Abstract
In a survey of microbial systems capable of generating unusual metabolite structural variability, Streptomyces venezuelae ATCC 15439 is notable in its ability to produce two distinct groups of macrolide antibiotics. Methymycin and neomethymycin are derived from the 12-membered ring macrolactone 10-deoxymethynolide, whereas narbomycin and pikromycin are derived from the 14-membered ring macrolactone, narbonolide. This report describes the cloning and characterization of the biosynthetic gene cluster for these antibiotics. Central to the cluster is a polyketide synthase locus (pikA) that encodes a six-module system comprised of four multifunctional proteins, in addition to a type II thioesterase (TEII). Immediately downstream is a set of genes for desosamine biosynthesis (des) and macrolide ring hydroxylation. The study suggests that Pik TEII plays a role in forming a metabolic branch through which polyketides of different chain length are generated, and the glycosyl transferase (encoded by desVII) has the ability to catalyze glycosylation of both the 12- and 14-membered ring macrolactones. Moreover, the pikC-encoded P450 hydroxylase provides yet another layer of structural variability by introducing regiochemical diversity into the macrolide ring systems. The data support the notion that the architecture of the pik gene cluster as well as the unusual substrate specificity of particular enzymes contributes to its ability to generate four macrolide antibiotics.
Keywords: polyketides/deoxysugar/ketolide
Combinatorial biosynthesis involves the genetic manipulation of multistep biosynthetic pathways to create molecular diversity in natural products for use in drug discovery. Polyketide synthases (PKSs) represent one of the most amenable systems for combinatorial technologies because of their inherent genetic organization and ability to produce polyketide metabolites, a large group of natural products generated by bacteria (primarily actinomycetes and myxobacteria) and fungi with diverse structures and biological activities. Complex polyketides are produced by multifunctional PKSs involving a mechanism similar to long-chain fatty acid synthesis in animals (1). Pioneering studies (2, 3) on the erythromycin PKS in Saccharopolyspora erythraea revealed a modular organization. Characterization of this multidomain protein system, followed by molecular analysis of rapamycin (4), FK506 (5), soraphen A (6), niddamycin (7), and rifamycin (8) PKSs, demonstrated a colinear relationship between modular structure of a multifunctional bacterial PKS and the structure of its polyketide product.
Complex polyketide synthesis follows a processive reaction mechanism, and each module within a PKS harbors a string of three to six enzymatic domains that catalyze reactions in nearly linear order as described in particular detail for the erythromycin-producing PKS (9–11). This mechanism combined with the requirement for a terminal thioesterase (TE) domain to release the nascent polyketide chain assures that only one product is generated by a modular PKS. Moreover, post-polyketide tailoring enzymes (e.g., glycosyl transferases, hydroxylases) normally have strict substrate specificity requirements that result in the generation of a single final product.
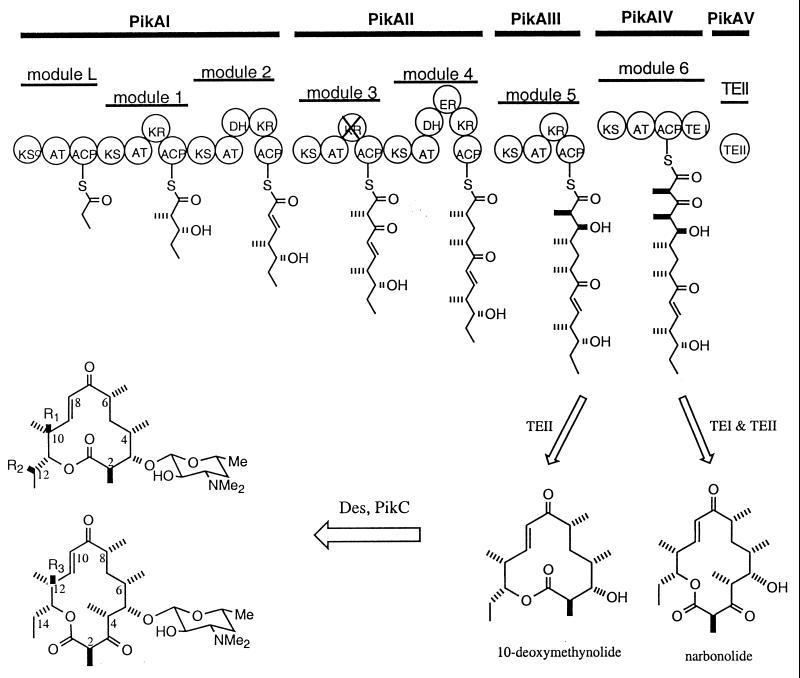
