Abstract
Phosphoglucose isomerase (PGI) catalyzes the isomerization of D-glucose-6-phosphate (G6P) and D-fructose-6-phosphate (F6P) in glycolysis and gluconeogenesis. Analysis of previously reported X-ray crystal structures of PGI without ligand, with the cyclic form of F6P, or with inhibitors that mimic the cis-enediol intermediate led to proposed mechanisms for the ring opening and isomerization steps in the multistep catalytic mechanism. To help complete our model of the overall mechanism, information is needed about the state of PGI between the ring opening and isomerization steps, in other words, a structure of the enzyme complexed with the open form of a substrate or an analog. Here, we report the crystal structure of rabbit PGI complexed with D-sorbitol-6-phosphate (S6P), an analog of the open chain form of G6P, at 2.0 Å resolution. As was seen in the PGI/F6P structure, a helix containing amino acid residues 512–520 is found in the “out” position, which provides sufficient space in the active site for a substrate in its cyclic form and which is probably the location of that helix just after ring opening (or just before ring closure). However, the S6P ligand is in an extended conformation, as was seen previously with ligands that mimic the cis-enediol intermediate. The extended conformation enables the ligand to interact with Glu357, which transfers a proton during the isomerization step. The PGI/S6P structure represents the conformation of the enzyme and substrate between the ring opening (or ring closing) step and the isomerization step and helps to complete the model for PGI’s catalytic mechanism.
Keywords: phosphoglucose isomerase, sorbitol-6-phosphate, catalytic mechanism
Phosphoglucose isomerase (PGI; E.C. 5.3.1.9) is a cytosolic enzyme that catalyzes the reversible isomerization of G6P and F6P. Its activity is important for glycolysis; gluconeogenesis; the pentose phosphate pathway; and the glycosylation of proteins, lipids, and other molecules. Recently, attention to PGI has grown because PGI is a moonlighting protein with a second function outside of the cell. PGI is the same protein as the extracellular growth factors neuroleukin (Chaput et al. 1988; Faik et al. 1988), autocrine motility factor (Watanabe et al. 1996), and differentiation and maturation mediator (Xu et al. 1996). It stimulates antibody secretion, promotes the survival of embryonic spinal neurons in vitro (Gurney et al. 1986a,b), affects an increase in tumor cell motility, and causes the differentiation of human myelogenous leukemia cells (Chiao et al. 1999) and a human myelocytic cell line (HL-60 cells) to terminal monocytic cells (Leung and Chiao 1985; Abolhassani et al. 1990). Recently, PGI also has been found to be the self-antigen in a mouse model of rheumatoid arthritis (Matsumoto et al. 1999; Schaller et al. 2001). One lab has identified antibodies to PGI in patients suffering from rheumatoid arthritis (Schaller et al. 2001).
The enzymatic activity of PGI was first described over 70 years ago (Lohmann 1933). Biochemical characterization of PGI enzyme activity by several labs led to a proposed multistep mechanism involving general acid–base catalysis (Rose and O’Connell 1961; Rose 1975, 1981; Willem et al. 1990). Both G6P and F6P are substrates for PGI activity. Because G6P and F6P exist predominantly in their cyclic forms in solution (Swenson and Barker 1971), it is believed that first the cyclic form of G6P or F6P binds to the active site. PGI catalyzes ring opening of the substrate to yield the open chain form. The reaction then proceeds through the isomerization step using acid–base catalysis with proton transfer.
PGI is a dimer with 557 amino acids in each subunit and has two equivalent active sites. X-ray crystal structures have been reported of PGI without bound ligand or with a substrate (F6P) or competitive inhibitors of catalytic activity bound in the active site pocket. X-ray crystal structures of mammalian (rabbit, pig, and human) PGI have been solved with 6PGA (Jeffery et al. 2000), 5PAA (Jeffery et al. 2001; Davies and Muirhead 2002; Davies et al. 2003), 5PAH (Arsenieva et al. 2002), or F6P (Lee et al. 2001), or with no bound ligand (Arsenieva and Jeffery 2002; Davies and Muirhead 2002, 2003). A crystal structure of human PGI with a sulfate ion was also reported (Read et al. 2001). Other crystal structures of PGI included two structures from a bacterium, Bacillus stearothermophilus, with no bound ligand (Sun et al. 1999) or complexed with 5PAA (Chou et al. 2000). Three crystal structures of PGI enzyme from Pyrococcus furiosus were also solved with no bound ligand (Berrisford et al. 2003), with 5PAA, and with 6PGA (Swan et al. 2003). Interestingly, the PGI enzyme from P. furiosus does not share sequence or structural homology with either mammalian or B. stearothermophilus PGI, and its activity is probably the result of convergent evolution.
