Abstract
The previously identified dendritic cell-derived ubiquitin-like protein (DC-UbP) was implicated in cellular differentiation and apoptosis. Sequence alignment suggested that it contains a ubiquitin-like (UbL) domain in the C terminus. Here, we present the solution NMR structure and backbone dynamics of the UbL domain of DC-UbP. The overall structure of the domain is very similar to that of Ub despite low similarity (<30%) in amino-acid sequence. One distinct feature of the domain structure is its highly positively charged surface that is different from the corresponding surfaces of the well-known UbL modifiers, Ub, NEDD8, and SUMO-1. The key amino-acid residues responsible for guiding polyubiquitinated proteins to proteasome degradation in Ub are not conserved in the UbL domain. This implies that the UbL domain of DC-UbP may have its own specific interaction partners with other yet unknown cellular functions related to the Ub pathway.
Keywords: solution structure, dynamics, ubiquitin-like domain, DC-UbP, NMR
Ubiquitin (Ub) is a small polypeptide (76 residues) that covalently modifies other cellular proteins and targets them to the proteasome for degradation (Hershko and Ciechanover 1998; Finley 2001; Pickart 2004). Ubiquitination is also a versatile regulatory signal controlling protein activity, function, stability, and intracellular localization (Finley et al. 2004). The ubiquitination process involves Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), Ub-ligase (E3), and the Ub-tagged substrates that are recognized and degraded by the 26S proteasome (Hershko and Ciechanover 1998; Pickart 2001).
It has now emerged that UbL proteins are involved in a variety of fundamental cellular processes, such as protein sorting, protein degradation, DNA repair, cell division, autophagy, and apoptosis (Jentsch and Pyrowolakis 2000). UbL proteins can be divided into two separate classes (Jentsch and Pyrowolakis 2000). The class I UBLs function as modifiers in a manner analogous to that of Ub (Schwartz and Hoschstrasser 2003). UBLs exist either in a free form or attached covalently to other proteins by their C termini. The conjugation pathways for UBLs closely resemble that for Ub. The class II UDPs contains a domain homologous to Ub as part of an often large, multidomain overall structure. In contrast to UBLs, UDPs are not conjugated to other proteins but implicated in interaction with other regulatory proteins (Jentsch and Pyrowolakis 2000).
Recently, studies about UBLs mainly focus on SUMO, NEDD8, Apg12, and UCRP/ISG15. Post-translational modification with the UbL protein SUMO is involved in transcriptional regulation and genome surveillance (Muller et al. 2004). NEDD8 modification is involved in cell cycle control and in embryogenesis by up-regulating the activities of cullin-based E3 ligases (Pan et al. 2004). Covalent attachment of Apg12 to Apg5 is essential for autophagy (Mizushima et al. 1998, 2003). ISG15 could be conjugated to a variety of proteins when cells are treated with type I interferon or lipopolysaccharide and may be associated with specialized functions in the innate immune system (Kim and Zhang 2003).
UDPs include a structurally and functionally heterogeneous group of proteins (Jentsch and Pyrowolakis 2000). Rad23 is involved in DNA repair and protein degradation. The UbL domain of Rad23 possibly mediates the binding of Rad23 to proteasomes (Schauber et al. 1998). Dsk2, another UDP, competes with Rad23 for proteasome binding through its UbL domain (Elsasser et al. 2002). Ubiquilin may be involved in endoplasmic reticulum stress-associated protein degradation through association of its UbL domain with the 19S proteasome subunits (Ko et al. 2004). UDPs are also involved in the Ub system. For example, UbP6 is a deubiquitinating enzyme (Wyndham et al. 1999), and Elongin B is a subunit of the CBC Ub ligase (Kamura et al. 2002). In addition to its UbL domain, parkin possesses a RBR domain composed of two RING fingers plus an IBR/DRIL domain, which may be part of cullin-containing Ub ligase complexes (Marin et al. 2004).