Streptomyces venezuelae ATCC 15439 produces four macrolide antibiotics that include the 12-membered ring macrolides methymycin and neomethymycin, and the 14-membered ring macrolides narbomycin and pikromycin (Fig. 1). Methymycin and neomethymycin are derived from 10-deoxymethynolide, and they differ only in hydroxyl group functionalization of the molecule after it has been fully assembled (12). However, pikromycin (and narbomycin) differ from methymycin in polyketide chain assembly. Specifically, the pikromycin aglycone, narbonolide (14-membered ring macrolide), requires an additional extension with methylmalonylCoA compared with the 12-membered ring aglycone, 10-deoxymethynolide. Because of the unusual ability of S. venezuelae to generate a diverse set of macrolide structures, we set out to answer the following question: are these macrolides generated by distinct biosynthetic pathways, or by a single pathway that includes a metabolic branch?
Figure 1.
The structure and biosynthesis of methymycin, neomethymycin, narbomycin, and pikromycin in S. venezuelae. Methymycin: R1 = OH, R2 = H; neomethymycin: R1 = H, R2 = OH; narbomycin R3 = H; pikromycin R3 = OH. Each circle represents an enzymatic domain in the PKS multifunctional protein. ACP, acyl carrier protein; KS, β-ketoacyl-ACP synthase; KSQ, a KS-like domain; AT, acyltransferase; KR, β-ketoacyl ACP reductase; KR with cross, inactive KR (same as KR0 in Table 1); DH, β-hydroxyl-thioester dehydratase; ER, enoyl reductase; TEI, thioesterase domain I; TEII, type II thioesterase. Des represents all eight enzymes in desosamine biosynthesis and transfer, which includes DesI, DesII, DesIII, DesIV, DesV, DesVI, DesVIII, and DesVII. For details see text and Table 1.
In this report, we describe the isolation, characterization, and analysis of the biosynthetic gene cluster for methymycin, neomethymycin, narbomycin, and pikromycin in S. venezuelae.
MATERIALS AND METHODS
Bacterial Strains and Media.
Escherichia coli DH5α was used throughout the study as a cloning host. E. coli LE392 was the host for a cosmid library derived from S. venezuelae genomic DNA. Luria–Bertani medium was used in E. coli propagation. S. venezuelae ATCC 15439 was obtained as a freeze-dried pellet from the American Type Culture Collection (ATCC). Media for vegetative growth and antibiotic production were used as described (12). Briefly, SGGP liquid medium was for propagation of S. venezuelae mycelia. Sporulation agar was used for production of S. venezuelae spores. Methymycin production was conducted in either SCM or vegetative medium, and pikromycin production was performed in Suzuki glucose-peptone medium.
Vectors, DNA Manipulation, and Cosmid Library Construction.
pUC119 was the routine cloning vector, and pNJ1 (13) was the cosmid vector used for genomic DNA library construction. Plasmid vectors for gene disruption were either pGM160 (14) or pKC1139 (15). Plasmid, cosmid, and genomic DNA preparation, restriction digestion, fragment isolation, and cloning were performed by using standard procedures (16). The cosmid library was made according to instructions from the Packagene λ-packaging system (Promega).
DNA Sequencing and Analysis.
An exonuclease III (Exo III) nested deletion series combined with PCR-based double-stranded DNA sequencing was employed to sequence the pik cluster. The Exo III procedure followed the Erase-a-Base protocol (Stratagene), and DNA sequencing reactions were performed by using the Dye Primer Cycle Sequencing Ready Reaction Kit (Applied Biosystems). The nucleotide sequences were read from an Applied Biosystems PRISM 377 sequencer on both DNA strands. DNA and deduced protein sequence analyses were performed by using geneworks and gcg sequence analysis packages. All analyses were performed by using the specific program default parameters.