The X-ray crystal structures of PGI have helped to identify active site amino acids, ordered water molecules, and conformational changes involved in the ligand binding, ring opening, and isomerization steps of the multistep catalytic mechanism. Comparison of a crystal structure of rabbit PGI without a bound ligand to structures with ligands provided information about induced fit of the enzyme upon substrate binding. A crystal structure of rabbit PGI complexed with F6P provided information about the roles of Glu216, His388, Lys518, and an ordered water molecule in the ring opening step (Lee et al. 2001). Crystal structures of rabbit PGI complexed with 5PAA and 5PAH were used to identify the role of Glu357 as the active site amino acid residue that transfers a proton between C1 and C2 during the isomerization step (Jeffery et al. 2001) and the role of a water molecule in transferring a proton between the hydroxyl groups on C1 and C2 (Arsenieva et al. 2002). Arg272 was proposed to have a role in helping stabilize the cis-enedio-l(ate) intermediate. Comparison of the PGI/5PAA, PGI/5PAH, and PGI/F6P complexes also indicated that the substrate undergoes a rotation about the C3–C4 bond between the ring opening and isomerization steps to position C1 and C2 near to Glu357. Also, a helix containing amino acid residues 512–520 moves in toward the substrate to help hold it in place for the isomerization step. However, less clear is the state of the active site and substrate between the ring opening and isomerization steps. To complete the details of the catalytic mechanism of PGI, a structure of PGI complexed with an open chain substrate, or an analog, is required. Here, we report an X-ray crystal structure of rabbit PGI complexed with an analog of the open chain form of G6P (S6P), bound in the active site. This structure was solved at 2.0 Å resolution.
Materials and methods
Crystallization of PGI
Rabbit skeletal muscle PGI was purchased from Sigma Chemical Co., desalted by ion exchange chromatography, and concentrated as described previously (Jeffery et al. 2000). The hanging drop vapor diffusion method was used to grow crystals at 22°C. The hanging drops contained an equal mixture of protein solution (20 mg/mL PGI, 10 mM imidazole at pH 7.5, and 50 mM KCl) and reservoir solution (16% PEG 8000, 250 mM magnesium acetate, and 100 mM sodium citrate at pH 5.4). The reservoir solution was modified from the reservoir solution used in previous reports of rabbit PGI structures (12%–14% PEG 8000, 250 mM magnesium acetate, and 100 mM sodium cacodylate at pH 6.5). The modified conditions helped to prevent formation of a sticky skin on the surface of the drops and also resulted in the decreased occurrence of deformed or twinned crystals. The drops were allowed to equilibrate with 1 mL of reservoir solution. The crystals grew as colorless hexagonal rods or hexagonal tablets. If crystals did not grow within 3–4 d, the drops were seeded with a crushed PGI crystal using the microseeding method.
Data collection
A diffraction data set from a single crystal of PGI complexed with S6P was collected at APS beamline 22-ID (SER-CAT) at Argonne National Laboratories using monochromatic X-rays. The crystal was ∼0.2 × 0.1 × 0.1 mm3 in size and had the symmetry of the orthorhombic space group C2221 (a = 82.77 Å, b = 115.19 Å, c = 271.79 Å). The data set was processed with the program HKL2000 (Otwinowski and Minor 1997). Following procedures similar to those used for previously reported structures of rabbit PGI with bound ligand (although using a new stabilization buffer), immediately before data collection, the crystal was soaked for ∼1 min in a stabilization solution containing the S6P ligand (18% PEG 8000, 250 mM magnesium acetate, 100 mM sodium citrate at pH 5.4, 5mM S6P, and 20% glycerol), and the crystal was flash frozen at −180°C. No damage to the crystal was visible after soaking. Attempts were also made to add in the G6P substrate after growing the crystals by including 5 mM G6P in the stabilization solution before freezing. Although several soaking times were tested for G6P, either no electron density or electron density that resembled F6P was seen in the active site pocket in difference electron density maps with coefficients of |Fo−Fc|. In addition, previous attempts to cocrystallize PGI with G6P resulted in F6P bound in the active site (Lee et al. 2001).
Structure refinement
Refinement consisted of alternating rounds of computational refinement using the CNS program package and manual fitting of the model to electron density maps using the program O (Jones et al. 1991). A previously reported rabbit PGI structure from which all the water molecules and the inhibitor molecules were removed (PDB entry 1KOJ (Arsenieva et al. 2002) was used as the initial model for refinement. Ten percent of the data set was set aside to compute Rfree (Brunger 1992). Simulated annealing was used to remove model bias. Data between 12 and 2.0 Å resolution were used in positional and B-factor refinement with a bulk solvent correction applied. During each round of refinement, the CNS program package was used to suggest potential water molecules using electron density maps with coefficients of |Fo−Fc| and contoured at 1.5 σ. Water molecules were added to the model based on those electron density maps, geometry, and refined B factors. During the final rounds of refinement, the S6P molecule was built into each active site in the PGI model. The stereochemistry of the model was checked with the programs CNS and PROCHECK (Collaborative Computational Project, No. 4 1994). The final R factor was 19.9 for all the data from 12 to 2.0 Å resolution with an Rfree factor of 21.9 (Table 1). Most of the amino acids were clearly visible in the electron density map, but some of the atoms in the side chains of several lysine, glutamine, glutamate, and leucine residues on the protein surface were disordered (amino acids 61, 72, 115, 123, 133, 146, 193, 230, 233, 251, 253, 417, 1115, 1193, 1233, 1253, 1260, and 1417). However, the side chains of these amino acids were included in the model for completeness.