In the previous study, a cDNA encoding a UbL protein was identified from dendritic cells (namely, DC-UbP) (Liu et al. 2003). The UbL domain of DC-UbP shares 28.6% identity and 55% similarity to Ub in the amino-acid sequence. In the present paper, we report the three-dimensional solution structure and backbone dynamics of the UbL domain of DC-UbP solved by NMR spectroscopy. Although the UbL domain and Ub share low sequence similarity, we find that the two proteins take a similar fold. The structure may provide clues for identification of its interaction partners and further exploration of the biological functions of DC-UbP protein.
Results
Structure determination
DC-UbP protein contains a UbL domain (residues 85– 190, numbered 1–106) in the C-terminal region. The sequence alignment of the UbL domain of DC-UbP with Ub, SUMO-1, and NEDD8 is shown in Figure 1 ▶. Circular dichroism (CD) study revealed that the UbL domain is compactly folded with possibly α/β structures (data not shown). We then used heteronuclear multidimensional NMR techniques to elucidate the three-dimensional structure of the UbL domain in solution.
Figure 1.
Sequence alignment of the UbL domain of DC-UbP, Ub, NEDD8, and SUMO-1. All the sequences are originated from Homo sapiens. The numerals are labeled according to the sequence of the UbL domain and the secondary structures are displayed based on Ub. T and η denote hydrogen-bonded turn and 310 helix, respectively. The figures were prepared by using the program ESPript (Gouet et al. 1999).
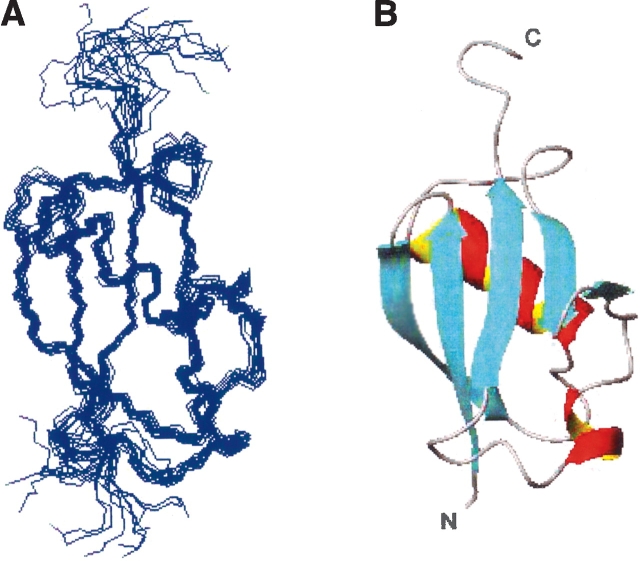
A summary of the NMR data used for structure calculation is presented in Table 1. All 15 final structures converged with no NOE or dihedral angle violation >0.3 Å or 5°, respectively. The average RMSD for the 15 structures within the secondary structure elements of residues Cys24–Val94 (including residues Cys24–Leu30, Lys34–Val40, Val46–Ala56, Arg65– Phe68, Arg71–Pro72, Lys78–Glu81, Val89–Ile93) for the backbone atoms and for all heavy atoms are 0.56 and 1.62 Å, respectively. Figure 2 ▶ depicts a superimposition of the 15 lowest energy structures and a ribbon representation of one of the 15 NMR structures. Ramachandran plot analysis (Table 1) of the domain within the secondary structural elements indicates 82.7% residues in most favored regions and no residues in disallowed regions. Moreover, both termini (residues Met1–Ser20 and Pro101–Asn106) are not included in the structural calculation process for their conformational flexibility. These flexible regions were identified based on diverse structural data: backbone {1H}-15N NOE (Jin et al. 2001; Huang et al. 2003), CSI (Wishart et al. 1992), NOE patterns (Cavanagh et al. 1995), and the Clean SEA-HSQC experiments (Lin et al. 2002). The peak intensity ratios in the Clean SEA-HSQC experiments indicate the amide solvent-exposed degrees of the UbL domain in DC-UbP (Fig. S1), suggesting