Gene Disruption.
A replicative plasmid-mediated homologous recombination approach was developed to conduct gene disruption in S. venezuelae. Plasmids for insertional inactivation were constructed by cloning a kanamycin resistance marker into target genes, and plasmid for gene deletion/replacement was constructed by replacing the target gene with a kanamycin or thiostrepton resistance gene in the plasmid. Disruption plasmids were introduced into S. venezuelae by either PEG-mediated protoplast transformation (17) or RK2-mediated conjugation (15). Then, spores from individual transformants or transconjugants were cultured on nonselective plates to induce recombination. The cycle was repeated three times to enhance the opportunity for recombination. Double crossovers yielding targeted gene disruption mutants were selected and screened by using the appropriate combination of antibiotics and finally confirmed by Southern blot hybridization.
Antibiotic Extraction and Analysis.
Methymycin, pikromycin, and related compounds were extracted following published procedures (18). Thin layer chromotography was routinely used to detect methymycin, neomethymycin, narbomycin, and pikromycin. Further purification was conducted by using flash column chromatography and HPLC, and the purified compounds were analyzed by 1H and 13C NMR spectroscopy and MS.
RESULTS
Cloning and Identification of the pik Cluster.
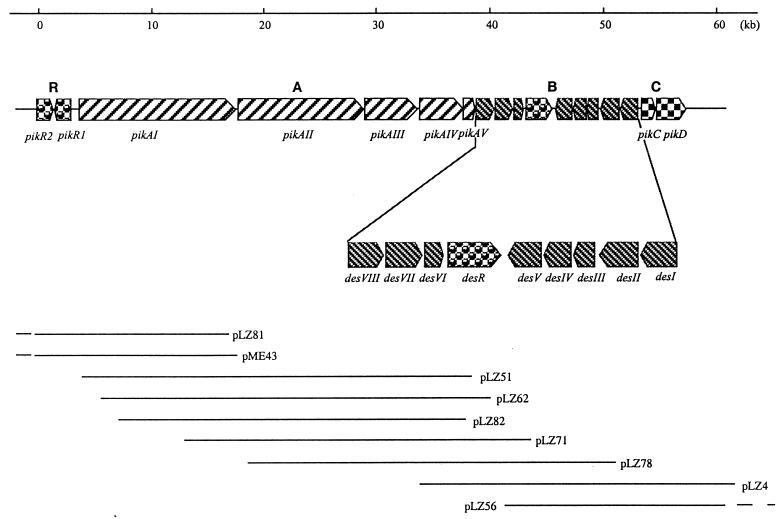
Heterologous hybridization was used to identify genes for methymycin, neomethymycin, narbomycin, and pikromycin biosynthesis in S. venezuelae. Initial Southern blot hybridization analysis with a type I PKS DNA probe revealed two multifunctional PKS clusters of uncharacterized function in the genome. Because these four antibiotics are all comprised of an identical desosamine residue, a tylAI α-d-glucose-1-phosphate thymidylyltransferase DNA probe (for mycaminose/mycorose/mycinose biosynthesis in the tylosin pathway) (19) was used to locate the corresponding biosynthetic gene cluster(s). This analysis established that only one of the PKS pathways contained a cluster of desosamine biosynthetic genes, and therefore work proceeded to isolate nine overlapping cosmid clones spanning over 80 kb on the bacterial chromosome that encompass the entire gene cluster (pik) for methymycin, neomethymycin, narbomycin, and pikromycin biosynthesis (Fig. 2). Through subsequent gene disruption the other PKS cluster (vep, devoid of linked desosamine biosynthetic genes) was found to play no role in production of methymycin, neomethymycin, narbomycin, or pikromycin (unpublished data).
Figure 2.
Organization of the pik cluster in S. venezuelae. Each arrow represents an ORF. The direction of transcription and relative sizes of the ORFs deduced from nucleotide sequence are indicated. The cluster is composed of four genetic loci: pikA, pikB (des), pikC, and pikR (see text and Table 2 for phenotype of the corresponding gene disruption mutants). Cosmid clones are denoted as overlapping lines.
Nucleotide Sequence of the pik Cluster.