Table 1.
Statistics for data collection and refinement
| Data collection | |
| Space group | C2221 |
| Cell dimensions (Å) | |
| a | 82.77 |
| b | 115.19 |
| c | 271.79 |
| Temperature | −180°C |
| Resolution range (Å) | 50–2.0 (2.09–2.02) |
| Observed reflections | |
| Total | 389,523 (33,700) |
| Unique | 84,679 (8425) |
| Completeness (%) | 99.1 (99.8) |
| Redundancy | 4.6 (4.0) |
| Rsym (% on I) | 0.073 (0.219) |
| Refinement | |
| Resolution range (Å) | 12–2.0 |
| R factor (%) | 19.9 |
| Rfree (%) | 21.9 |
| Ordered water molecules | 479 |
| R.m.s. deviations from ideal geometry | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.50 |
| Average B factor (Å2) | |
| Protein | 24.9 |
| S6P ligand | 27.6 |
| Solvent molecules | 30.0 |
Rsym = ∑ |Ii − 〈I〉|/∑ Ii R factor = ∑ ||Fo−Fc||/∑|Fo|. Rfree is the equivalent of R factor, but it is calculated for 10% of randomly chosen reflections that were omitted from the refinement process.
Results and Discussion
The structure of rabbit PGI complexed with S6P has been solved at 2.0 Å resolution by X-ray crystallography with an R factor of 19.9 and an Rfree factor of 21.9 (Table 1). The asymmetric unit contains a dimer of PGI, two S6P molecules, and 479 water molecules. Each protein subunit is made up of 557 amino acids, but amino acid 557 is disordered in the electron density maps and is not included in our model. The conformations of the amino acid residues in the model were checked with the program PROCHECK (Collaborative Computational Project, No. 4 1994), and all were in the allowed region of the Ramachandran Plot with good geometry. The crystal structure of rabbit PGI with bound S6P has the same overall fold as the previous crystal structures of rabbit and human PGI complexed with different ligands (inhibitors or substrates) or without ligand (Jeffery et al. 2000, 2001; Lee et al. 2001; Read et al. 2001; Arsenieva and Jeffery 2002; Arsenieva et al. 2002; Davies and Muirhead 2003; Davies et al. 2003). Each subunit contains two α/β sandwich domains and a C-terminal region that extends across the other subunit in the dimer. The root mean square deviation of α carbon positions was 0.34 Å compared with the structure of rabbit PGI with F6P bound and 0.44 Å compared with the structure of rabbit PGI with 5PAH bound.
S6P binding in the active site
Immediately before data collection, the PGI crystal was soaked for ∼1 min in a solution containing 5 mM S6P. S6P is an analog of the open chain form of the substrate (Fig. 1 ▶). Like G6P, it is a hexose with a phosphoryl group on C6. The orientation of the hydroxyl groups on C2, C3, C4, and C5 are the same in both molecules. However, G6P has an aldehyde group on C1, where S6P has a hydroxyl group. Because of this difference, S6P does not undergo cyclization and is a good analog of the open chain form of G6P.
Figure 1.