that, besides N- and C-terminal flexible residues, the residues within the UbL domain such as Thr32, Thr45, Glu60, Ser69–Gly70, Leu73–Asp75, and Glu81 might be also exposed to solvent. These data are in good agreement with the structure determined. The overall fold of the UbL domain in DC-UbP is very similar to that of Ub (Cornilescu et al. 1998), NEDD8 (Whitby et al. 1998), and SUMO-1 (Bayer et al. 1998; Jin et al. 2001) despite low similarity in amino-acid sequences. Like Ub, the UbL domain contains a five-strand β-sheet and two helices. Four strands (β1, β2, β3, and β5) form a main plane, but the β4 strand is twisted against the main plane. Strands β1 and β2 as well as β3 and β5 are anti-parallel aligned, while strands β1 and β5 are parallel. The first α-helix is long (11 residues) and rotates ~45° relative to the β-sheet formed by β1 and β2. The second helix is short with only four residues. This arrangement also appears in SUMO-1 and NEDD8. In the UbL domain of DC-UbP, residues Thr32–Gly33 and Glu60–Gly62 are located in loops, which are flexible, as few long- or medium-range NOEs were obtained. Residues Pro85–Asp87 are also flexible, probably due to lack of resonance assignments for residues Pro85 and Lys86.
Table 1.
Structural restraints used and structural statistics
| No. of experimental restraints | ||
| Total unambiguous distance restraints | 851 | |
| Intraresidual | 301 | |
| Sequential (| i – j | = 1) | 247 | |
| Medium (2≤|I–j|≤4) | 149 | |
| Long range (|i–j|≥5) | 148 | |
| Hydrogen bond restraints | 60 | |
| Dihedral angle restraints | 83 | |
| φ: | 44 | |
| Ψ: | 39 | |
| Structural statistics | ||
| Coordinate precision (Å)a, secondary structure | ||
| Backbone (N, Ca, C′) | .56 | ±0.11 |
| Heavy atoms | 1.62 | ±0.17 |
| Coordinate precision (Å), residues 24–94 | ||
| Backbone (N, Ca, C′) | 0.87 | ±0.20 |
| Heavy atoms | 1.82 | ±0.26 |
| RMSD from experimental restraints | ||
| NOE distances (Å) | 0.0250 | ±0.0012 |
| Dihedral angles (deg.) | 0.3788 | ±0.0290 |
| RMSD from idealized geometry | ||
| Bonds (Å) | 0.0030 | ±0.00014 |
| Angles (deg.) | 0.4802 | ±0.0118 |
| Impropers (deg.) | 0.4309 | ±0.0201 |
| Ramachandran analysisb | Secondary structure | Residues 24–94 |
| Residues in most favored regions | 82.7% | 66.0% |
| Residues in additionally allowed regions | 17.3% | 28.2% |
| Residues in generously allowed regions | 0.0% | 2.9% |
| Residues in disallowed regions | 0.0% | 2.8% |
aThe coordinate precision is defined as the average RMS deviation among the 15 final structures.
b Exception of Pro and Gly residues.
Figure 2.
Three-dimensional structure of the UbL domain of DC-UbP (residues 21–100). (A) Backbone atom superposition of the final 15 structures. The structures are superimposed adopting the residues 24– 94. (B) Ribbon diagram representation of the UbL domain of DC-UbP. Figures were prepared using the program MOLMOL (Koradi et al. 1996).
