Abstract
Among the many PWWP-containing proteins, the largest group of homologous proteins is related to hepatoma-derived growth factor (HDGF). Within a well-conserved region at the extreme N-terminus, HDGF and five HDGF-related proteins (HRPs) always have a PWWP domain, which is a module found in many chromatin-associated proteins. In this study, we determined the solution structure of the PWWP domain of HDGF-related protein-3 (HRP-3) by NMR spectroscopy. The structure consists of a five-stranded β-barrel with a PWWP-specific long loop connecting β2 and β3 (PR-loop), followed by a helical region including two α-helices. Its structure was found to have a characteristic solvent-exposed hydrophobic cavity, which is composed of an abundance of aromatic residues in the β1/β2 loop (β-β arch) and the β3/β4 loop. A similar ligand binding cavity occurs at the corresponding position in the Tudor, chromo, and MBT domains, which have structural and probable evolutionary relationships with PWWP domains. These findings suggest that the PWWP domains of the HDGF family bind to some component of chromatin via the cavity.
Keywords: NMR, HDGF, HATH region, PR-loop, β-β arch, cavity, protein structure
The PWWP domain, which was named after the relatively well-conserved residues, Pro-Trp-Trp-Pro, is composed of ~90 residues and is present within all eukaryotes (Stec et al. 1998). PWWP domains have been found in >60 proteins, which are involved in the processes of transcriptional regulation, DNA repair, and DNA methylation (Stec et al. 2000).
Recently, the function of the PWWP domain as a chromatin targeting module has been indicated in the mammalian DNA methyltransferases, Dnmt3a and Dnmt3b, which methylate genomic DNA during gametogenesis and early embryonic development (Ge et al. 2004). Experiments using mammalian cells have shown that the PWWP domain is required for the association of Dnmt3a and Dnmt3b with chromatin, suggesting that the PWWP-mediated chromatin targeting is essential for the enzyme function during development. The functional importance of the PWWP-mediated chromatin targeting was also suggested by the fact that a missense mutation in the PWWP domain causes ICF (immunodeficiency, centromeric heterochromatin instability, and facial anomalies) syndrome, which is characterized by hypomethylation in classical satellite DNA (Shirohzu et al. 2002). Experiments revealed that the mutant protein had completely lost its chromatin targeting capacity (Ge et al. 2004). However, the ligand and its functional interaction mechanism are still unknown.
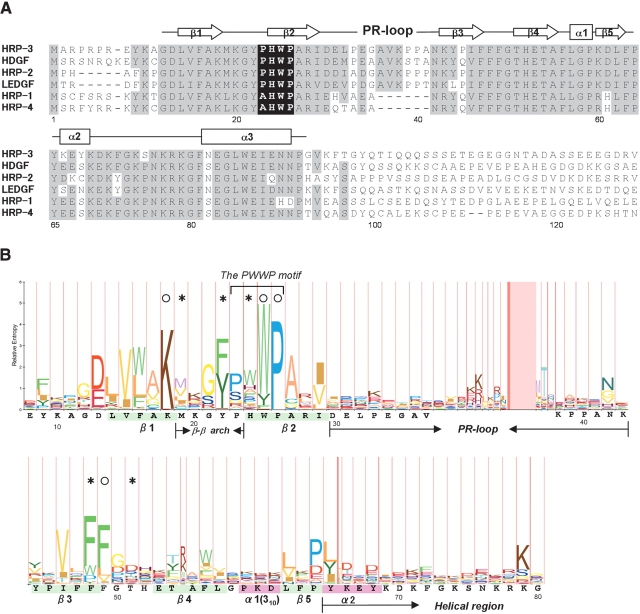
Among the PWWP-containing proteins, the largest homologous group is related to the hepatoma-derived growth factor (HDGF), which has growth-stimulating activity in several specific cells (Stec et al. 2000). The PWWP domain always occurs at the N-termini of HDGF and the HDGF-related proteins (HRPs). Its sequence is well conserved in the HDGF family (Fig. 1A ▶), and is also termed the HATH (homologous to amino terminus of HDGF) region (Izumoto et al. 1997). In contrast, the region outside the PWWP domain has different sequences in the HDGF family, and is termed a gene-specific region.
Figure 1.
(A) Sequence alignment of the N-terminal regions of the HDGF family members. Accession codes used are as follows: in Mus musculus, HRP-3, NP_038914; HDGF, BAA09838; HRP-2, NP_032259; LEDGF, NP_598709; HRP-1, NP_032258; in Bos taurus, HRP-4, CAB40348. Gray-colored residues indicate that more than half of the six residues in the same position are identical [the following residues in parentheses are grouped into the same type: (I, L, M, V) and (E, D)]. The PWWP motif is indicated by a black background. (B) Alignment of the Pfam PWWP domains (Bateman et al. 2004) shown in the HMM-Logo format, where the tallest residues are invariant (Schuster-Böckler et al. 2004). The sequence of the PWWP domain of HRP-3 is shown below with the secondary structural elements colored. Circles and asterisks indicate highly conserved residues involved in the interactions between α3 and the β-barrel substructure, and the formation of the cavity, respectively, as described in the text.