The nucleotide sequence of the pik cluster was completely determined and shown to contain 18 ORFs that span ≈60 kb. Central to the cluster are four large ORFs, pikAI, pikAII, pikAIII, and pikAIV, encoding a multifunctional PKS (Fig. 2). Analysis of the six modules comprising the pik PKS immediately indicated that it would specify production of narbonolide, the 14-membered ring aglycone precursor of narbomycin and pikromycin (Fig. 1). Initial analysis unveiled two significant architectural differences in the pikA-encoded PKS. First, compared with eryA (20) and oleA (21), two PKS clusters that produce 14-membered ring macrolides erythromycin and oleadomycin similar to pikromycin, the presence of separate ORFs, pikAIII and pikAIV, encoding Pik module 5 and Pik module 6 (as individual modules) as opposed to one bimodular protein as in eryAIII and ole ORFIII is striking. Second, the presence of a TEII immediately downstream of the type I PKS cluster is also unprecedented (Fig. 2). These two characteristics suggest that pikA may produce the 12-membered ring macrolactone 10-deoxymethynolide as well. Indeed, the domain organization of PikAI–AIII (module L-5) is consistent with the predicted biosynthesis of 10-deoxymethynolide (Fig. 1), except for the absence of a TE function at the C terminus of Pik module 5 (PikAIII). We propose that lack of a TE domain in PikAIII may be compensated by the TEII (encoded by pikAV) immediately downstream of pikAIV. Consistent with the supposition that two distinct polyketide ring systems are assembled from the pik PKS, two macrolide-lincosamide-streptogramin B type resistant genes, pikR1 and pikR2, are found upstream of the pik PKS (Fig. 2), which presumably provide cellular self-protection for S. venezuelae.
The genetic locus for desosamine biosynthesis and glycosyl transfer are immediately downstream of pikA. Seven genes, desI, desII, desIII, desIV, desV, desVI, and desVIII, are responsible for the biosynthesis of the deoxysugar, and the eighth gene, desVII, encodes a glycosyltransferase that apparently catalyzes transfer of desosamine onto the alternate (12- and 14-membered ring) polyketide aglycones (Fig. 2). The existence of only one set of desosamine genes indicates that DesVII can accept both 10-deoxymethynolide and narbonolide as substrates, a valuable feature of this class of enzyme (22). The largest ORF in the des locus, desR, encodes a β-glucosidase that was shown to be involved in a drug inactivation-reactivation cycle for bacterial self-protection (23).
Just downstream of the des locus is a gene (pikC) encoding a cytochrome P450 hydroxylase similar to eryF (24), and eryK (25), PikC, and a gene (pikD) encoding a putative transcriptional activator, PikD. Interestingly, PikC is the only P450 hydroxylase identified in the entire pik cluster, suggesting that the enzyme can accept both 12- and 14-membered ring macrolide substrates and, more remarkably, it is active on both C-10 and C-12 of the YC-17 (12-membered ring) intermediate to produce methymycin and neomethymycin (unpublished data). PikD is a putative regulatory protein similar to ORF H in the rapamycin gene cluster (26). However, the site and mechanism of PikD regulation is currently not clear.
The combined functionality coded by the 18 genes in the pik cluster predicts biosynthesis of methymycin, neomethymycin, narbomycin, and pikromycin (Table 1). Flanking the pik cluster locus are genes presumably involved in primary metabolism and genes that may be involved in both primary and secondary metabolism. A S-adenosyl-methionine synthase gene is located downstream of pikD that may help to provide the methyl group in desosamine synthesis. A threonine dehydratase gene was identified upstream of pikR2 that may provide precursors for polyketide biosynthesis. Presently it is not apparent that any of these genes are dedicated to antibiotic biosynthesis and, therefore, we consider them outside the limits of the pik cluster.
Table 1.
