogenetic analysis
Pairwise sequence identities between the 99 non-redundant ectodomain sequences used in the screen (although both Musk FL and Musk SV1 splice variants were included) were determined by optimal local alignments using the EMBOSS implementation of the Smith–Waterman algorithm (Rice et al. 2000). A multiple alignment was created using MAFFT (Katoh et al. 2005), and a phylogenetic tree was calculated using the maximum-likelihood algorithm implementation from the PHYLIP package (Felsenstein 2005). Networks were rewired by implementing the edge swapping algorithm as described in Maslov and Sneppen (2002).
Zebrafish husbandry
All animal experiments were performed under United Kingdom Home Office and Institutional regulation and guidelines. Zebrafish were maintained at 28.5°C on a 14 h/10 h light/dark cycle, and embryos were collected from spontaneous spawning according to Kimmel et al. (1995).
In situ hybridization
Probe templates were made by amplifying by PCR the entire extracellular regions (including the signal peptide) from the expression clones, using an antisense oligonucleotide that contained a T7 RNA polymerase binding site. DIG-labeled antisense probes were made, and the in situ hybridizations were performed by standard procedures (Thisse and Thisse 1998). Embryos were mounted in glycerol, and images were captured using a Zeiss AxioCam HRc color digital camera mounted on a Zeiss Axio Imager M1 microscope; images were processed to adjust brightness and contrast in Adobe Photoshop 7.0.
Antibodies and immunohistochemistry
Polyclonal goat antibodies were raised by Everest Biotech using the purified His-tagged ectodomain proteins of Mpzl2, Mpzl3, and Cssl:d179. The sera were subsequently affinity purified on an antigen column made by immobilizing the appropriate biotinylated ectodomain protein onto streptavidin-coated Sepharose (Pharmacia). Affinity-purified sera were used at 1:4 to 1:10 dilutions as determined by experiment. Zebrafish embryos were processed for cryosectioning essentially as described (Haddon and Lewis 1996), and antibodies were detected using donkey anti-goat Alexa 564 (Invitrogen). Sections were counterstained using Slow-Fade Gold-DAPI and analyzed on a Zeiss LSM510 Meta confocal microscope. Whole mounts were analyzed on a Zeiss AxioPlan 2 microscope fitted with an OptiGrid system (Improvision).
Acknowledgments
We thank A. Neil Barclay and Marion H. Brown for reagents; the Sanger Institute cORF program and Nick Bockett for gene cloning; and David Goulding for help with confocal microscopy. This work was supported by the Wellcome Trust and a Marie Curie and Sanger postdoctoral fellowship to C.S.
Footnotes
[Supplemental material is available online at www.genome.org. The sequences of all the genes cloned for this study have been submitted to GenBank, and their accession numbers are listed in Supplemental Table S1. All protein–protein interaction data have been submitted to the IntAct database under accession nos. EBI-1578837 and EBI-1578841 for AVEXIS and SPR validation data, respectively.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7187808.
References
- Aricescu A.R., Hon W.C., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Hon W.C., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Siebold C., Lu W., van der Merwe P.A., Jones E.Y., Lu W., van der Merwe P.A., Jones E.Y., van der Merwe P.A., Jones E.Y., Jones E.Y. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. EMBO J. 2006;25:701–712. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A.N. Membrane proteins with immunoglobulin-like domains—A master superfamily of interaction molecules. Semin. Immunol. 2003;15:215–223. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Brown M.H., Barclay A.N., Barclay A.N. Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Protein Eng. 1994;7:515–521. doi: 10.1093/protein/7.4.515. [DOI] [PubMed] [Google Scholar]
- Brown M.H., Boles K., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., Boles K., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., van der Merwe P.A., Kumar V., Mathew P.A., Barclay A.N., Kumar V., Mathew P.A., Barclay A.N., Mathew P.A., Barclay A.N., Barclay A.N. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 1998;188:2083–2090. doi: 10.1084/jem.188.