Abstract
The BTB domain (also known as the POZ domain) is an evolutionarily conserved protein–protein interaction motif found at the N terminus of 5–10% of C2H2-type zinc-finger transcription factors, as well as in some actin-associated proteins bearing the kelch motif. Many BTB proteins are transcriptional regulators that mediate gene expression through the control of chromatin conformation. In the human promyelocytic leukemia zinc finger (PLZF) protein, the BTB domain has transcriptional repression activity, directs the protein to a nuclear punctate pattern, and interacts with components of the histone deacetylase complex. The association of the PLZF BTB domain with the histone deacetylase complex provides a mechanism of linking the transcription factor with enzymatic activities that regulate chromatin conformation. The crystal structure of the BTB domain of PLZF was determined at 1.9 Å resolution and reveals a tightly intertwined dimer with an extensive hydrophobic interface. Approximately one-quarter of the monomer surface area is involved in the dimer intermolecular contact. These features are typical of obligate homodimers, and we expect the full-length PLZF protein to exist as a branched transcription factor with two C-terminal DNA-binding regions. A surface-exposed groove lined with conserved amino acids is formed at the dimer interface, suggestive of a peptide-binding site. This groove may represent the site of interaction of the PLZF BTB domain with nuclear corepressors or other nuclear proteins.
The BTB domain (Broad-Complex, Tramtrack, and Bric à brac) (1, 2), also known as POZ (poxvirus and zinc finger) (3), is an evolutionarily conserved protein–protein interaction domain often found in developmentally regulated transcription factors. The domain is strongly implicated in the regulation of gene expression through the local control of chromatin conformation (4). The domain was first identified in a set of Drosophila and poxvirus genes (5), and examples of BTB domain genes have since been found in organisms ranging from yeast to man.
A search of the current publicly available sequence databases reveals 56 distinct human BTB entries, of which 22 correspond to named, full-length genes, whereas the remaining entries are known only as tentative human consensus (THC) sequences, or expressed sequence tags (EST), (a tabulation of these genes can be found at http://xtal.oci.utoronto.ca/prive/btbtable.html). Approximately two-thirds of the full-length human BTB genes also encode C2H2 zinc finger modules, whereas approximately one-half of the remaining entries contain the kelch motif (4, 6). None of the known human BTB genes contain more than one single copy of the domain, making it likely that most, if not all, of the known BTB tentative human consensus (THC) and expressed sequence tag (EST) entries correspond to distinct genes. Based on the projection that there are 300–700 zinc finger proteins in man (7), we estimate that 5–10% of zinc finger proteins also contain BTB domains. The domain is known to form homomeric and heteromeric associations with other BTB domains (3), and given the large number of BTB domain proteins in the genomes of higher eukaryotes, an attractive possibility is that the domain generates a combinatorial diversity of complexes within a family of BTB domain proteins.
The organization of the human promyelocytic leukemia zinc finger (PLZF) protein is typical of BTB domain proteins: the 120 amino acid BTB domain is found at the N terminus of the protein, followed by a central region of several hundred amino acids, and ending with a series of C2H2 Kruppel-type zinc fingers. The PLZF protein is a potent transcriptional repressor implicated in embryonic development and in hematopoesis (8, 9). It occurs as a fusion protein with the retinoic acid receptor α (RARα) in the rare t(11;17) form of acute promyelocytic leukemia (8). In this acute promyelocytic leukemia, the BTB domain from PLZF is responsible for the dominant negative properties of the PLZF-RARα fusion against wild-type RARα-retinoic acid X receptor (RXRα) function (10). Cells from the more common t(15;17) PML-RARα acute promyelocytic leukemia can be induced to differentiate with treatment with retinoic acid (RA), but PLZF-RARα cells are responsive to RA only if potentiated with inhibitors of the histone deacetylase complex (11–14). Because the BTB domain from PLZF can associate with mSin3A, histone deacetylase 1 (14), SMRT (silencing mediator for retinoid and thyroid hormone receptors) (13), and N-CoR (nuclear receptor corepressor) (12), the PLZF–RARα fusion protein has been proposed to repress transcription at RA-sensitive sites through the BTB-mediated recruitment of the histone deacetylase complex (11–14).