Backbone dynamics
For the UbL domain of DC-UBP, the relaxation rates for 70 residues (excluding prolines and the resonances that were severely overlapped) out of the 80 residues were obtained (Fig. 3 ▶). The backbone 15N resonances of flexible residues Met1–Ser20 and Pro101–His114 (including histidine tag) show negative NOE values, which are not shown in the figure. The average heteronuclear NOE value for residues Gly21–Asn100 is 0.82 (±0.07), and the average R1 and R2 values are 1.55 (±0.05) sec−1 and 14.40 (±1.46) sec−1, respectively. To better illustrate the backbone dynamics, the plot of R2/R1 ratio as a function of residue number is also shown in Figure 3 ▶. By combining the relaxation data and the structural coordinates (Tjandra et al. 1995), the ratio of two principal axes (D||/D⊥) of the diffusion tensor for the UbL domain was estimated to be 1.32, suggesting that the domain has the attribute of anisotropic tumbling.
Figure 3.

Plot of NOE, R1, R2, and R2/R1 parameters for the UbL domain of DC-UbP as a function of residue number. Only residues whose {1H}-15N cross-peaks are resolved enough to permit accurate measurements of their intensities are included.
The backbone 15N relaxation data provide useful information about the internal conformational dynamics of a protein (Bruschweiler 2003). Residues Gly21, Tyr22, and Gln96 show smaller NOE values than the average. In addition, these residues also exhibit lower R2 values and higher R1 values than the respective average. These data indicate that both termini of the UbL domain are conformationally flexible. Within the core structure of the UbL domain, some residues like Cys24, Ser42, Arg51, Gly58, Glu81, and Leu82 exhibit higher R2 values (also high R2/R1) than the average, which are often associated with conformational exchange broadening due to intermediate timescale dynamics. Structurally, residues Cys24, Ser42, Glu81, and Leu82 form a cluster on the surface of the structure of the UbL domain. These cluster residues reside mainly in the N-terminal α2-helix (residues Glu81 and Leu82), the loop linking β2-strand and α1-helix (Ser42), and the N-terminal β1-strand (Cys24). This kind of dynamically specific cluster was also revealed in a cold-shock adaptation protein, RbfA (Huang et al. 2003).
Discussion
Although having an overall fold similar to that of Ub, the UbL domain of DC-UbP appears different from Ub in respect to biological function. Being a class II UbL protein, DC-UbP does not possess two conserved Gly residues in its C-terminal UbL domain as that in Ub. In addition, the domain lacks the conserved Lys residues corresponding to Lys29 and Lys48 in Ub, which are responsible for the formation of Ub polymers. The Lys86 residue of the UbL domain is conserved, corresponding to Lys63 in Ub (Fig. 1 ▶), which has recently been found to be involved in activation of NF-κB transcription factor (Chen and Fuchs 2004). The three residues (Leu8, Ile44, and Val70) of Ub that have been demonstrated to be important for binding to the proteasome are not conserved in the UbL domain of DC-UbP. Instead, these residues are replaced by Ser31, Phe67, and Ile93 in the domain. The difference in the key residues between the UbL domain and Ub implies that DC-UbP protein may own specific interaction partners for its function. Our in vitro pull-down experiments showed that the UbL domain of DC-UbP does not interact with some Ub binding domains, such as UBA (Ub-associated domain) and UIM (Ub interacting motif) (data not shown). The NMR structure of the UbL domain of DC-UbP may provide structural basis for functional annotation of the DC-UbP protein.
Many UbLs and UDPs possess a UbL fold (McNally et al. 2003; Mueller and Feigon 2003; Ramelot et al. 2003; Sakata et al. 2003). However, they may have different binding partners and diverse biological function. One distinct feature of the UbL domain of DC-UbP is the positively charged surface that is different from Ub, NEDD8, and SUMO-1. As shown in Figure 4 ▶, the first surface view of the UbL domain (A) displays a large hydrophobic patch surrounded by positively charged epitopes (contributed by Arg27, Arg29, Lys37, Arg65, Arg71, and Lys86), which is similar to Ub (B) and NEDD8 (C), but dissimilar to SUMO-1 (D). The region is also the binding interface of Ub and NEDD8 with their specific enzymes or interaction factors (Goettsch and Bayer 2002), suggesting that this region is also a binding interface of the UbL domain in DC-UbP with specific interaction factors. Opposite the positively charged region, on the other side of the β-sheet of the UbL domain (E), there is a large positively charged surface formed by Arg41, His48, Lys50, Arg51, Arg52, His54, and Lys76 in the domain, which is absent on the corresponding surfaces of Ub (F), NEDD8 (G), and SUMO-1 (H). This charge topology difference might implicate an alternative interaction pattern or biological function for the UbL domain of DC-UbP, which remains to be discovered.