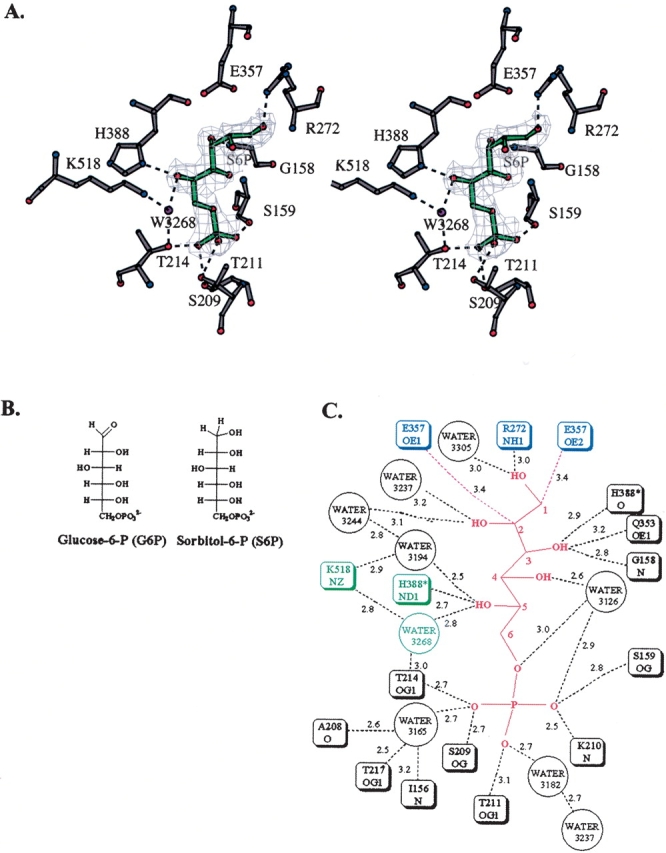
The PGI active site containing bound S6P. (A) Stereo diagram of the PGI active site. Ball-and-stick models of the inhibitor S6P (green); active site amino acid residues His388, Lys518, Glu216, Glu357, and Arg272 (gray); and ordered water molecules (purple) are shown. A simulated annealing composite omit map was calculated and is shown at a 1.5-σ contour level. This figure and Figure 2 ▶ were made using the program BOBSCRIPT (Esnouf 1997). Hydrogen bonds between active site amino acid residues and S6P are shown (dashed lines). (B) Both G6P and S6P are hexoses with a phosphoryl group on C6. The orientation of the hydroxyl groups on C2, C3, C4, and C5 are the same. However, G6P has an aldehyde group at C1 and S6P has a hydroxyl group at C1. S6P is unable to cyclize and is a good analog of the open chain form of G6P. This figure and Figure 3 ▶ were made using the program ChemDraw (CambridgeSoft). (C) A schematic diagram of interactions between PGI and the inhibitor S6P is shown. The side chain of Glu357 is near C1 and C2 so that it can abstract a proton at the start of the isomerization step (these distances are indicated by red dashed lines). Several other amino acid residues form hydrogen bonds (black dashed lines) with the S6P molecule. The side chain of Arg272 forms hydrogen bonds with the hydroxyl group on C1. The side chain of His388 forms a hydrogen bond with the hydroxyl group on C5. The side chain of Lys518 forms a hydrogen bond with an ordered water molecule, 3268, which forms a hydrogen bond with the hydroxyl group on C5. The phosphoryl group of S6P forms hydrogen bonds with the side chains of amino acid residues Thr214, Ser209, Thr211, and Ser159 and three water molecules. Lengths of potential hydrogen bonds are shown (in Angstroms) next to the dashed lines. Distances shown are from subunit 1. Corresponding distances in subunit 2 are within 0.2 Å.
After data collection and processing, a composite simulated annealing omit map was calculated for the entire structure and contoured at 1.5 σ. The electron density map for each of the two equivalent active sites in the dimer contained clear electron density corresponding to a bound S6P molecule (Fig. 1 ▶). The overall orientation of S6P in each active site corresponds to that of the ligand in each of the previous crystal structures of rabbit and human PGI with bound F6P, 6PGA, 5PAH, or 5PAA (Jeffery et al. 2000, 2001; Lee et al. 2001; Arsenieva et al. 2002; Davies et al. 2003). The position of the phosphoryl group of each of these ligands also corresponds to the position of a bound sulfate ion in another crystal structure of human PGI (Read et al. 2001). The phosphoryl group of S6P forms hydrogen bonds with the side chains of amino acid residues Thr214, Ser209, Thr211, and Ser159 and the backbone amide of residue Lys210 (Fig. 1 ▶). The phosphoryl group of S6P also forms hydrogen bonds with a water molecule, 3165, which also interacts with the side chain hydroxyl group of Thr217, the carbonyl oxygen of Ala208, and the backbone amide of Ile156.
The C1–C2 region of S6P is near amino acid residues Glu357 and Arg272, which are involved in the isomerization step. This conformation of S6P is similar to the conformation of the 5PAH inhibitor (Fig. 2B ▶) in the structure containing that inhibitor. The structure of PGI/5PAH mimics the enzyme with the cis-enediol(ate) intermediate of the isomerization step bound in the active site. Although the hydroxamate moiety of 5PAH is planar, and the S6P C1–C2 region is not, both groups are positioned similarly in the active site pocket. The carboxylate side chain of Glu357 is shifted and rotated, although only a small amount, so that it is approximately the same distance to C1 and C2 in S6P as to the corresponding atoms, N1 and C1, in the PGI/5PAH structure. S6P C1 is 3.4 Å from Glu357 Oɛ2, and C2 is 3.4 Å from Glu357 Oɛ1. This arrangement of S6P and the Glu357 side chain supports the model that the PGI/S6P structure corresponds to the enzyme–substrate complex with the open chain form of the substrate G6P before the isomerization step, with Glu357 poised to abstract a proton from C1. In addition to interactions with Glu357, the hydroxyl group on C1 forms hydrogen bonds with the side chain of Arg272, which helps stabilize the cis-enediol(ate) intermediate during the isomerization step, and there is an ordered water molecule, 3305, between the two hydroxyl groups on C1 and C2. From analysis of the PGI/5PAH structure, a water molecule in this position was proposed to play a role in transferring a proton between the hydroxyl groups on C1 and C2 during the isomerization step (Arsenieva et al. 2002). However, there are two differences between the PGI/S6P structure and the PGI/5PAH structure: The helix containing amino acid residues 512–520 is positioned in the “in” position (close to the bound inhibitor), and the side chain of Lys518 makes a direct interaction with the ligand in the PGI/5PAH structure.