Deduced function of ORFs in the pik cluster
| Polypeptide (ORF) | Amino acids, no. | Proposed function or sequence similarity detected | ||||||
|---|---|---|---|---|---|---|---|---|
| PikAI | 4,613 | PKS | ||||||
| Loading module | KSQ | AT(P) | ACP | |||||
| Module 1 | KS | AT(P) | KR | ACP | ||||
| Module 2 | KS | AT(A) | DH | KR | ACP | |||
| PikAII | 3,739 | PKS | ||||||
| Module 3 | KS | AT(P) | KR0 | ACP | ||||
| Module 4 | KS | AT(P) | DH | ER | KR | ACP | ||
| PikAIII | 1,562 | PKS | ||||||
| Module 5 | KS | AT(P) | KR | ACP | ||||
| PikAIV | 1,346 | PKS | ||||||
| Module 6 | KS | AT(P) | ACP | TE | ||||
| PikAV | 281 | TEII | ||||||
| DesI | 415 | 4-Dehydrase | ||||||
| DesII | 485 | Reductase? | ||||||
| DesIII | 292 | α-d-Glucose-1-phosphate thymidylyltransferase | ||||||
| DesIV | 337 | TDP-glucose 4,6-dehydratase | ||||||
| DesV | 379 | Transaminase | ||||||
| DesVI | 237 | N,N-Dimethyltransferase | ||||||
| DesVII | 426 | Glycosyl transferase | ||||||
| DesVIII | 402 | Tautomerase? | ||||||
| DesR | 809 | β-Glucosidase (involved in resistance mechanism) | ||||||
| PikC | 416 | P450 hydroxylase | ||||||
| PikD | 928 | Putative regulator | ||||||
| PikR1 | 336 | rRNA methyltransferase (mls resistance) | ||||||
| PikR2 | 322 | rRNA methyltransferase (mls resistance) | ||||||
Mutational Analyses of the pik Cluster.
Extensive disruption of genes in the pik cluster were carried out to address the role of key enzymes in antibiotic production (Table 2). First, PikAI, the first putative enzyme involved in the biosynthesis of 10-deoxymethynolide and narbonolide was inactivated by insertional mutagenesis. The resulting mutant, AX903, did not produced methymycin, neomethymycin, or narbomycin, pikromycin, indicating that pikA encodes a PKS required for both 12- and 14-membered ring macrolactone formation.
Table 2.
Summary of mutational analyses of the pik cluster
| Mutant | Type of mutation | Target gene | Antibiotic production/intermediate accumulation
|
|
|---|---|---|---|---|
| Methymycin and neomethymycin | Pikromycin | |||
| AX903 | Insertion | pikAI | No/no | No/no |
| LZ3001 | Deletion/replacement | desVI | No/10-deoxymethynolide | No/narbonolide |
| LZ4001 | Deletion/replacement | desV | No/10-deoxymethynolide | No/narbonolide |
| AX905 | Deletion/replacement | pikAV | <5%/no | <5%/no |
| AX906 | Insertion | pikC | No/YC-17 | No/narbomycin |
Second, deletion of both desVI and desV abolished methymycin, neomethymycin, narbomycin, and pikromycin production, and the resulting mutants, LZ3001 and LZ4001, accumulate 10-deoxymethynolide and narbonolide in their culture broth, indicating that enzymes for desosamine synthesis and transfer are also shared by the 12- and 14-membered ring macrolides.
To understand the mechanism of polyketide chain termination at PikAIII (PikAIII (module 5) is presumed to be the termination point in construction of 10-deoxymethynolide), the pik TEII gene, pikAV, was deleted. The deletion/replacement mutant, AX905, produces <5% of methymycin, neomethymycin, and <5% of pikromycin compared with wild-type S. venezuelae. This abrogation in product formation occurs without significant accumulation of the expected aglycone intermediates, suggesting that pik TEII is involved in the termination of 12- as well as 14-membered ring macrolides at PikAIII and PikAIV, respectively. Although we considered that polar effects may influence the observed phenotype in AX905, this has been ruled out after the consideration of mutant LZ3001, in which mutation in a gene downstream of pikAV accumulated 10-deoxymethynolide and narbonolide. The fact that mutant AX905 failed to accumulate these intermediates suggested that the polyketide chains were not efficiently released from the PKS protein in the absence of Pik TEII. Therefore, Pik TEII presumably plays a crucial role in polyketide chain release and cyclization, and it may be part of the mechanism for alternative termination in pik polyketide biosynthesis.
Finally, disruption of pikC confirmed that PikC is the sole enzyme catalyzing hydroxylation of both YC-17 (at C-10 and C-12) and narbomycin (at C-12). The relaxed substrate specificity of PikC and its regional specificity at C-10 and C-12 provide another layer of metabolite diversity in the pik-encoded biosynthetic system.