The first member, HDGF, was initially isolated as a heparin binding growth factor from the conditioned medium of HuH-7, a hepatoma cell line. It is expressed in various normal tissues and is amplified in several tumor cell lines (Wanschura et al. 1996; Everett 2001). HDGF participates in many cellular processes, such as astrocyte proliferation (Matsuyama et al. 2001), renal development (Oliver and Al-Awqati 1998), vascular lesion formation (Everett et al. 2000), and cardiovascular differentiation (Everett 2001). HDGF has mitogenetic activity for the growth of a variety of cells, including fibroblasts, hepatoma cells, vascular smooth muscle cells, and endothelial cells (Nakamura et al. 1994; Oliver and Al-Awqati 1998; Everett et al. 2000). HDGF contains two bipartite nuclear localization signals, but lacks a hydrophobic signal sequence for secretion (Everett et al. 2001; Kishima et al. 2002). The nuclear localization signals were shown to be required for HDGF to stimulate DNA replication (Everett et al. 2001; Kishima et al. 2002).
Five HDGF-related proteins (HRPs) have been identified thus far: mouse HRP-1 (Izumoto et al. 1997), mouse HRP-2 (Izumoto et al. 1997), human and mouse HRP-3 (Ikegame et al. 1999; Abouzied et al. 2004), bovine HRP-4 (Dietz et al. 2002), and human LEDGF (lens epithelium-derived growth factor) (Singh et al. 2000a). In addition to the PWWP domain at the N-terminus, these proteins always have putative bipartite nuclear localization signals. The mitogenic activity was confirmed with HRP-3 in fibroblasts and renal epithelial cells, HRP-4 in fibroblasts, and LEDGF in lens epithelial cells, keratinocytes, and fibroblasts. The mRNA expression shows a broad tissue distribution of HRP-2 as well as HDGF, whereas those of HRP-1 and HRP-4 are restricted to testis and that of HRP-3 is restricted to brain. All of the HRPs, as well as HDGF, were detected in the nuclei. Presumably, the HDGF family proteins are primarily targeted to the nucleus via the nuclear localization signals. Except for their growth factor activities, the functions of HRPs 1–4 are largely unknown, while the different functions of LEDGF have recently been revealed. By virtue of its homology to HDGF, the protein was originally found to be a novel growth factor on lens epithelial cells, keratinocytes, and fibroblasts, and hence it was termed LEDGF (Singh et al. 2000b). In addition, LEDGF has been shown to be a survival factor and a transcriptional activator, which rescues cells under stress by activating the expression of stress-related genes via its binding to their promoter elements (Singh et al. 2000b; Shinohara et al. 2002).
To obtain insights into the functions of the PWWP domains of the HDGF family, we determined the solution structure of the PWWP domain of mouse HRP-3 by heteronuclear NMR methods. Detailed structural comparisons with several domains having probable evolutionary relationships revealed a characteristic cavity as a putative binding site of the PWWP domain as well as the distinguishing structural features.
Results and Discussion
Resonance assignments and structural description
Samples of the 13C/15N-labeled PWWP domain of HRP-3, composed of 96 residues, were prepared for structure determination by a cell-free protein expression system. The protein sample has tag sequences (14 residues in total) at the N-terminus (GSSGSSG) and the C-terminus (SGPSSG), which are both derived from the expression vector. NMR resonances were assigned using conventional heteronuclear methods with the 13C/15N-labeled protein. The backbone resonance assignments were complete, with the exception of the first three residues in the tag sequence region at the N-terminus. Tertiary structures were calculated using the CYANA software package (Güntert et al. 1997; Herrmann et al. 2002), based on a total of 1377 NOE-derived distance restraints and 47 backbone torsion angle restraints (Table 1). A best-fit superposition of the ensemble of the 20 lowest-energy calculated structures is shown in Figure 2A ▶. The root mean square deviation (RMSD) from the mean structure was 0.30 ± 0.03 Å for the backbone (N, Cα, C′) atoms and 0.76 ± 0.03 Å for all heavy (nonproton) atoms in the well-ordered region (residues 13–29, 44–68, and 81–90). The statistics of the structures are summarized in Table 1.
Table 1.
Summary of conformational constraints and statistics of the final 20 best structures of the PWWP domain of mouse HRP-3
| NOE upper distance restraints | |
| Intraresidual (|i - j| = 0) | 349 |
| Medium-range (1 ≤ |i - j| ≤ 4) | 547 |
| Long-range (|i - j| > 4) | 481 |
| Total | 1377 |
| Dihedral angle restraints (φ and ψ) | 47 |
| CYANA target function value (Å2) | 0.70 ± 0.09 |
| Number of restraint violations | |
| Distance restraint violations (> 0.20 Å) | 0 |
| Dihedral angle restraint violations (> 5.0°) | 0 |
| AMBER energies (kcal/mol) | |
| Total | −2954 ± 71 |
| van der Waals | −297 ± 12 |
| Electrostatic | −3375 ± 64 |
| RMSD from ideal geometry | |
| Bond length (Å) | 0.0074 ± 0.0001 |
| Bond angles (°) | 1.87 ± 0.03 |
| Ramachandran plot (%) (residues 8–90) | |
| Residues in most favored regions | 81.7 |
| Residues in additional allowed regions | 16.9 |
| Residues in generously allowed regions | 0.9 |
| Residues in disallowed regions | 0.4 |
| RMSD deviation from the averaged coordinates (Å) | |
| Backbone atoms (residues 8–90) | 0.57 ± 0.12 |
| Heavy atom | 1.04 ± 0.10 |
| Backbone atoms (residues 13–29, 44–68, and 81–90) | 0.30 ± 0.03 |
| Heavy atom | 0.76 ± 0.03 |
Figure 2.