Figure 2.
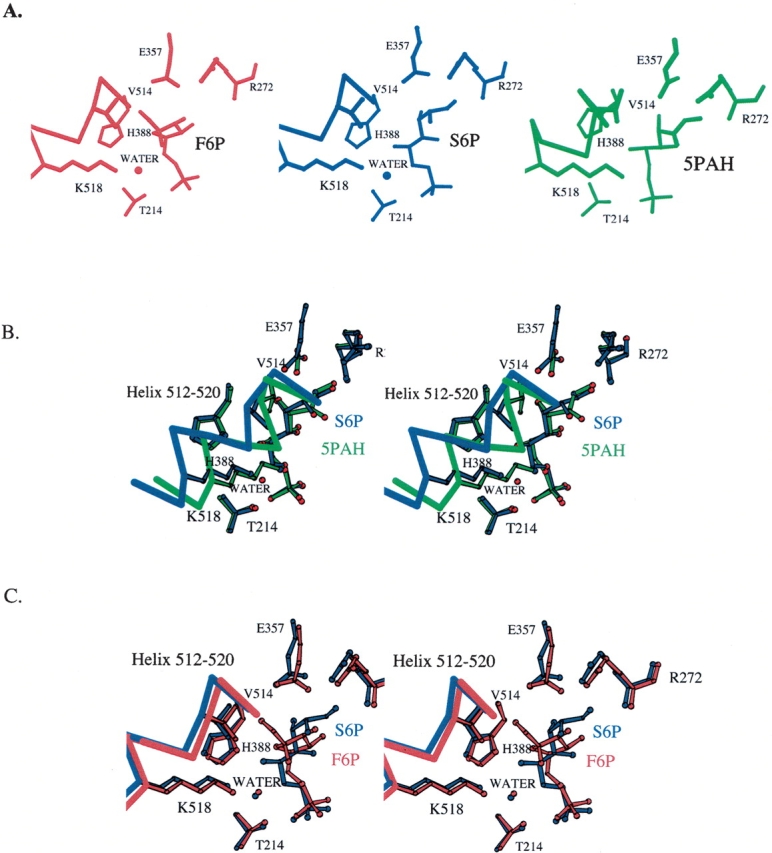
The active sites of the PGI/S6P, PGI/F6P, and PGI/5PAH complexes (A). There are three main differences in the active sites of the PGI/S6P, PGI/F6P, and PGI/5PAH complexes: (1) A helix containing amino acids 512–520 is in the “in” position in the PGI/5PAH complex (green) and it is in the “out” position in the PGI/F6P (red) and the PGI/S6P (blue) complexes. (2) An ordered water molecule is found between Lys518 and the ligand in the PGI/F6P (W333) and PGI/S6P (Water3268) complexes but not in the PGI/5PAH complex. The side chains of Lys518 and Thr214 help position the ordered water molecule. (3) The ligand is in an extended conformation in the PGI/S6P and PGI/5PAH complexes, but it is in a cyclic conformation in the PGI/F6P complex. Stereo diagrams of the superposition of the PGI/5PAH (B) or PGI/F6P (C) structures onto the PGI/S6P structure. The PGI/F6P (red) complex structure or the PGI/5PAH (green) complex structure was superposed onto the entire PGI/S6P (blue) complex structure (all 556 α-carbon positions). Only those regions of the structures in or near the active site pockets are shown. There are only minor differences in the positions of most amino acid residues in the active sites of PGI/S6P and PGI/F6P. The superpositions enable a clearer view of the differences among the three structures: (1) position of the helix containing amino acid residues 512–520, (2) presence or absence of an ordered water molecule between Lys518 and the ligand, and (3) conformation of the ligand.
Instead, in the PGI/S6P structure, the helix is in the “out” position (i.e., farther away from the ligand than in structures with 5PAA and 5PAH), and there is a water molecule between the ligand and Lys518 in the PGI/S6P structure. The “out” position of the helix and the presence of the water molecule are also seen in the crystal structure of PGI with the cyclic form of F6P (Fig. 2C ▶). The PGI/F6P structure represents the conformation of the ligand and protein just before the ring opening step (or just after the ring closing step). This “out” position of the helix prevents a steric clash between the side chain of Val514 and the hydroxyl group on C1 of the substrate.