Because the domain is widely represented in eukaryotic genomes, features common to BTB proteins provide crucial hints as to the function of the domain. Punctate nuclear staining patterns are commonly seen in zinc-finger BTB proteins (4), and for ZID, PLZF, and BCL6 (or LAZ3), this has been shown to require an intact BTB domain (3, 10, 15). Many BTB domain proteins, including Tramtrack, PLZF, and BCL6, are transcriptional repressors, although some, such as GAGA factor, can counteract repression by chromatin remodeling (16). As with other conserved domains, we expect that the domain carries out a common underyling molecular role, which can be used for a variety of biological functions. Our structure of the PLZF BTB domain reveals that the BTB domain is a tightly interwound dimer and identifies sites at the dimer interface, which may be important for BTB–BTB domain interactions, as well as surface features in the dimer, which may be involved in interactions with other proteins.
METHODS
Codons 1–132 from a human PLZF cDNA (gift from J. D. Licht, Mount Sinai School of Medicine, New York) were amplified by PCR and subcloned into the plasmid pET-32(a), producing an expression vector for an N-terminal thioredoxin domain followed by a 6X His tag sequence, a 40-aa linker, and amino acids 1–132 of PLZF. The selenomethionyl-substituted fusion protein was overexpressed in the Escherichia coli methionine auxotroph strain B834(DE3). After cell lysis and protein purification by nickel chelate affinity chromatography, the peak fractions containing the fusion protein were pooled and dialyzed against 100 mM NaCl, 50 mM Tris⋅HCl (pH 7.5), 2.5 mM CaCl2, and 0.5 mM Tris [2-carboxyethyl]phosphine hydrochloride (TCEP), and trypsin was added to the fusion protein in digest buffer at a ratio of 1:1000 (wt/wt) to remove the sequences N-terminal to the BTB domain. Under these conditions, the domain was resistant to trypsin treatment for 24 hr at room temperature. The PLZF BTB domain was purified from the digest mixture by anion exchange chromatography and size exclusion chromatography. The purified protein was concentrated to 8 mg/ml in 300 mM NaCl, 50 mM Tris⋅HCl (pH 7.5), and 1 mM TCEP. The final protein fragment consists of amino acids 6–132 of PLZF, as confirmed by Edman N-terminal amino acid sequencing and ion spray mass spectrometry. Crystals of PLZF(6–132) were grown at 4°C in hanging drops by mixing 2 μl of protein with 2 μl of reservoir buffer (0.15 M CaCl2, 0.1 M Hepes (pH 7.5), and 4% isopropanol), and equilibrating with 1 ml of reservoir buffer for 48 hr. The space group was I4122 (a = 72.63 Å, b = 72.63 Å, and c = 168.98 Å) with one BTB monomer in the asymmetric unit. Crystals were cryoprotected in reservoir solution enriched in CaCl2 to 0.18 M and MPD to 20.0% before flash-freezing to 100 K. A multiwavelength anomalous diffraction experiment was performed at the Cornell High Energy Synchrotron Source (CHESS) on beamline F-2, using an Area Detector Systems Quantum-4 Charge-Coupled Device (CCD) detector (17). Data processing and reduction were done with mosflm and scala (18). The positions of 4 of the 6 expected selenium atoms were determined by both Patterson and Direct methods by using shelxs-97. The selenium positions were refined with sharp (19), and three more selenium sites were found by examining residual log-likelihood gradient maps after initial refinement runs. Met-58 displays multiple side chain conformations, accounting for the extra selenium site. The final multiwavelength anomalous diffraction phase set had an overall figure of merit of 0.72 for the 17,622 reflections in the resolution range from 20 to 1.9 Å. The resulting electron density maps were solvent flattened with dm (18), producing maps with clear, connected density for the main chain atoms 6–126 except residue 66 and for 113 of the remaining 120 side chains. An atomic model was built into the density with the program O (20) and refined with refmac (21). The final model contains all atoms for residues 6–126 of PLZF. The last six residues (amino acids 127–132) are disordered in the crystal and are not visible in the electron density map.
RESULTS
General Description of the Structure.