Figure 4.
Comparison of the electrostatic surfaces of the UbL domain of DC-UbP (A,E), Ub (B,F), NEDD8 (C,G), and SUMO-1 (D,H). The first surface view of the UbL domain of DC-UbP (A) corresponds to the front view shown in Figure 2 ▶. The corresponding surface views of Ub (B), NEDD8 (C), and SUMO-1 (D) are shown in the upper right part of the figure as a comparison. The second surface view of the UbL domain of DC-UbP (E) corresponds to the opposite view shown in Figure 2 ▶ that is rotated around the longitudinal axis by 180°. The corresponding surface views of Ub (F), NEDD8 (G), and SUMO-1 (H) are shown in the lower right part of the figure. The positive charges are shown in blue, and the negative charges are in red. The PDB accession codes of Ub, NEDD8, and SUMO-1 are 1D3Z, 1NDD, and 1A5R, respectively. The electrostatic surfaces were calculated and colored using the program MOLMOL (Koradi et al. 1996).
In summary, the solution structure of the UbL domain of DC-UbP shares a common topology of UbL fold. The distinct charged surfaces and internal dynamics suggest its specific binding partners with biological significance.
Materials and methods
Protein expression and purification
The DNA fragment encoding the UbL domain of DC-UbP (residues 85–190, numbered 1–106) from human dendritic cell was cloned into the pET22b (+) vector. The plasmid was transformed into Escherichia coli BL21(DE3) cells. The fragment protein was separated by affinity chromatography using Ni-NTA agarose resin (QIAGEN) and further purified by FPLC gel-filtration on a Superose 12 column (Amersham Biosciences). Uniformly labeled proteins with 15N and/or 13C were prepared in M9 medium with 15NH4Cl (1 g/L, Cambridge Isotope Laboratories, Inc.) and/or [U-13C]-glucose (2 g/L, Cambridge Isotope Laboratories, Inc.) as sole nitrogen and carbon sources. The purified protein was concentrated to 1.2 mM in a sodium acetate buffer (10 mM NaAc, 100 mM NaCl, 5 mM DTT, and 0.05% w/v NaN3 [pH 5.6]) (92% H2O/8% D2O or 100% D2O) for NMR experiments.
Circular dichroism measurements
The far-UV CD experiments were performed on a Jasco-J715 spectropolarimeter at room temperature. The spectra of 190- to 250-nm wavelength range were recorded with a bandwidth of 1 nm, a speed of 20 nm/min, and a response time of 0.25 sec. Each spectrum was the average of three consecutive scans, followed by subtracting the baseline for buffer solutions (20 mM phosphate, 100 mM NaCl [pH 7.0]). The protein concentration for far-UV CD measurements was about 0.2 mg/mL.
NMR spectroscopy
All NMR measurements were carried out at 25°C on a Varian INOVA 600 spectrometer equipped with three RF channels and a triple-resonance pulsed-field gradient probe. The sequential backbone assignment of the UbL domain of DC-UbP was obtained from standard heteronuclear correlation experiments including 3D HNCA, HN (CO) CA, HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH. The side-chain assignments were obtained from a 3D HCCH-TOCSY experiment. Distance information between protons was derived from 3D 15N-edited and 13C-edited NOESY recorded on the 15N- or 13C/15N-labeled sample. Torsion angles were derived from 3J (HN, Hα) coupling constants measured by HNHA experiment. Solvent-exposed amides were mapped using Clean SEA-HSQC experiments (Lin et al. 2002).