(A) Stereo view illustrating a trace of the backbone atoms for the ensemble of the 20 lowest energy structures of the mouse HRP-3 PWWP domain (residues 8–90). (B) Ribbon diagrams of the PWWP domains from the mouse HRP-3, the S. pombe protein SPBC215.07c (PDB code 1H3Z), and the mouse DNA methyltransferase Dnmt3b (1KHC) in the same view. Four residues in parentheses indicate those of the PWWP motif of each PWWP domain. The α-helices, β-strands and the PR-loop are colored pink, green, and dark orange, respectively.
The NMR results show that the PWWP domain has a β-barrel followed by a helical region, with the N- and C-termini on the same side of the molecule (Fig. 2B ▶). The β-barrel consists of five antiparallel strands (β1: 14–18, β2: 24–29, β3: 44–48, β4: 53–57; β5: 62–64). Between β4 and β5 occurs a 310-helix (α1:59–61). The helical region consists of a one-turn α-helix (α2: 65–68), a coil region (69–72), and a 10-residue α-helix (α3: 81–90).
The common topology for the PWWP structures
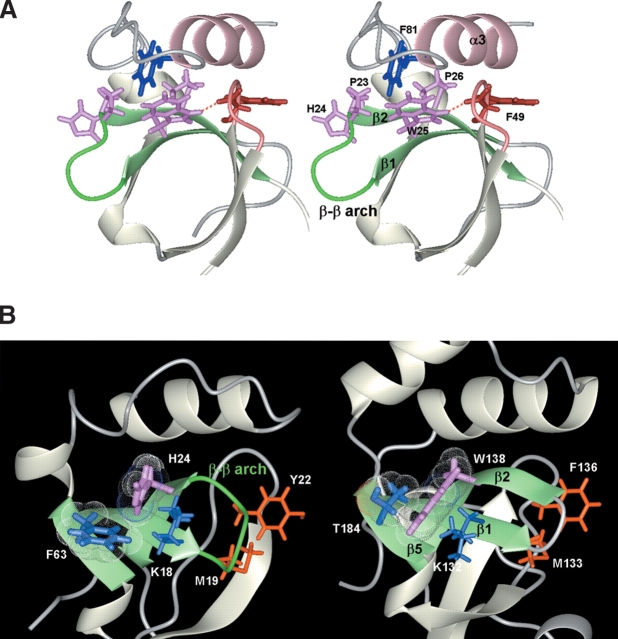
The structures of the PWWP domains were reported from mouse DNA methyltransferase Dnmt3b (Qiu et al. 2002) and from a Schizosaccharomyces pombe protein, SPBC215.07c, of unknown function (Slater et al. 2003). Structural comparisons showed significant similarities in the β-barrel region, and variations in the following helical region, revealing the structural essence of the PWWP domains (Fig. 2B ▶). In the helical region, only the positioning of an α-helix equivalent to α3 (the α3 equivalent) is virtually identical among the three PWWP structures, despite the apparent lack of sequence conservation (Fig. 1B ▶). This common α-helix is always placed in a groove between β2 and the β3/β4 loop, in a fixed direction (Fig. 2B ▶). The interactions involve highly conserved residues, including the third (Trp25) and fourth (Pro26) residues of the PWWP motif in β2 (Fig. 3A ▶). The aromatic ring of Trp25 vertically stacks with that of Phe81 (Tyr in the Dnmt3b domain and Leu in the S. pombe domain) at the beginning of α3, and the opposite side is partially solvent accessible. The Hɛ1 of the indole ring of Trp25 hydrogen-bonds with the O of Phe49 in the β3/β4 loop. The fourth residue, Pro26, located in the middle of β2, is buried with α3. In addition, the aromatic ring of Phe49 in the β3/β4 loop is snugly sandwiched between α2 and α3. This PWWP architecture involving the highly conserved residues, Trp25, Pro26, and Phe49, is essentially the same among the three PWWP structures. The positioning of α3, β2, and the β3/β4 loop is probably retained in all of the PWWP domains, and is involved in forming a cavity as a putative binding site, as described later. It thus appears that the common topology for the PWWP structures is β-β-β-β-310-β-the α3 equivalent.
Figure 3.
(A) Stereo view illustrating the interactions between α3 and the β-barrel substructure. β1, β2 and the connecting loop (β-β arch) are depicted in green, while α3 is pink. Shown are the PHWP residues (hot pink), Phe49 and the β3/β4 loop (brown), and Phe81 (navy). A hydrogen bond is depicted by a red dotted line. (B) Comparison of the second residue of the PWWP motif between the HRP-3 and S. pombe domains. β1, β2 and β5 are depicted in green. van der Waals surfaces are shown for the side chain atoms of His24 (hot pink) and Phe63 (navy) in the HRP-3 domain, and for those of Trp138 (hot pink) and Thr184 (navy) in the S. pombe domain. Residues in the β-β arch are colored orange.