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark H.F., Gurney A.L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Gurney A.L., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Abaya E., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Baker K., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Baldwin D., Brush J., Chen J., Chow B., Chui C., Crowley C., Brush J., Chen J., Chow B., Chui C., Crowley C., Chen J., Chow B., Chui C., Crowley C., Chow B., Chui C., Crowley C., Chui C., Crowley C., Crowley C., et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: A bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.E., Wright C.L., Edwards C.A., Davis M.P., Grinham J.A., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Wright C.L., Edwards C.A., Davis M.P., Grinham J.A., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Edwards C.A., Davis M.P., Grinham J.A., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Davis M.P., Grinham J.A., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Grinham J.A., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Cole C.G., Goward M.E., Aguado B., Mallya M., Mokrab Y., Goward M.E., Aguado B., Mallya M., Mokrab Y., Aguado B., Mallya M., Mokrab Y., Mallya M., Mokrab Y., Mokrab Y., et al. A genome annotation-driven approach to cloning the human ORFeome. Genome Biol. 2004;5:R84. doi: 10.1186/gb-2004-5-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Kinetz E., Compton D.L., Rojas E., Park J.S., Compton D.L., Rojas E., Park J.S., Rojas E., Park J.S., Park J.S., et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Durocher Y., Perret S., Kamen A., Perret S., Kamen A., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:e9. doi: 10.1093/nar/30.2.e9. http://nar.oxfordjournals.org/cgi/content/full/30/2/e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco M., Marcenaro E., Romeo E., Bellora F., Marras D., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Marcenaro E., Romeo E., Bellora F., Marras D., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Romeo E., Bellora F., Marras D., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Bellora F., Marras D., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Marras D., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Vely F., Ferracci G., Moretta L., Moretta A., Bottino C., Ferracci G., Moretta L., Moretta A., Bottino C., Moretta L., Moretta A., Bottino C., Moretta A., Bottino C., Bottino C. Homophilic interaction of NTBA, a member of the CD2 molecular family: induction of cytotoxicity and cytokine release in human NK cells. Eur. J. Immunol. 2004;34:1663–1672. doi: 10.1002/eji.200424886. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.6. 2005. [Google Scholar]
- Flaig R.M., Stark S., Watzl C., Stark S., Watzl C., Watzl C. Cutting edge: NTB-A activates NK cells via homophilic interaction. J. Immunol. 2004;172:6524–6527. doi: 10.4049/jimmunol.172.11.6524. [DOI] [PubMed] [Google Scholar]
- Futschik M.E., Chaurasia G., Herzel H., Chaurasia G., Herzel H., Herzel H. Comparison of human protein–protein interaction maps. Bioinformatics. 2007;23:605–611. doi: 10.1093/bioinformatics/btl683. [DOI] [PubMed] [Google Scholar]
- George R.A., Heringa J., Heringa J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- Gill S.C., von Hippel P.H., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez L.C., Loyet K.M., Calemine-Fenaux J., Chauhan V., Wranik B., Ouyang W., Eaton D.L., Loyet K.M., Calemine-Fenaux J., Chauhan V., Wranik B., Ouyang W., Eaton D.L., Calemine-Fenaux J., Chauhan V., Wranik B., Ouyang W., Eaton D.L., Chauhan V., Wranik B., Ouyang W., Eaton D.L., Wranik B., Ouyang W., Eaton D.L., Ouyang W., Eaton D.L., Eaton D.L. A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc. Natl. Acad. Sci. 2005;102:1116–1121. doi: 10.1073/pnas.0409071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger M., Sutti F., Panigada M., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F., Sutti F., Panigada M., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F., Panigada M., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F., Porcellini S., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F., Merati B., Mariani M., Teesalu T., Consalez G.G., Grassi F., Mariani M., Teesalu T., Consalez G.G., Grassi F., Teesalu T., Consalez G.G., Grassi F., Consalez G.G., Grassi F., Grassi F. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J. Cell Biol. 1998;141:1061–1071. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon C., Lewis J., Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J. Comp. Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hall C., Flores M.V., Murison G., Crosier K., Crosier P., Flores M.V., Murison G., Crosier K., Crosier P., Murison G., Crosier K., Crosier P., Crosier K., Crosier P., Crosier P. An essential role for zebrafish Fgfrl1 during gill cartilage development. Mech. Dev. 2006;123:925–940. doi: 10.1016/j.mod.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Holler N., Kataoka T., Bodmer J.L., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P., Kataoka T., Bodmer J.L., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P., Bodmer J.L., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P., Deperthes D., Engel J., Tschopp J., Schneider P., Engel J., Tschopp J., Schneider P., Tschopp J., Schneider P., Schneider P. Development of improved soluble inhibitors of FasL and CD40L based on oligomerized receptors. J. Immunol. Methods. 2000;237:159–173. doi: 10.1016/s0022-1759(99)00239-2. [DOI] [PubMed] [Google Scholar]