Structure calculation
The NMR Pipe software suite (Delaglio et al. 1995) was applied to process the NMR data, and NMR View (Johnson and Blevins 1994) software packages were used for picking resonance peaks and for data analysis. The CNS program (Brünger et al. 1998) with the ARIA module (Nilges et al. 1997) was adopted to assign NOE peaks and to calculate structures. The protein structures were displayed by using the MOLMOL program (Koradi et al. 1996). Hydrogen bond restraints (two per hydrogen bond) were generated by a combination of H/D exchange data, medium-range NOEs, and CSI. Backbone dihedral angle restraints (φ and Ψ) were derived from the 3J(HN, Hα) coupling constants measured with 3D HNHA experiment (Vuister et al. 1993) and the chemical shifts by using TALOS program (Cornilescu et al. 1999). The restraint range was extended to ±50° for the residues located in β-strands and to ±30° for the residues located in other regions. The restraints used for structural calculation are summarized in Table 1.
The structural calculation in combination with iterative NOE peak assignments were performed for nine cycles and a total of 150 structures were finally obtained. The solution NMR structure generated from the calculation is represented by the 15 lowest energy structures out of the 150 structures. The 1H, 15N, and 13C chemical shifts have been deposited in the BioMagResBank (accession no. 6609). The coordinates of the UbL domain of DC-UbP (residues 21–100) and the supporting data have been deposited in the Protein Data Bank (accession code 1TTN).
Determination of backbone relaxation parameters
All 15N relaxation data were acquired at 25°C using two-dimensional proton-detected heteronuclear NMR experiments implementing the standard pulse sequences (Farrow et al. 1994). Spin-lattice relaxation rates (R1) were measured in a series of spectra with relaxation delays of 10, 200, 400 (×2), 600, 800, 1000, 1200, 1400, 1600 (×2), and 1800 msec. Spin– spin relaxation rates (R2) were determined with relaxation delays of 10, 30 (×2), 50, 70, 90, 110 (×2), 130, 150, 170, and 190 msec. A recycle delay of 1.5 sec was used for measuring R1 and R2 relaxation rates. The spectra for {1H}-15N heteronuclear NOE measurements were recorded with a 2-sec relaxation delay followed by a 3-sec period of proton saturation. The spectra recorded in the absence of proton saturation employed a relaxation delay of 5 sec. All spectra were processed using NMRPipe and NMRView software. The relaxation rates, R1 and R2, were obtained by fitting measured peak intensity to a two-parameter function using a nonlinear least-square fitting algorithm (Press et al. 1989). Uncertainties in peak intensity were determined from duplicated spectra. The steady-state {1H}-15N NOE enhancements were calculated as the ratio of peak intensity in spectra recorded with or without proton saturation. The standard errors were determined from two data sets.
Acknowledgments
We thank Dr. Wang W.N. and Miss Lin X.J. for technical help. This work was supported by grants from the 863 Hi-Tech program (2002BA711A13), the Chinese Academy of Sciences (KSCX1-SW-17), Shanghai Commission of Science and Technology (03JC14081), and One Hundred Talents Program of the Chinese Academy of Sciences.
Abbreviations
CSI, chemical shift index
DC-UbP, dendritic cell-derived ubiquitin-like protein
HSQC, heteronuclear single quantum coherence
IPTG, isopropyl-β,D-thiogalactopyranoside
NOESY, nuclear Overhauser effect spectroscopy
ORF, open reading frame
ppm, parts per million
RMSD, root-mean-square deviation
SA, simulated annealing
Ub, ubiquitin
UbL, ubiquitin-like
UBLs, ubiquitin-like modifiers
UDP, ubiquitin-domain protein
VdW, van der Waals
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051455505.