Role of the unique His residue among the PHWP residues
Although the third (Trp) and fourth (Pro) residues of the PWWP motif are highly conserved, the other residues vary (Fig. 1B ▶). The first residue of the PWWP motif is located at the end of a loop, with the side chain accessible to the solvent, and thus its position could be occupied by several different residues. The second residue position is usually occupied by aromatic residues. The His residue at the second position is unique to the PWWP domains of the HDGF family. The PWWP structure with the PHWP residues shows that the imidazole ring of His24 in β2 is located over β1, and vertically contacts the aromatic ring of Phe63 in β5 (Fig. 3B ▶). Instead of the His and Phe residues, the PWWP domain of the S. pombe SPBC215.07c protein with the PWWP residues has Trp138 and Thr184 at the corresponding regions, respectively, and the indole ring of Trp is located over β1 and stacks well with the methyl of Thr in β5 (Slater et al. 2003). Hence, by comparison between the two PWWP domains, it follows that the indole ring of Trp, which is larger than that of His by a phenyl ring, compensates well for the space created by the replacement of Phe by Thr in β5. One side of each of the aromatic rings of His24 and Trp138 in the HRP-3 and S. pombe domains interacts with the aliphatic portions of the invariant Lys residues in β1, Lys18 and Lys132, respectively (Fig. 3B ▶). A similar conformation also exists in the Dnmt3b domain, with Trp and Val in the corresponding positions (Qiu et al. 2002). Hence, despite these residue differences, the structural role of the second residue of the PWWP motif is virtually identical among the three PWWP structures. As an exception to the aromatic residues, Ala, Ser, or Pro occurs at the second residue position in a small number of PWWP domains (Fig. 1B ▶). It will be interesting to examine the PWWP structures with these aliphatic residues at the second position of the PWWP motif.
β-β Arch
The loop connecting β1 and β2 is composed of five residues, 19MKGYP23, in the region neighboring the conserved Lys18 through the first residue, Pro23, of the PWWP motif (Fig. 3B ▶). According to the Ramachandran nomenclature (Efimov 1993), this region adopts a ββαLββ conformation. Its conformation provides a reverse turn in the polypeptide chain and facilitates its transition from one layer to the other. Thus, this loop is called a β-β arch, but it is not a canonical β-turn (Efimov 1993). The arch formation allows the side chains of Met19 and Tyr22 to be directed to the β3/β4 loop, and to participate as components of the cavity, as described later. A similar ββαLββ conformation is seen in the range of 133MSGFP137 in the S. pombe domain (Slater et al. 2003) (in the X-ray structure of the Dnmt3b domain, the electron density is not observed for Phe243, corresponding to Tyr22 in the HRP-3 domain). In all of the PWWP domains, the β-β arch region always contains five residues, and the predominant residue pattern is M/L/V-K-G-F/Y/H-P/S/A (Fig. 1B ▶), suggesting the conservation of the β-β arch conformation among all of the domains, presumably also to allow the cavity formation.
The proline-rich loop (PR-loop) connecting β2 and β3
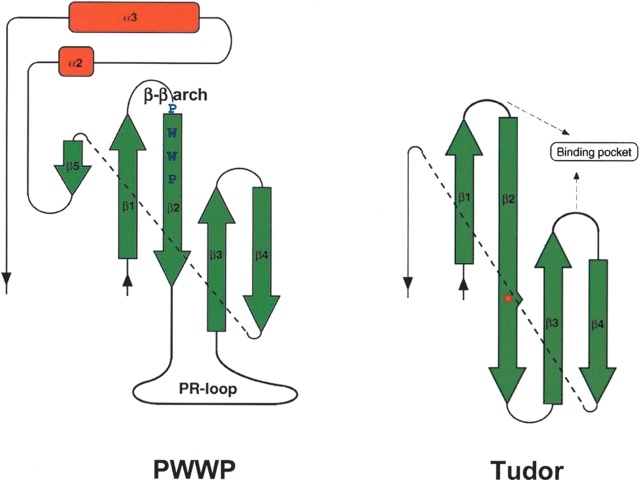
Within the β-barrel substructure of the three PWWP domains, the only apparent differences were found in a loop connecting β2 and β3, which includes short helices in two domains (Fig. 2B ▶). The sequence and length (8 ~ 20 residues) of this region vary among all of the PWWP domains (Fig. 1B ▶). This loop is relatively rich in prolines, and thus we call it the proline-rich loop, PR-loop, in this paper. The characteristic feature of the PR-loop is highlighted by comparison with the corresponding loop in the Tudor domain, which has the most significant similarity to the β-barrel substructure of the PWWP domain (Qiu et al. 2002; Maurer-Stroh et al. 2003; Fig. 4 ▶). In the Tudor domain, the presence of a β-bulge in β2 allows the β-strand to bend sufficiently to form a β-barrel. Consequently, β2 and β3 form a longer β-sheet than that found in the PWWP domain, and the region connecting β2 and β3 forms a short loop or a turn. In contrast, in the PWWP domain, the lack of a β-bulge in β2 prevents the formation of such a long β-sheet, and at the position equivalent to the β-bulge, the PR-loop begins. Accordingly, β2 and β3 cross each other like the character “λ,” so that the connecting loop, the PR-loop, is stretched in a characteristic form.
Figure 4.
Topological comparisons of the PWWP and Tudor domains. Note that the region depicted by a broken line region corresponds to a 310-helix in the two domains. An asterisk indicates a β-bulge.