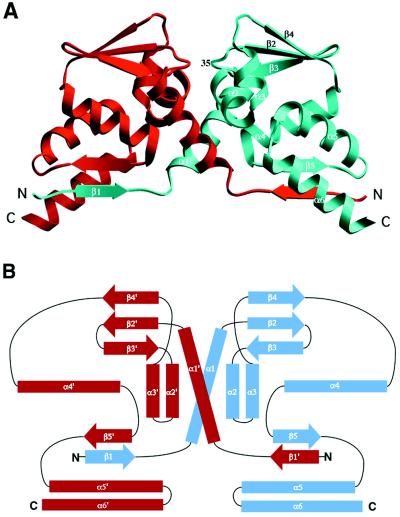
We have expressed and crystallized the BTB domain from PLZF as a selenomethionine-substituted protein and the structure was determined by multiwavelength anomalous diffraction phasing (Tables 1 and 2). The structure reveals a tightly intertwined homodimer, consistent with hydrodynamic studies (22). The dimer can be described roughly as a prolate ellipsoid with overall dimensions of ≈60 Å by 26 Å by 32 Å. Because of the flattened shape of the molecule, most residues are at or near the surface of the protein. The central scaffolding of the protein is made up of a cluster of α-helices flanked by short β-sheets at both the top and bottom of the molecule (Figs. 1 and 2). By using the dali server (25), a search for proteins with similar three-dimensional (3D) structures revealed no significant entries in the Brookhaven Protein Data Bank (PDB). The N terminus of each chain is associated with the main body of the other chain, generating a two-stranded antiparallel β-sheet between strand β1 of one monomer and strand β5′ of the other. (For ease of discussion, positions on the symmetry-related molecule will be labeled with primes. In addition, equivalent symmetry-related pairs of interactions, such as β1/β5′ and β1′/β5 will not be explicitly mentioned.) Helix α6 and the β1/β5′-sheet, along with the symmetry-related elements, form an extended, concave surface on the underside of the protein dimer. Both chains begin and end at the base of the dimer, but because of the swapped amino termini, the N terminus of one chain is next to the C terminus of the other (Fig. 2). This places the two C termini at opposite ends of the base of the dimer, separated by a distance of >58 Å. In intact PLZF, the remaining C-terminal regions of the protein would extend from these points.
Table 1.
Diffraction data
| Wavelength, Å | Resolution, Å | Reflections, total/unique | Completeness, % | Rsym*, % | 〈I〉/〈σ(I)〉† |
|---|---|---|---|---|---|
| λ1 = 0.9793 | 20.0–1.90 | 204897/18253 | 99.3 (96.7) | 4.4 (24.8) | 11.2 (3.0) |
| λ2 = 0.9790 | 20.0–1.90 | 189702/18254 | 99.4 (97.3) | 4.1 (26.9) | 12.0 (2.9) |
| λ3 = 0.9638 | 20.0–1.90 | 156050/17398 | 94.7 (92.2) | 4.3 (25.7) | 11.4 (2.8) |
Parentheses denote statistics in the 2.0–1.9 Å resolution shell.
Rsym = 100 × (ΣhΣi(Ih,i − Ih))/ΣhΣiIh,i for the intensity I of i observations of reflection h. Ih is the mean intensity of the reflection.
〈I〉/〈σ(I)〉 = mean intensity/mean standard deviation.
Table 2.
Data used in the refinement and refinement statistics
| Resolution, Å | 20.0–1.9 |
| Data cutoff F/σ(F) | 0.0 |
| Number of Reflections | 17,266 |
| Completeness, % | 94.0 |
| R value* | 21.2 |
| Rfree† | 25.2 |
| Number of water molecules | 130 |
| rmsd, bond distances, Å | 0.021 |
| rmsd, angle distances, Å | 0.042 |
| 〈B〉 (Å2) main chain protein atoms | 28.9 |
| 〈B〉 (Å2) all protein atoms | 33.4 |
| 〈B〉 (Å2) water atoms | 44.5 |
R value = 100 × Σ|Fobs − Fcalc|/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor magnitudes, respectively.
Rfree is the same as the R value but is calculated from 5% of the reflection data excluded from the refinement.
Figure 1.
Sequence alignment of selected BTB domains and the observed secondary structure of the PLZF BTB domain. TTK (Tramtrack) and GAGA factor are Drosophila proteins, and the others are human proteins. The numbering is according to PLZF, and the crystal structure consists of residues 6–126. Black and gray backgrounds are used to indicate identical and/or conserved residues found in at least 50% of the proteins at a given position. Residues in a 310 helix conformation are indicated by hatching. • indicates amino acids that are classified as buried in the PLZF BTB monomer according to the criteria of Rice and Eisenberg (23). Colored numbers represent the percentage of contribution of the residue to the buried interface surface in the dimer, rounded to the nearest integer (i.e., a value of “0” represents a contribution from 0.1 to 0.49%, values of 1 represent a contribution of 0.5–1.49%, etc.). Residues indicated with red numbers participate in the swapped or closed interface, and amino acids involved with the central open interface are labeled in blue. Residues 16–19 contribute to both and are not classified.