Therefore, the PGI/S6P structure contains features of both the PGI/F6P and PGI/5PAH complexes: The ligand is in the extended conformation, but the helix containing amino acid residues 512–520 is in the “out” position seen in the PGI/F6P structure. The difference in the position of the 512–520 helix between the PGI/5PAH and PGI/S6P structures appears to affect the conformation of the inhibitors. Although the positions of the two ends of S6P in the active site are similar to the positions of the ends of 5PAH, there are small differences along the middle of the molecule. Small rotations around the C2–C3 and C4–C5 bonds in S6P relative to the equivalent bonds in 5PAH place C3, C4, C5, and C6 of S6P in slightly different positions from the corresponding atoms in 5PAH (C2, C3, C4, and C5 of 5PAH, respectively). The largest difference is a 1.3 Å shift in the position of C6 in S6P relative to the corresponding position of C5 in 5PAH. The shifts in relative atom positions are smaller toward the ends of the two molecules. Also, the distances from the hydroxyl groups on C3 and C4 in S6P to the active site amino acids suggest the equivalent hydrogen bonds form in both enzyme–inhibitor complexes. The hydroxyl group on C3 forms hydrogen bonds with the carbonyl oxygen of amino acid residue His388, the backbone amide group of Gly158, and the side chain of Gln353. The hydroxyl group on C4 interacts with a water molecule, 3126. However, while the hydroxyl group on C5 forms a hydrogen bond with the side chain of His388 in both structures, a second hydrogen bond in the PGI/S6P structure is with the water molecule (3268) that is held in place by Lys518 and Thr214. In the PGI/5PAH structure, the water molecule is absent, and the side chain of Lys518 interacts directly with the hydroxyl group on C5. This lack of direct interaction of S6P with Lys518 helps explain the shift of the position of the C6 atom in S6P relative to the corresponding atom in 5PAH and supports the model that the helix moves in to interact with and help position the ligand before the isomerization step.
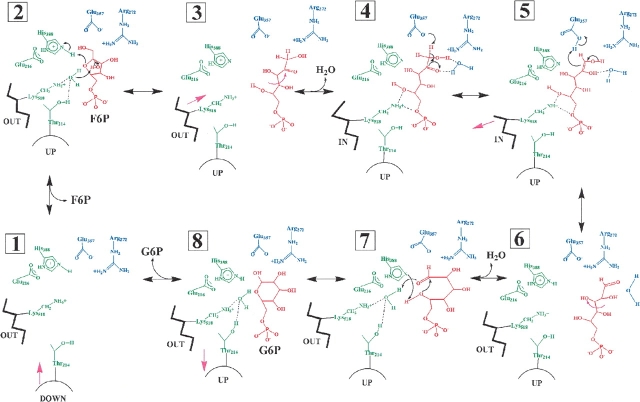
Model for mechanism of reaction
Previously reported biochemical studies and X-ray crystal structures led to a detailed model for catalysis by PGI, and the new PGI/S6P structure helps provide more details of the mechanism (Fig. 3 ▶). Upon binding of F6P in the active site of PGI, the enzyme undergoes induced fit with the loop containing amino acid residues 210–214 moving “up” to interact with the phosphoryl group of the ligand. The side chain of His388, which is held in position by Glu216, then promotes ring opening by protonating the ring oxygen of the substrate, and Lys518 and Thr214 position a water molecule to deprotonate the hydroxyl group on C2 of F6P.
Figure 3.
The proposed catalytic mechanism for PGI. The proposed multistep catalytic mechanism is shown, including the ligand binding, ring opening, isomerization, ring closing, and ligand release steps. A helix containing amino acid residues 512–520 moves between an “in” position, in which it interacts directly with the bound ligand, and an “out” position, in which there is a water molecule located between Lys518 and the ligand. A loop containing amino acid residues 210–214 moves “up” to interact with the phosphoryl group of the ligand upon ligand binding and “down” upon release of the ligand. The numbering of the steps refers to the F6P to G6P direction of catalysis. Amino acid residues involved in the ring-opening step are shown in green. Amino acid residues involved in the isomerization step are shown in blue. The cyclic and open chain forms of the substrates are shown in red. The dashed lines indicate hydrogen bond interactions. Purple arrows represent conformational changes of the ligand or enzyme. (1) Ligand binding: The F6P substrate binds in the active site, and the 210–214 loop moves “up.” (2) Ring opening: His388 and a water molecule held by Lys518 and Thr214 catalyze the ring opening step. (3) Conformational changes: The substrate undergoes rotation about its C3–C4 bond and extends so that C1–C2 approaches Glu357. The 512–520 helix moves to the “in” position. An ordered water molecule located between the 512–520 helix and Lys518 is lost. (4) Isomerization: Glu357 abstracts a proton to yield the cis-enediol(ate) intermediate. (5) Glu357 transfers a proton to the intermediate to yield the open chain form of G6P. (6) Conformational changes: The 512–520 helix moves to the “out” position and an ordered water molecule inserts between the helix and the open chain form of G6P. The G6P undergoes rotation about its C3–C4 bond to approach its cyclic conformation. (7) Ring closure: His388, Lys518, and Thr214 assist in ring closure. (8) Product release: The G6P product is released, and the 210–214 loop moves “down.”