DISCUSSION
The work described in this report has established that methymycin, neomethymycin, narbomycin, and pikromycin biosynthesis is prescribed by the pik cluster in S. venezuelae. Three key enzymes as well as the unique architecture of the cluster enable this relatively compact system to produce multiple macrolide antibiotics. Foremost, the presence of pik modules 5 and 6 as separate proteins, PikAIII and PikAIV, and activity of pik TEII enable the bacterium to terminate the polyketide chain at two different points of assembly, thereby producing two macrolactones of different ring size. Second, DesVII, the glycosyltransferase in the pik cluster, can accept both 12- and 14-membered ring macrolactones as substrates. Finally, PikC, the P450 hydroxylase, has a remarkable substrate and regiochemical specificity that introduces another aspect of diversity into the system.
It is interesting to consider that pikA evolved in a line analogous to eryA and oleA because each of these PKSs specify the synthesis of 14-membered ring macrolactones. Therefore, pik may have acquired the capacity to generate methymycin when a mutation in the primordial pikAIII–pikAIV linker region caused splitting of Pik modules 5 and 6 into two separate gene products. This notion is raised by two features of the nucleotide sequence. First, the intergenic region between pikAIII and pikAIV is 105 bp, which may be the remnant of an intramodular linker peptide of 35 amino acids. Moreover, the potential for independently regulated expression of pikAIV is implied by the presence of a 100-nucleotide region at the 5′ end of the gene that is relatively AT-rich (62% as comparing 74% G+C content in coding region). Thus, as the mutation in an original ORF encoding the bimodular multifunctional protein (PikAIII–PikAIV) occurred so too may have evolved a mechanism for regulated synthesis of the new gene product (PikAIV).
The apparent role of Pik TEII in alternative termination of polyketide chain elongation intermediates provides a means of diversity generation in natural product biosynthesis. Engineered polyketides of different chain length are typically generated by moving the TE catalytic domain to alternate positions in a modular PKS (27). Repositioning of the TE domain necessarily abolishes production of the original full-length polyketide so only one macrolide is produced each time. In contrast to the fixed-position TE domain, the independent Pik TEII polypeptide seems to have the flexibility to catalyze termination at different stages of polyketide assembly, therefore, enabling the system to produce multiple products of variant chain length. Combinatorial biology technologies can now exploit this system for generating molecular diversity through construction of novel PKS systems with TEIIs for simultaneous production of several new molecules as opposed to the TE domains alone that limit catalysis to a single termination step.
It is noteworthy that sequences similar to Pik TEII are found in almost all known polyketide and nonribosomal polypeptide biosynthetic systems (28). Currently the pik TEII is the first to be characterized in a modular PKS. However, recent work on a TEII gene in the lipopeptide surfactin biosynthetic cluster (29) demonstrated that srf-TEII plays an important role in polypeptide chain release, and may suggest that srf-TEII reacts at multiple stages in peptide assembly as well (28). It is clear that the precise function of Pik TEII, and TEIIs from polyketide and polypeptide biosynthetic systems in general, need further investigation.
The enzymes involved in post-polyketide assembly of 10-deoxymethynolide and narbonolide are particularly intriguing, especially the glycosyltransferase, DesVII, and P450 hydroxylase, PikC. Both have the remarkable ability to accept substrates with significant structural variability. Moreover, disruption of desVI demonstrated that DesVII also tolerates variations in deoxysugar structure (30). Likewise, PikC has recently been shown to convert YC-17 to methymycin/neomethymycin and narbomycin to pikromycin in vitro (unpublished data).
With 18 genes spanning less than 60 kb of DNA capable of producing four active macrolide antibiotics, the pik cluster represents the least complex yet most versatile modular PKS system so far investigated. This simplicity provides the basis for a compelling expression system in which active ketolide products are engineered and produced with considerable facility for discovery of a diverse range of new biologically active compounds.