A characteristic cavity as a putative binding site
A possible functional role of the PWWP domain is suggested by its structural characteristics, which are related to its putative evolutionary relationships. Extensive structural and sequence analyses revealed a family of homologous motifs containing a three-β-stranded core element, which includes the PWWP, Tudor, chromo (chromatin-binding), and MBT (malignant brain tumor) domains (Maurer-Stroh et al. 2003). The last three domains have a ligand interaction site located at the corresponding position, where a richly aromatic site involving the β-β arch and the β3/β4 loop plays a major role (in the chromodomain, the β2 equivalent is missing) (Sathyamurthy et al. 2003; Wang et al. 2003; Fig. 5A ▶). The binding sites seem to be sandwiched by a clothespin made of the two sticks of β1/β2 and β3/β4. As for the ligand, the Tudor domain of the survival motor neuron (SMN) protein binds to methylated Arg residues in the C-terminal Arg-Gly-Gly rich tails of the Sm proteins (Selenko et al. 2001). The chromodomain binds to the methylated lysine peptide from the histone H3 tail (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002). Although the function of the MBT repeats is unknown, one ligand binding pocket per MBT repeat seems to exist at the corresponding position, which in the X-ray structure can accommodate the morpholino ring of 2-(N-morpholino) ethanesulfonic acid (MES) or the proline ring of the C-terminal peptide segment (Wang et al. 2003).
Figure 5.
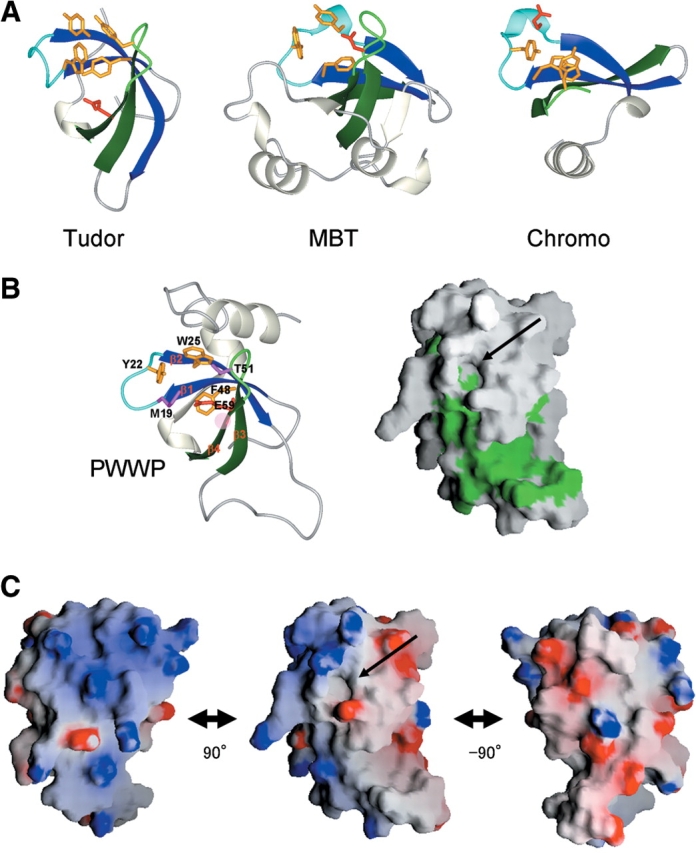
(A) Binding sites of the Tudor (PDB code 1G5V), MBT (1OZ3) and chromo (1KNA) domains. In the MBT domain, the region encompassing residues 349–44 is shown. The equivalents to β1 and β2 in the PWWP domain are colored blue, while those corresponding to β3 and β4 are dark green. Aromatic residues and negatively charged residues involved in ligand binding are colored dark orange and red, respectively. (B) A cavity as a putative binding site in the PWWP domain (left) and molecular surface representations showing hydrophobic residues in green (right). The figure on the right was prepared with GRASP (Nicholls et al. 1991). Residues involved in cavity formation are shown. Arrows indicate the cavity as a putative binding site. The red hatched circle indicates the position corresponding to that in which a missense mutation causes ICF syndrome in the human Dnmt3b PWWP domain, as described in the text. (C) Molecular surface representations of the electrostatic potential (blue, positive; red, negative) of the PWWP domain, calculated by GRASP. The middle view is in the same orientation as those in (B). On the left and right, the views are rotated by 90° around the vertical axis.
The corresponding position of the PWWP structure has a solvent-exposed hydrophobic cavity, which is composed of Trp25 in β2 of the PWWP motif, Met19 and Tyr22 in the β-β arch (the β1/β2 loop), Thr51 in the β3/β4 loop and Phe48 in β3 (Fig. 5B ▶). Many PWWP domains possess M/L/V at position 19, Y/F/H at 22, and T/S/D/H at 51; and the more conserved residues, W (in a few, Y) at 25 and F (W) at 48 (Fig. 1B ▶). In addition to these residues in the β-barrel region, only in the PWWP domains, the α3 equivalent, which interacts with β2 (particularly, Trp25) and the β3/β4 loop, appears to contribute to cavity formation. At the side of the cavity, a negatively charged residue, Glu53, exists in β4, which is an interaction site in the Tudor and chromo domains (Fig. 5A ▶). Its probable evolutionary relationship to those domains that bind to the modified residues suggests that the PWWP domain binds to a modified residue of some chromatin component via the cavity. In the PWWP domains of the HDGF family, the cavity residues are all identical, suggesting the same ligand for a minimal element. The possibility cannot be excluded that other elements of the ligand, for example the flanking residues, differ depending on the proteins. It is notable that this cavity is followed by a hydrophobic patch composed mainly of residues in the PR-loop (Fig. 5B ▶), which is the only region that differs in sequence and length among the PWWP domains of the HDGF family (Fig. 1A ▶). The PR-loop may play specific roles in the interaction of other elements of the ligand, conferring ligand variations to each PWWP domain.