After ring opening, a rotation around the C3–C4 bond enables the ligand to extend toward Glu357. An ordered water molecule located between the helix containing amino acid residues 512–520 and the substrate is lost, and the helix moves closer to the open chain form of the substrate, forming a hydrogen bond between Lys518 and the substrate C5 hydroxyl group. In the isomerization step, Glu357 abstracts a proton from C1 to yield the cis-enediol(ate) intermediate. The proton removed from the substrate is then transferred back to the cis-enediol(ate) intermediate to yield the open chain form of the product, G6P. Then the helix containing amino acid residues 512–520 moves “out”, away from the product, a water molecule inserts in between the helix and the product, and the product changes conformation to approach its cyclic form.
In a ring closure step, His388 deprotonates the incipient ring oxygen and Lys518 protonates the hydroxyl group on C1 through an ordered water molecule to yield the cyclic form of G6P. Upon release of the product, the loop containing amino acid residue Thr214 moves “down,” away from the ligand binding site.
Conclusion
The X-ray crystal structure of rabbit PGI with S6P has been solved at 2.0 Å resolution. The overall crystal structure of PGI with S6P has similarities and differences to structures of PGI complexed with cyclic substrate or analogs of the (extended) cis-enediol(ate) intermediate (Jeffery et al. 2000; Lee et al. 2001; Arsenieva et al. 2002; Davies et al. 2003). The position of the helix containing amino acid residues 512–520 is the same as in the crystal structure of PGI with the cyclic form of F6P; however, the conformation of the ligand is more extended, poised to undergo proton abstraction by Glu357. Small differences in the center of the S6P molecule compared with the 5PAH inhibitor suggests that movement of the helix containing amino acid residues 512–520 to the “in” position to interact directly with the substrate helps to position the substrate for the isomerization step.
Coordinates
Coordinates for the structure of phosphoglucose isomerase complexed with the inhibitor D-sorbitol-6-phosphate have been deposited in the Protein Data Bank with the ID code 1XTB.
Acknowledgments
Research was supported by grants from the American Cancer Society and the American Heart Association to C.J.J. Data were collected at the Southeast Regional Collaborative Access Team (SET-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No.W-31–109-Eng-38.
Abbreviations
PGI, phosphoglucose isomerase
PDB, Protein Data Bank
6PGA, 6-phospho-D-gluconate
5PAH, 5-phospho-D-arabinonhydroxamate
5PAA, 5-phospho-D-arabinonate
G6P, D-glucose-6-phosphate
F6P, D-fructose-6-phosphate
S6P, D-sorbitol-6-phosphate
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041070205.
References
- Abolhassani, M., Riley, W.M., Feldman, E., Arlin, Z., and Chiao, J.W. 1990. Two separate differentiation inducing proteins for human myeloid leukemia cells and their isolation from normal lymphocytes. Proc. Soc. Exp. Biol. Med. 195 288–291. [DOI] [PubMed] [Google Scholar]
- Arsenieva, D. and Jeffery, C.J. 2002. Conformational changes in phosphoglucose isomerase induced by ligand binding. J. Mol. Biol. 323 77–84. [DOI] [PubMed] [Google Scholar]
- Arsenieva, D., Hardre, R., Salmon, L., and Jeffery, C.J. 2002. The crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonohydroxamic acid. Proc. Natl. Acad. Sci. 99 5872–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrisford, J.M., Akerboom, J., Turnbull, A.P., de Geus, D., Sedelnikova, S.E., Staton, I., McLeod, C.W., Verhees, C.H., van der Oost, J., Rice, D.W., et al. 2003. Crystal structure of Pyrococcus furiosus phosphoglucose isomerase. Implications for substrate binding and catalysis. J. Biol. Chem. 278 33290–33297. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355 472–474. [DOI] [PubMed] [Google Scholar]
- Chaput, M., Claes, V., Portetelle, D., Cludts, I., Cravador, A., Burny, A., Gras, H., and Tartar, A. 1988. The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase. Nature 332 454–455. [DOI] [PubMed] [Google Scholar]
- Chiao, J.W., Zu, W., Seiter, K., Feldman, E., and Ahmed, T. 1999. Neuroleukin mediated differentiation induction of myelogenous leukemia cells. Leuk. Res. 23 13–18. [DOI] [PubMed] [Google Scholar]
- Chou, C.C., Sun, Y.J., Meng, M., and Hsiao, C.D. 2000. The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition. J. Biol. Chem. 275 23154–23160. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, No. 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Davies, C. and Muirhead, H. 2002. Crystal structure of phosphoglucose isomerase from pig muscle and its complex with 5-phosphoarabinonate. Proteins 50 577–579. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Structure of native phosphoglucose isomerase from rabbit: Conformational changes associated with catalytic function. Acta Crystallogr. D Biol. Crystallogr. 59 453–465. [DOI] [PubMed] [Google Scholar]