Acknowledgments
This work represents an equal collaboration from both laboratories. We thank Leonard Katz (Abbott Laboratories) for authentic pikromycin, and David E. Cane (Brown University) for standards samples of methymycin and YC-17. The expert technical assistance of Daniel Wilson and Liang Zhou are gratefully acknowledged. This work was supported by National Institutes of Health Grants GM48562 (to D.H.S.), GM35906, and GM54346 (to H.-w.L.) and a grant from the Office of Naval Research (to D.H.S.). Y.X. is a recipient of the Dennis W. Watson Fellowship (University of Minnesota).
ABBREVIATIONS
- PKS
polyketide synthase
- TE
thioesterase
Footnotes
References
- 1. Hopwood D A, Sherman D H. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 2.Cortes J, Haydock S F, Roberts G A, Bevitt D J, Leadlay P F. Nature (London) 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 3.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio J F, Molnar I, Schwecke T, Konig A, Haydock S F, Khaw L E, Staunton J, Leadlay P F. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 5.Motamedi H, Cai S J, Shafiee A, Elliston K O. Eur J Biochem. 1997;244:74–80. doi: 10.1111/j.1432-1033.1997.00074.x. [DOI] [PubMed] [Google Scholar]
- 6.Schupp T, Toupet C, Cluzel B, Neff S, Hill S, Beck J J, Ligon J M. J Bacteriol. 1995;177:3673–3679. doi: 10.1128/jb.177.13.3673-3679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakavas S J, Katz L, Stassi D. J Bacteriol. 1997;179:7515–7522. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.August P R, Tang L, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffmann D, Kim C G, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 9.Katz L. Chem Rev. 1997;97:2557–2575. doi: 10.1021/cr960025+. [DOI] [PubMed] [Google Scholar]
- 10.Khosla C. Chem Rev. 1997;97:2577–2590. doi: 10.1021/cr960027u. [DOI] [PubMed] [Google Scholar]
- 11.Staunton J, Wilkinson B. Chem Rev. 1997;97:2611–2629. doi: 10.1021/cr9600316. [DOI] [PubMed] [Google Scholar]
- 12.Lambalot R H, Cane D E. J Antibiot. 1992;45:1981–1982. doi: 10.7164/antibiotics.45.1981. [DOI] [PubMed] [Google Scholar]
- 13.Tuan J S, Weber J M, Staver M J, Leung J O, Donadio S, Katz L. Gene. 1990;90:21–29. doi: 10.1016/0378-1119(90)90435-t. [DOI] [PubMed] [Google Scholar]
- 14.Muth G, Nubhaumer B, Wohlleben W, Puhler A. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 15.Bierman M, Logan R, O’Brien K, Seno E T, Nagaraja Rao R, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K J, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. U.K.: The John Innes Foundation; 1985. [Google Scholar]
- 18.Cane D, E, Lambalot R, H, Prabhakaran P, C, Ott W, R. J Am Chem Soc. 1993;115:522–526. [Google Scholar]
- 19.Merson-Davies L A, Cundliffe E. Mol Microbiol. 1994;13:347–355. doi: 10.1111/j.1365-2958.1994.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 20.Donadio S, Katz L. Gene. 1992;111:51–60. doi: 10.1016/0378-1119(92)90602-l. [DOI] [PubMed] [Google Scholar]
- 21.Swan D G, Rodriguez A M, Vilches C, Mendez C, Salas J A. Mol Gen Genet. 1994;242:358–362. doi: 10.1007/BF00280426. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen J R, Hutchinson C R, Cane D E, Khosla C. Science. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Sherman D H, Liu H-w. J Am Chem Soc. 1998;120:9374–9375. [Google Scholar]
- 24.Andersen J F, Hutchinson C R. J Bacteriol. 1992;174:725–735. doi: 10.1128/jb.174.3.725-735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stassi D, Donadio S, Staver M J, Katz L. J Bacteriol. 1993;175:182–189. doi: 10.1128/jb.175.1.182-189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, et al. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes J, Wiesmann K E, Roberts G A, Brown M J, Staunton J, Leadlay P F. Science. 1995;268:1487–1489. doi: 10.1126/science.7770773. [DOI] [PubMed] [Google Scholar]
- 28.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 29.Schneider A, Marahiel M A. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, L., Sherman, D. H. & Liu, H.-w. (1998) J. Am. Chem. Soc., in press.