- Ispolatov I., Yuryev A., Mazo I., Maslov S., Yuryev A., Mazo I., Maslov S., Mazo I., Maslov S., Maslov S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T., Kuma K., Toh H., Miyata T., Toh H., Miyata T., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F., Kimmel S.R., Ullmann B., Schilling T.F., Ullmann B., Schilling T.F., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kumaresan P.R., Lai W.C., Chuang S.S., Bennett M., Mathew P.A., Lai W.C., Chuang S.S., Bennett M., Mathew P.A., Chuang S.S., Bennett M., Mathew P.A., Bennett M., Mathew P.A., Mathew P.A. CS1, a novel member of the CD2 family, is homophilic and regulates NK cell function. Mol. Immunol. 2002;39:1–8. doi: 10.1016/s0161-5890(02)00094-9. [DOI] [PubMed] [Google Scholar]
- Letarte M., Voulgaraki D., Hatherley D., Foster-Cuevas M., Saunders N.J., Barclay A.N., Voulgaraki D., Hatherley D., Foster-Cuevas M., Saunders N.J., Barclay A.N., Hatherley D., Foster-Cuevas M., Saunders N.J., Barclay A.N., Foster-Cuevas M., Saunders N.J., Barclay A.N., Saunders N.J., Barclay A.N., Barclay A.N. Analysis of leukocyte membrane protein interactions using protein microarrays. BMC Biochem. 2005;6:2. doi: 10.1186/1471-2091-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.C., Ho W.H., Gurney A., Rosenthal A., Ho W.H., Gurney A., Rosenthal A., Gurney A., Rosenthal A., Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat. Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- Martin M., Romero X., de la Fuente M.A., Tovar V., Zapater N., Esplugues E., Pizcueta P., Bosch J., Engel P., Romero X., de la Fuente M.A., Tovar V., Zapater N., Esplugues E., Pizcueta P., Bosch J., Engel P., de la Fuente M.A., Tovar V., Zapater N., Esplugues E., Pizcueta P., Bosch J., Engel P., Tovar V., Zapater N., Esplugues E., Pizcueta P., Bosch J., Engel P., Zapater N., Esplugues E., Pizcueta P., Bosch J., Engel P., Esplugues E., Pizcueta P., Bosch J., Engel P., Pizcueta P., Bosch J., Engel P., Bosch J., Engel P., Engel P. CD84 functions as a homophilic adhesion molecule and enhances IFN-gamma secretion: Adhesion is mediated by Ig-like domain 1. J. Immunol. 2001;167:3668–3676. doi: 10.4049/jimmunol.167.7.3668. [DOI] [PubMed] [Google Scholar]
- Maslov S., Sneppen K., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Mavaddat N., Mason D.W., Atkinson P.D., Evans E.J., Gilbert R.J., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Mason D.W., Atkinson P.D., Evans E.J., Gilbert R.J., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Atkinson P.D., Evans E.J., Gilbert R.J., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Evans E.J., Gilbert R.J., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Gilbert R.J., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Stuart D.I., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Fennelly J.A., Barclay A.N., Davis S.J., Brown M.H., Barclay A.N., Davis S.J., Brown M.H., Davis S.J., Brown M.H., Brown M.H. Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity. J. Biol. Chem. 2000;275:28100–28109. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- Parrott M.B., Barry M.A., Barry M.A. Metabolic biotinylation of secreted and cell surface proteins from mammalian cells. Biochem. Biophys. Res. Commun. 2001;281:993–1000. doi: 10.1006/bbrc.2001.4437. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J.B., Levy E.D., Kamp C., Teichmann S.A., Levy E.D., Kamp C., Teichmann S.A., Kamp C., Teichmann S.A., Teichmann S.A. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo-Parada L., Bose C.M., Landmesser L.T., Bose C.M., Landmesser L.T., Landmesser L.T. Alterations in transmission, vesicle dynamics, and transmitter release machinery at NCAM-deficient neuromuscular junctions. Neuron. 2001;32:815–828. doi: 10.1016/s0896-6273(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Quarles R.H. Myelin sheaths: glycoproteins involved in their formation, maintenance and degeneration. Cell. Mol. Life Sci. 2002;59:1851–1871. doi: 10.1007/PL00012510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J., McNamee C., Rackstraw S., Jenkins J., Moss D., McNamee C., Rackstraw S., Jenkins J., Moss D., Rackstraw S., Jenkins J., Moss D., Jenkins J., Moss D., Moss D. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J. Cell Sci. 2004;117:3961–3973. doi: 10.1242/jcs.01261. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Romero X., Zapater N., Calvo M., Kalko S.G., de la Fuente M.A., Tovar V., Ockeloen C., Pizcueta P., Engel P., Zapater N., Calvo M., Kalko S.G., de la Fuente M.A., Tovar V., Ockeloen C., Pizcueta P., Engel P., Calvo M., Kalko S.G., de la Fuente M.A., Tovar V., Ockeloen C., Pizcueta P., Engel P., Kalko S.G., de la Fuente M.A., Tovar V., Ockeloen C., Pizcueta P., Engel P., de la Fuente M.A., Tovar V., Ockeloen C., Pizcueta P., Engel P., Tovar V., Ockeloen C., Pizcueta P., Engel P., Ockeloen C., Pizcueta P., Engel P., Pizcueta P., Engel P., Engel P. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J. Immunol. 2005;174:7033–7042. doi: 10.4049/jimmunol.174.11.7033. [DOI] [PubMed] [Google Scholar]
- Rougon G., Hobert O., Hobert O. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu. Rev. Neurosci. 2003;26:207–238. doi: 10.1146/annurev.neuro.26.041002.131014. [DOI] [PubMed] [Google Scholar]
- Schatz P.J. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Sidorenko S.P., Clark E.A., Clark E.A. The dual-function CD150 receptor subfamily: The viral attraction. Nat. Immunol. 2003;4:19–24. doi: 10.1038/ni0103-19. [DOI] [PubMed] [Google Scholar]
- Stagljar I., Korostensky C., Johnsson N., te Heesen S., Korostensky C., Johnsson N., te Heesen S., Johnsson N., te Heesen S., te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B., Thisse B. The Zebrafish Sci. Mon. Vol. 5. 1998. High resolution whole-mount in situ hybridization; pp. 8–9. [Google Scholar]
- Thisse B., Thisse C., Weston J.A., Thisse C., Weston J.A., Weston J.A. Novel FGF receptor (Z-FGFR4) is dynamically expressed in mesoderm and neurectoderm during early zebrafish embryogenesis. Dev. Dyn. 1995;203:377–391. doi: 10.1002/aja.1002030309. [DOI] [PubMed] [Google Scholar]
- Tomschy A., Fauser C., Landwehr R., Engel J., Fauser C., Landwehr R., Engel J., Landwehr R., Engel J., Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P.A., Barclay A.N., Barclay A.N. Transient intercellular adhesion: The importance of weak protein–protein interactions. Trends Biochem. Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- van der Merwe P.A., Barclay A.N., Barclay A.N. Analysis of cell-adhesion molecule interactions using surface plasmon resonance. Curr. Opin. Immunol. 1996;8:257–261. doi: 10.1016/s0952-7915(96)80065-3. [DOI] [PubMed] [Google Scholar]
- van der Merwe P.A., Barclay A.N., Mason D.W., Davies E.A., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Barclay A.N., Mason D.W., Davies E.A., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Mason D.W., Davies E.A., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Davies E.A., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Tone M., Krishnam A.K., Ianelli C., Davis S.J., Krishnam A.K., Ianelli C., Davis S.J., Ianelli C., Davis S.J., Davis S.J. Human cell-adhesion molecule CD2 binds CD58 (LFA-3) with a very low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- Voulgaraki D., Mitnacht-Kraus R., Letarte M., Foster-Cuevas M., Brown M.H., Barclay A.N., Mitnacht-Kraus R., Letarte M., Foster-Cuevas M., Brown M.H., Barclay A.N., Letarte M., Foster-Cuevas M., Brown M.H., Barclay A.N., Foster-Cuevas M., Brown M.H., Barclay A.N., Brown M.H., Barclay A.N., Barclay A.N. Multivalent recombinant proteins for probing functions of leucocyte surface proteins such as the CD200 receptor. Immunology. 2005;115:337–346. doi: 10.1111/j.1365-2567.2005.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.F., Barclay A.N., Barclay A.N. The immunoglobulin superfamily—Domains for cell surface recognition. Annu. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Wright G.J., Puklavec M.J., Willis A.C., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N., Puklavec M.J., Willis A.C., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N., Willis A.C., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N., Hoek R.M., Sedgwick J.D., Brown M.H., Barclay A.N., Sedgwick J.D., Brown M.H., Barclay A.N., Brown M.H., Barclay A.N., Barclay A.N. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity. 2000;13:233–242. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- Zhang J., Lefebvre J.L., Zhao S., Granato M., Lefebvre J.L., Zhao S., Granato M., Zhao S., Granato M., Granato M. Zebrafish unplugged reveals a role for muscle-specific kinase homologs in axonal pathway choice. Nat. Neurosci. 2004;7:1303–1309. doi: 10.1038/nn1350. [DOI] [PubMed] [Google Scholar]