In the human Dnmt3b PWWP domain, a missense mutation (S282P, the corresponding position is Ala55 in β4 of the PWWP domain in this study) causes ICF syndrome (Shirohzu et al. 2002). Intriguingly, this position is quite close to the cavity, or could be one component of the cavity structure (Fig. 5B ▶). This mutation may alter the structure or the volume of the cavity so significantly as to affect the ligand binding affinity and/or specificity.
The possible chromatin binding property of the PWWP domain is supported by its characteristic electrostatic charge distribution (Fig. 5C ▶). The charged residues are clustered in distinctly polarized patches, which surround the putative binding cavity. A similar surface electrostatic charge distribution exists in the bromodomain, which binds selectively to multiply acetylated histone H4 peptides (Jacobson et al. 2000). Upon binding, the charged surfaces may make non-specific contacts with the nucleic acid on the basic side, and contact the core histone molecules along the acidic surface.
Concluding remarks
Recent in vivo experiments have demonstrated that the PWWP domain of DNA methyltransferases is essential for the chromatin targeting of the enzymes, suggesting a direct interaction between the PWWP domain and chromatin (Ge et al. 2004). It is quite likely that some chromatin modifications, such as modified residues, are recognized by the PWWP-containing proteins via the PWWP domain, in which the cavity shown in this study plays a key role in the interaction.
Among the HDGF family members, LEDGF preferentially associates with condensed chromatin areas (Nishizawa et al. 2001). LEDGF confers cell protection against stress-induced apoptosis by binding to the promoter elements of heat shock and stress-related genes for activation (Singh et al. 2000b; Shinohara et al. 2002), and the PWWP domain may simultaneously bind to the chromatin surrounding the genes to facilitate or regulate the DNA binding. During apoptosis, interestingly, LEDGF is reportedly cleaved at three sites, by caspases-3 and -7, into two fragments lacking the PWWP domain (65 and 58 kDa), and this cleavage abrogates its pro-survival function (Wu et al. 2002). One of the cleavage sites (-DEVPD↑G-) is located in the PR-loop of LEDGF. These findings suggest the importance of the PWWP-mediated chromatin targeting for the protein function, as also seen with Dnmt3b. Similarly, the other HRPs and HDGF would be translocated to the nucleus by the nuclear localization signals, where they would associate via the PWWP domains with chromatin for their specific functions, which presumably depend on each gene-specific region in the C-terminus. Thus, the chromatin binding of these proteins seems to affect transcriptional processes, and consequently to regulate various cellular processes and development. The structural features of the PWWP domains provide insights for further experiments in the search for a ligand.
Materials and methods
Protein expression and purification
The DNA encoding the PWWP domain of mouse HRP-3 (Glu9-Asn90) was subcloned by PCR from the mouse full-length cDNA clone, with the ID RIKEN cDNA 4632410H03 (Okazaki et al. 2002). This DNA fragment was cloned into the expression vector pCR2.1 (Invitrogen) as a fusion with an N-terminal 6-His affinity tag and a TEV protease cleavage site. The 13C/15N-labeled fusion protein was synthesized by a cell-free protein expression system, as described (Kigawa et al. 1999, 2004). The solution was first adsorbed to a HiTrap Chelating column (Amersham Biosciences), which was washed with buffer A (50 mM sodium phosphate buffer, pH 8.0, containing 500 mM sodium chloride and 20 mM imidazole) and was eluted with buffer B (50 mM sodium phosphate buffer,pH 8.0, containing 500 mM sodium chloride and 500 mM imidazole). To remove the His-tag, the eluted protein was incubated at 30°C for 1 h with the TEV protease. After dialysis against buffer A without imidazole, the dialysate was mixed with imidazole (20 mM final concentration) and then applied to a HiTrap Chelating column, which was washed with buffer A. The flow through fraction was dialyzed against buffer C (20 mM sodium phosphate buffer, pH 7.2, containing 1 mM PMSF). The dialysate was fractionated on a HiTrap SP column by a concentration gradient of buffer C and buffer D (20 mM sodium phosphate buffer, pH 7.2, containing 1 M sodium chloride and 1 mM PMSF). The PWWP-containing fractions were collected.
For NMR measurements, the purified protein was concentrated to 0.7 mM in 1H2O/2H2O (9:1) 20 mM sodium phosphate buffer (pH 6.0), containing 100 mM NaCl, 1 mM 1,4-dl-dithiothreitol-d10 (d-DTT), and 0.02% NaN3. It was stable for at least 6 mo, when stored at 4°C.
NMR spectroscopy, structure determination, and analysis
All NMR measurements were performed at 25°C on Bruker AVANCE 600 and AVANCE 800 spectrometers. Sequence-specific backbone chemical shift assignments (Wüthrich 1986) were made with the 13C/15N-labeled sample, using standard triple-resonance experiments (Bax 1994; Cavanagh et al. 1996). Assignments of side chains were obtained from HBHACONH, HCCCONNH, CCCONNH, HCCH-TOCSY, and HCCH-COSY spectra. 15N- and 13C-edited NOESY spectra with 80- and 40-msec mixing times were used to determine the distance restraints. The spectra were processed with the program NMRpipe (Delaglio et al. 1995). The program KUJIRA (N. Kobayashi, pers. comm.), created on the basis of NMRView (Johnson and Blevins 1994), was employed for optimal visualization and spectral analysis.