- Davies, C., Muirhead, H., and Chirgwin, J. 2003. The structure of human phosphoglucose isomerase complexed with a transition-state analogue. Acta Crystallogr. D Biol. Crystallogr. 59 1111–1113. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15 132–134. [DOI] [PubMed] [Google Scholar]
- Faik, P., Walker, J.I., Redmill, A.A., and Morgan, M.J. 1988. Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3′ sequences. Nature 332 455–457. [DOI] [PubMed] [Google Scholar]
- Gurney, M.E., Apatoff, B.R., Spear, G.T., Baumel, M.J., Antel, J.P., Bania, M.B., and Reder, A.T. 1986a. Neuroleukin: A lymphokine product of lectin-stimulated T cells. Science 234 574–581. [DOI] [PubMed] [Google Scholar]
- Gurney, M.E., Heinrich, S.P., Lee, M.R., and Yin, H.S. 1986b. Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science 234 566–574. [DOI] [PubMed] [Google Scholar]
- Jeffery, C.J., Bahnson, B.J., Chien, W., Ringe, D., and Petsko, G.A. 2000. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry 39 955–964. [DOI] [PubMed] [Google Scholar]
- Jeffery, C.J., Hardré, R., and Salmon, L. 2001. Crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonate identifies the role of Glu357 in catalysis. Biochemistry 40 1560–1566. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Chang, K.Z., Patel, V., and Jeffery, C.J. 2001. Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate D-fructose 6-phosphate. Biochemistry 40 7799–7805. [DOI] [PubMed] [Google Scholar]
- Leung, K., and Chiao, J.W. 1985. Human leukemia cell maturation induced by a T-cell lymphokine isolated from medium conditioned by normal lymphocytes. Proc. Natl. Acad. Sci. 82 1209–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann, K. 1933. Uber Phosphorylierung and Dephosphorylierung. Bildung der naturlichen Hexosemonophosphorsaure aus ihren Komponenten (German). Biochem. Z. 262 137. [Google Scholar]
- Matsumoto, I., Staub, A., Benoist, C., and Mathis, D. 1999. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science 286 1732–1735 [DOI] [PubMed] [Google Scholar]
- Otwinowski, A. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Read, J., Pearce, J., Li, X., Muirhead, H., Chirgwin, J., and Davies, C. 2001. The crystal structure of human phosphoglucose isomerase at 1.6 Å resolution: Implications for catalytic mechanism, cytokine activity and haemolytic anaemia. J. Mol. Biol. 309 447–463. [DOI] [PubMed] [Google Scholar]
- Rose, I.A. 1975. Mechanism of the aldose-ketose isomerase reactions. Adv. Enzymol. Relat. Areas Mol. Biol. 43 491–517. [DOI] [PubMed] [Google Scholar]
- ———. 1981. Chemistry of proton abstraction by glycolytic enzymes (aldolase, isomerases and pyruvate kinase). Philos. Trans. R. Soc. Lond. B Biol. Sci. 293 131–143. [DOI] [PubMed] [Google Scholar]
- Rose, I.A. and O’Connell, E.L. 1961. Intramolecular hydrogen transfer in the phosphoglucose isomerase reaction. J. Biol. Chem. 236 3086–3092. [PubMed] [Google Scholar]
- Schaller, M., Burton, D.R., and Ditzel, H.J. 2001. Autoantibodies to GPI in rheumatoid arthritis: Linkage between an animal model and human disease. Nat. Immunol. 8 746–53. [DOI] [PubMed] [Google Scholar]
- Sun, Y.J., Chou, C.C., Chen, W.S., Wu, R.T., Meng, M., and Hsiao, C.D. 1999. The crystal structure of a multifunctional protein: Phosphoglucose isomerase/autocrine motility factor/neuroleukin. Proc. Natl. Acad. Sci. 96 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, M.K., Solomons, J.T., Beeson, C.C., Hansen, T., Schonheit, P., and Davies, C. 2003. Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus. J. Biol. Chem. 278 47261–47268. [DOI] [PubMed] [Google Scholar]
- Swenson, C.A. and Barker, R. 1971. Proportion of keto and aldehydo forms in solutions of sugars and sugar phosphates. Biochemistry 10 3151–3154. [DOI] [PubMed] [Google Scholar]
- Watanabe, H., Takehana, K., Date, M., Shinozaki, T., and Raz, A. 1996. Tumor cell autocrine motility factor is the neuroleukin/phosphohexose isomerase polypeptide. Cancer Res. 56 2960–2963. [PubMed] [Google Scholar]
- Willem, R., Malaisse-Lagae, F., Ottinger, R., and Malaisse, W.J. 1990. Phosphoglucoisomerase-catalysed interconversion of hexose phosphates. Kinetic study by 13C N. M. R. of the phosphoglucoisomerase reaction in 2H2O. Biochem. J. 265 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Seiter, K., Feldman, E., Ahmed, T., and Chiao J.W. 1996. The, differentiation and maturation mediator for human myeloid leukemia cells shares homology with neuroleukin or phosphoglucose isomerase. Blood 87 4502–4506. [PubMed] [Google Scholar]