Supplemental material: see www.proteinscience.org
References
- Bayer, P., Arndt, A., Metzger, S., Mahajan, R., Melchior, F., Jaenicke, R., and Becker, J. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280 275–286. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Bruschweiler, R. 2003. New approaches to the dynamic interpretation and prediction of NMR relaxation data from proteins. Curr. Opin. Struct. Biol. 13 175–183. [DOI] [PubMed] [Google Scholar]
- Cavanagh, J., Fairbrother, W., Palmer, A., and Skelton, N. 1995. Protein NMR spectroscopy: Principles and practice. Academic Press, San Diego, CA.
- Chen, Z.J. and Fuchs, S.Y. 2004. Ubiquitin-dependent activation of NF-κB: K63-linked ubiquitin chains: A link to cancer? Cancer Biol. Ther. 3 286–288. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Marquardt, J.L., Ottinger, M., and Bax, A. 1998. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 120 6836–6837. [Google Scholar]
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Gali, R.R., Schwickart, M., Larsen, C.N., Leggett, D.S., Muller, B., Feng, M.T., Tubing, F., Dittmar, G.A., and Finley, D. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4 725–730. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33 5984–6003. [DOI] [PubMed] [Google Scholar]
- Finley, D. 2001. Signal transduction. An alternative to destruction. Nature 412 283–286. [DOI] [PubMed] [Google Scholar]
- Finley, D., Ciechanover, A., and Varshavsky, A. 2004. Ubiquitin as a central cellular regulator. Cell S116 S29–S32. [DOI] [PubMed] [Google Scholar]
- Goettsch, S. and Bayer, P. 2002. Structural attributes in the conjugation of ubiquitin, SUMO and RUB to protein substrates. Front. Biosci. 7 A148–162. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15 305–308. [DOI] [PubMed] [Google Scholar]
- Hershko, A. and Ciechanover, A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67 425–479. [DOI] [PubMed] [Google Scholar]
- Huang, Y.J., Swapna, G.V., Rajan, P.K., Ke, H., Xia, B., Shukla, K., Inouye, M., and Montelione, G.T. 2003. Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J. Mol. Biol. 327 521–536. [DOI] [PubMed] [Google Scholar]
- Jentsch, S. and Pyrowolakis, G. 2000. Ubiquitin and its kin: How close are the family ties? Trends Cell Biol. 10 335–342. [DOI] [PubMed] [Google Scholar]
- Jin, C., Shiyanova, T., Shen, Z., and Liao, X. 2001. Heteronuclear nuclear magnetic resonance assignments, structure and dynamics of SUMO-1, a human ubiquitin-like protein. Int. J. Biol. Macromol. 28 227–234. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Blevins, R.A. 1994. A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Brower, C.S., Conaway, R.C., and Conaway, J.W. 2002. A molecular basis for stabilization of the von Hippel-Lindau (VHL) tumor suppressor protein by components of the VHL ubiquitin ligase. J. Biol. Chem. 277 30388–30393. [DOI] [PubMed] [Google Scholar]
- Kim, K.I. and Zhang, D.E. 2003. ISG15, not just another ubiquitin-like protein. Biochem. Biophys. Res. Commun. 307 431–434. [DOI] [PubMed] [Google Scholar]
- Ko, S.H., Uehara, T., Tsuruma, K., and Nomura, Y. 2004. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 566 110–114. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J.Mol.Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Lin, D., Sze, K.H., Cui, Y., and Zhu, G. 2002. Clean SEA-HSQC: A method to map solvent exposed amides in large non-deuterated proteins with gradient-enhanced HSQC. J. Biomol. NMR 23 317–322. [DOI] [PubMed] [Google Scholar]