Figure 2.
(A) Ribbon diagram of the PLZF BTB domain dimer. One monomer is colored red and the other blue. The secondary structure elements of one monomer are indicated, and the position of the conserved residue Asp-35 is labeled. In this view, the two halves of the dimer are related by a vertical twofold symmetry axis. This figure was generated with the graphical program setor (24). (B) Schematic representation of the topology of the dimer. α-Helices are indicated by rectangles, and β-sheets by thick arrows. The symmetry related secondary structure elements are indicated with primes.
A crystal contact between the lower sheets of two symmetry related BTB dimers generates a short 4-stranded antiparallel β-sheet involving four different peptide chains. This association is probably not biologically important because in solution we have only observed dimers at all protein concentrations. This result is consistent with the findings of Li et al. (22) on a PLZF molecule having only slightly different termini than our construct.
Because the BTB domain self-associates into a dimer and forms complexes with other proteins, we discuss three structural classes of residues in the molecule: those buried within a monomer, those buried at the dimer interface, and residues which are exposed on the surface.
Monomer Core.
As expected, most of the strongly conserved residues are located within the core of the monomer (Fig. 1). Especially conserved are His-48, Leu-52, Ser-56, and Tyr-88, and these buried residues are all in contact with each other and not involved in either the dimer interface, nor exposed to the protein surface.
Dimer Interface.
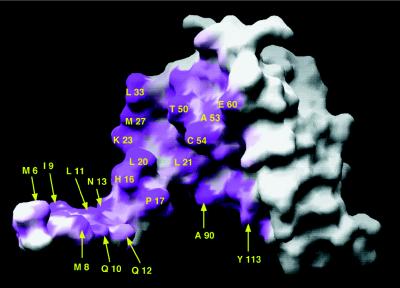
The residues at the intermolecular interface determine the specificity and stability of the dimer and are of particular interest because of the possibility of the formation of heterodimers between BTB domain proteins. Interface amino acids are distributed throughout the entire length of the protein (Figs. 1 and 3). Of the 8,153 Å2 of solvent accessible surface area in the monomer, 1,999 Å2 are involved in the dimer surface contact. Because of the interchanged β1 segment of the monomers, the structure is suggestive of a 3D domain-swapped dimer (27). Using the terminology used for 3D domain-swapped proteins, the intersubunit contacts can be grouped into two classes (27): a pair of domain-swapped “closed interfaces” involving β1/β5′/α6′ and β1′/β5/α6 and a central “open interface” with contributions mainly from α1, α1′, α2, and α2′. We emphasize that there is no evidence of exchange between monomeric and dimeric forms of the protein (22), and confirmation that the BTB dimer fold is the product of 3D domain-swapping in an ancient precursor will require the knowledge of a monomeric protein with the same fold but with the swapped β1 element as part of its main domain. Although this has yet to be described for this fold, the general organization of the dimer is very similar to that of other systems, which have met this criteria for 3D domain-swapping. The probable “hinge-loop” region that would exist in a different conformation in the case of a monomeric BTB fold consists of amino acids 16–19, and these are well ordered and form multiple bridging contacts in the PLZF BTB dimer.
Figure 3.
View of one monomer displayed as a solvent accessible surface in grasp (26). The orientation is roughly the same as for the blue monomer in Fig. 2A. The surface buried upon dimer formation is indicated in magenta, and residues that contribute at least 2% of the buried surface of the dimer (Fig. 1) are labeled. Residues from the closed interface are indicated with arrows. Not shown in the diagram are the residues from β5 and α6, which form a groove on the underside of the monomer that accommodates β1′ from the adjoining monomer. The entrance to this groove is lined with Ala-90 and Tyr-113.