Automated NOE cross-peak assignments (Herrmann et al. 2002) and structure calculations with torsion angle dynamics (Güntert et al. 1997) were performed using the software package CYANA1.0.7 (Güntert 2003). Peak lists of the two NOESY spectra were generated as input with the program NMRView (Johnson and Blevins 1994). The input further contained the chemical shift list corresponding to the sequence-specific assignments. Dihedral angle restraints were derived using the program TALOS (Cornilescu et al. 1999). No hydrogen bond constraints were used.
A total of 100 structures were independently calculated. The 20 conformers of the CYANA cycle 7 with the lowest final CYANA target function values were energy-minimized in a water shell with the program OPALp (Koradi et al. 2000), using the AMBER force field (Cornell et al. 1995). The structures were validated using PROCHECK-NMR (Laskowski et al. 1996). The program MOLMOL (Koradi et al. 1996) was used to analyze the resulting 20 conformers, and to prepare drawings of the structures, unless otherwise noted in the legends.
The atomic coordinates of the 20 energy-minimized CYANA conformers of the mouse HRP-3 PWWP domain have been deposited at the RCSB Protein Data Bank (www.rcsb.org) under the PDB Accession code 1N27.
Acknowledgments
We are indebted to Dr. Yutaka Muto for valuable suggestions and comments on the structural analysis. We thank Emi Nunokawa, Natsuko Matsuda, Yoko Motoda, Yukako Miyata, Atsuo Kobayashi, Noriko Hirakawa, Noriko Sakagami, Fumiko Hiroyasu, Yasuko Tomo, Takushi Harada, Miyuki Sato, Mari Hirato, Satoko Yasuda, and Takashi Osanai for their technical assistance. We also thank Tomoko Nakayama and Kiyomi Yajima for expert secretarial assistance. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04975305.
References
- Abouzied, M.M., Baader, S.L., Dietz, F., Kappler, J., Gieselmann, V., and Franken, S. 2004. Expression patterns and different subcellular localization of the growth factors HDGF (hepatoma-derived growth factor) and HRP-3 (HDGF-related protein-3) suggest functions in addition to their mitogenic activity. Biochem. J. 378 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32 D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax, A. 1994. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 4 738–744. [Google Scholar]
- Cavanagh, J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1996. Protein NMR spectroscopy, principles and practice. Academic Press, San Diego, CA.
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117 5179–5197. [Google Scholar]
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Dietz, F., Franken, S., Yoshida, K., Nakamura, H., Kappler, J., and Gieselmann, V. 2002. The family of hepatoma-derived growth factor proteins: Characterization of a new member HRP-4 and classification of its subfamilies. Biochem. J. 366 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov, A.V. 1993. Standard structures in proteins. Prog. Biophys. Mol. Biol. 60 201–239. [DOI] [PubMed] [Google Scholar]
- Everett, A.D. 2001. Identification, cloning, and developmental expression of hepatoma-derived growth factor in the developing rat heart. Dev. Dyn. 222 450–458. [DOI] [PubMed] [Google Scholar]
- Everett, A.D., Lobe, D.R., Matsumura, M.E., Nakamura, H., and McNamara, C.A. 2000. Hepatoma-derived growth factor stimulates smooth muscle cell growth and is expressed in vascular development. J. Clin. Invest. 105 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett, A.D., Stoops, T., and McNamara, C.A. 2001. Nuclear targeting is required for hepatoma-derived growth factor-stimulated mitogenesis in vascular smooth muscle cells. J. Biol. Chem. 276 37564–37568. [DOI] [PubMed] [Google Scholar]
- Ge, Y.Z., Pu, M.T., Gowher, H., Wu, H.P., Ding, J.P., Jeltsch, A., and Xu, G.L. 2004. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279 25447–25454. [DOI] [PubMed] [Google Scholar]
- Güntert, P. 2003. Automated NMR protein structure calculation. Prog. NMR Spectrosc. 43 105–125. [Google Scholar]
- Güntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Herrmann, T., Güntert, P., and Wüthrich, K. 2002. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24 171–189. [DOI] [PubMed] [Google Scholar]
- Ikegame, K., Yamamoto, M., Kishima, Y., Enomoto, H., Yoshida, K., Suemura, M., Kishimoto, T., and Nakamura, H. 1999. A new member of a hepatoma-derived growth factor gene family can translocate to the nucleus. Biochem. Biophys. Res. Commun. 266 81–87. [DOI] [PubMed] [Google Scholar]
- Izumoto, Y., Kuroda, T., Harada, H., Kishimoto, T., and Nakamura, H. 1997. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 238 26–32. [DOI] [PubMed] [Google Scholar]
- Jacobs, S.A. and Khorasanizadeh, S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295 2080–2083. [DOI] [PubMed] [Google Scholar]
- Jacobson, R.H., Ladurner, A.G., King, D.S., and Tjian, R. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288 1422–1425. [DOI] [PubMed] [Google Scholar]
- Johnson, B. and Blevins, R. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Kigawa, T., Yabuki, T., Yoshida, Y., Tsutsui, M., Ito, Y., Shibata, T., and Yokoyama, S. 1999. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 442 15–19. [DOI] [PubMed] [Google Scholar]