- Liu, S., An, H., Li, N., Yu, Y., Lin, N., Wan, T., Zhang, M., Wang, W., and Cao, X. 2003. Cloning and identification of a novel human ubiquitin-like protein, DC-UbP, from dendritic cells. Biochem. Biophys. Res. Commun. 300 800–805. [DOI] [PubMed] [Google Scholar]
- Marin, I., Lucas, J.I., Gradilla, A.C., and Ferrus, A. 2004. Parkin and relatives: The RBR family of ubiquitin ligases. Physiol. Genomics 17 253–263. [DOI] [PubMed] [Google Scholar]
- McNally, T., Huang, Q., Janis, R.S., Liu, Z., Olejniczak, E.T., and Reilly, R.M. 2003. Structural analysis of UBL5, a novel ubiquitin-like modifier. Protein Sci. 12 1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M.D., Klionsky, D.J., Ohsumi, M., and Ohsumi, Y. 1998. A protein conjugation system essential for autophagy. Nature 395 395–398. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yoshimori, T., and Ohsumi, Y. 2003. Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 35 553–561. [DOI] [PubMed] [Google Scholar]
- Mueller, T.D. and Feigon, J. 2003. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 22 4634–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., Ledl, A., and Schmidt, D. 2004. SUMO: A regulator of gene expression and genome integrity. Oncogene 23 1998–2008. [DOI] [PubMed] [Google Scholar]
- Nilges, M., Macias, M.J., O’Donoghue, S.I., and Oschkinat, H. 1997. Automated NOESY interpretation with ambiguous distance restraints: The refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J. Mol. Biol. 269 408–422. [DOI] [PubMed] [Google Scholar]
- Pan, Z.Q., Kentsis, A., Dias, D.C., Yamoah, K., and Wu, K. 2004. Nedd8 on cullin: Building an expressway to protein destruction. Oncogene 23 1985–1997. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Back to the future with ubiquitin. Cell 116 181–190. [DOI] [PubMed] [Google Scholar]
- Press, W.H., Flannery, B.P., Teukolsky, S.A., and Vetterling, W.T. 1989. Numerical recipes in C. Cambridge University Press, Cambridge, UK.
- Ramelot, T.A., Cort, J.R., Yee, A.A., Semesi, A., Edwards, A.M., Arrowsmith, C.H., and Kennedy, M.A. 2003. Solution structure of the yeast ubiquitin-like modifier protein Hub1. J. Struct. Funct. Genomics 4 25–30. [DOI] [PubMed] [Google Scholar]
- Sakata, E., Yamaguchi, Y., Kurimoto, E., Kikuchi, J., Yokoyama, S., Yamada, S., Kawahara, H., Yokosawa, H., Hattori, N., Mizuno, Y., et al. 2003. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 4 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber, C., Chen, L., Tongaonkar, P., Vega, I., Lambertson, D., Potts, W., and Madura, K. 1998. Rad23 links DNA repair to the ubiquitin/ proteasome pathway. Nature 391 715–718. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.C. and Hochstrasser, M. 2003. A superfamily of protein tags: Ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28 321– 328. [DOI] [PubMed] [Google Scholar]
- Tjandra, N., Feller, S.E., Pastor, R.W., and Bax, A. 1995. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J. Am. Chem. Soc. 117 12562–12566. [Google Scholar]
- Vuister, G.W., Delaglio, F., and Bax, A. 1993. The use of 1JCα-Hα coupling constants as a probe for protein backbone conformation. J. Biomol. NMR 3 67–80. [DOI] [PubMed] [Google Scholar]
- Whitby, F.G., Xia, G., Pickart, C.M., and Hill, C.P. 1998. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273 34983–34991. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Sykes, B.D., and Richards, F.M. 1992. The chemical shift index: A fast and simple method of the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wyndham, A.M., Baker, R.T., and Chelvanayagam, G. 1999. The Ubp6 family of deubiquitinating enzymes contains a ubiquitin-like domain: SUb. Protein Sci. 8 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]