- Kigawa, T., Yabuki, T., Matsuda, N., Matsuda, T., Nakajima, R., Tanaka, A., and Yokoyama, S. 2004. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics 5 63–68. [DOI] [PubMed] [Google Scholar]
- Kishima, Y., Yamamoto, H., Izumoto, Y., Yoshida, K., Enomoto, H., Yamamoto, M., Kuroda, T., Ito, H., Yoshizaki, K., and Nakamura, H. 2002. Hepatoma-derived growth factor stimulates cell growth after translocation to the nucleus by nuclear localization signals. J. Biol. Chem. 277 10315–10322. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Güntert, P. 2000. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 124 139–147. [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Matsuyama, A., Inoue, H., Shibuta, K., Tanaka, Y., Barnard, G.F., Sugimachi, K., and Mori, M. 2001. Hepatoma-derived growth factor is associated with reduced sensitivity to irradiation in esophageal cancer. Cancer Res. 61 5714–5717. [PubMed] [Google Scholar]
- Maurer-Stroh, S., Dickens, N.J., Hughes-Davies, L., Kouzarides, T., Eisenhaber, F., and Ponting, C.P. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28 69–74. [DOI] [PubMed] [Google Scholar]
- Nakamura, H., Izumoto, Y., Kambe, H., Kuroda, T., Mori, T., Kawamura, K., Yamamoto, H., and Kishimoto, T. 1994. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor. Its homology with high mobility group-1 protein. J. Biol. Chem. 269 25143–25149. [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Nielsen, P.R., Nietlispach, D., Mott, H.R., Callaghan, J., Bannister, A., Kouzari-des, T., Murzin, A.G., Murzina, N.V., and Laue, E.D. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416 103–107. [DOI] [PubMed] [Google Scholar]
- Nishizawa, Y., Usukura, J., Singh, D.P., Chylack Jr., L.T., and Shinohara, T. 2001. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 305 107–114. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563–573. [DOI] [PubMed] [Google Scholar]
- Oliver, J.A. and Al-Awqati, Q. 1998. An endothelial growth factor involved in rat renal development. J. Clin. Invest. 102 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, C., Sawada, K., Zhang, X., and Cheng, X. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 9 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamurthy, A., Allen, M.D., Murzin, A.G., and Bycroft, M. 2003. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2). J. Biol. Chem. 278 46968–46973. [DOI] [PubMed] [Google Scholar]
- Schuster-Böckler, B., Schultz, J., and Rahmann, S. 2004. HMM Logos for visualization of protein families. BMC Bioinformatics 5 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko, P., Sprangers, R., Stier, G., Bühler, D., Fischer, U., and Sattler, M. 2001. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8 27–31. [DOI] [PubMed] [Google Scholar]
- Shinohara, T., Singh, D.P., and Fatma, N. 2002. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 21 341–358. [DOI] [PubMed] [Google Scholar]
- Shirohzu, H., Kubota, T., Kumazawa, A., Sado, T., Chijiwa, T., Inagaki, K., Suetake, I., Tajima, S., Wakui, K., Miki, Y., et al. 2002. Three novel DNMT3B mutations in Japanese patients with ICF syndrome. Am. J. Med. Genet. 112 31–37. [DOI] [PubMed] [Google Scholar]
- Singh, D.P., Kimura, A., Chylack Jr., L.T., and Shinohara, T. 2000a. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene 242 265–273. [DOI] [PubMed] [Google Scholar]
- Singh, D.P., Ohguro, N., Kikuchi, T., Sueno, T., Reddy, V.N., Yuge, K., Chylack Jr., L.T., and Shinohara, T. 2000b. Lens epithelium-derived growth factor: Effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem. Biophys. Res. Commun. 267 373–381. [DOI] [PubMed] [Google Scholar]
- Slater, L.M., Allen, M.D., and Bycroft, M. 2003. Structural variation in PWWP domains. J. Mol. Biol. 330 571–576. [DOI] [PubMed] [Google Scholar]
- Stec, I., Wright, T.J., van Ommen, G.J., de Boer, P.A., van Haeringen, A., Moorman, A.F., Altherr, M.R., and den Dunnen, J.T. 1998. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum. Mol. Genet. 7 1071–1082. [DOI] [PubMed] [Google Scholar]
- Stec, I., Nagl, S.B., van Ommen, G.J., and den Dunnen, J.T. 2000. The PWWP domain: A potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473 1–5. [DOI] [PubMed] [Google Scholar]
- Wang, W.K., Tereshko, V., Boccuni, P., MacGrogan, D., Nimer, S.D., and Patel, D.J. 2003. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure 11 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanschura, S., Schoenmakers, E.F., Huysmans, C., Bartnitzke, S., Van de Ven, W.J., and Bullerdiek, J. 1996. Mapping of the gene encoding the human hepatoma-derived growth factor (HDGF) with homology to the high-mobility group (HMG)-1 protein to Xq25. Genomics 32 298–300. [DOI] [PubMed] [Google Scholar]
- Wu, X., Daniels, T., Molinaro, C., Lilly, M.B., and Casiano, C.A. 2002. Caspase cleavage of the nuclear autoantigen LEDGF/p75 abrogates its pro-survival function: Implications for autoimmunity in atopic disorders. Cell Death Differ. 9 915–925. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.