In the closed interface, one side of the β1/β5′ sheet is exposed to solvent, and the other side is packed against helix α6′. Here, the five main chain H-bonds of the antiparallel β-sheet contribute to the dimer stability, as well as the burial of the hydrophobic residues Ile-9, Leu-11, Leu-92′, and Ala-94′ that point into the protein interior. It is notable that a few BTB protein such as Miz-1 (28) and most of the poxvirus proteins (5) have a shortened N terminus and thus cannot form strand β1. The sequence conservation is preserved in the β5 region of these proteins, even though the lower β-sheet cannot exist in these domains. It remains to be seen what the effects of this shortened N terminus are on the dimer.
The central open interface has a total buried surface of 891 Å2 and has contributions mainly from α1 and α2. Interface residues His-16, Pro-17, Leu-20, Leu-21, Lys-23, Ala-24, and Met-27 are all located on one face of α1, but most of the contacts are located in the first half of the helix because the second half is mostly associated with the main body of the individual monomer fold. Two short helices, α2 and α3, form a hairpin with helix α2 packed against the lower portion of α1′, such that His-16′ through Leu-20′ are sandwiched between helices α1 and α2 (Fig. 2). The sequence of the short α2 helix is one of the most conserved regions in the BTB family, and residues from this region make contributions to the monomer core, the dimer interface, and the external surface. This region of the dimer interface is predominately nonpolar in nature, involving mostly contacts between hydrophobic groups. For example, the γ-CH2 of residue Glu-60 is packed against the ɛ-CH2 of Met-27, leaving its charged carboxylate group fully exposed to the exterior solvent.
The contacts of the central interface form a closed cavity with an interior volume of 213 Å3 in the center of the dimer. A total of three ordered water molecules have been located in this largely apolar cavity, two of which hydrogen bond to each other but make no polar contacts with protein atoms. The rim of this cavity is formed by residues 16, 17, 20, 21, 23, 27, 33, 50, and 54 and sequesters residues Ala-24/24′ and Val-51/51′ from the external solvent. Such cavities are common in intersubunit contacts (29). The only polar protein atoms in this cavity are the hydroxyl groups of Thr-50 and Thr-50′, which hydrogen bond to each other, and the main chain carbonyl oxygens of Pro-17 and 17′, which are bridged by the third interior water. Remarkably, the Thr-50 interaction is the only direct polar bond across this central interface, and yet this amino acid is not conserved in the family of BTB domains (Fig. 1).
Potential Ligand-Binding Groove.
A grouping of conserved residues at a protein surface is a common feature of protein functional sites, and next we consider residues that are exposed to the bulk solvent and have sequence conservation. A cluster of conserved, exposed residues from three different loops is found at the top of the dimer (Fig. 4). The floor of this groove is made up of residues in the α1/β2 and β3/α2 loops (Leu-33, Cys-34, Asp-35, and Arg-49), and the walls of the cleft are formed by the α3/β4 loop (Phe-63, His-64, Asn-66, Ser-67, and Gln-68). The groove is centered about the twofold axis of the dimer, and the equivalent residues from both chains contribute to the formation of this site. Thus, the groove exists only in the dimer. Asp-35 is adjacent to Arg-49, but the two residues do not form a salt bridge. Instead, the guanidinium group of the arginine forms hydrogen bonds to the main chain carbonyl atoms of Asp-35 and Ser-67 from the same monomer. Asp-35 is 37% exposed to solvent, but Arg-49 has only 10% of its surface area exposed and is classified as a buried residue by the criteria of Rice and Eisenberg (23). The groove is largely electroneutral as a result of the close proximity of Asp-35 and Arg-49. From over 100 BTB domains known to date, Asp-35 is absolutely conserved, whereas position 49 is either a lysine or an arginine in all but two examples.
Figure 4.
Sequence conservation in the exposed surface of the BTB dimer. (A) View in the same orientation as in Fig. 2A. (B) View looking directly down the twofold axis of the dimer. A multiple sequence alignment was constructed as follows: residues 6–126 of PLZF were used in a fasta3 (38) search of the swall database (Nonredundant Protein sequence database including Swissprot, Trembl, and TremblNew) at the EMBL- European Bioinformatics Institute server (http://www2.ebi.ac.uk/fasta3). Entries with E score <0.1 were used for further processing. Identical and closely matching sequences were removed from the set, so that the no two pairs in the final set of 42 sequences had >88% sequence identity. The set was then aligned, and the sequence variability was calculated as the number of different amino acids present at each residue position. The sequence variability was then displayed on the solvent accessible surface of the dimer by using grasp (26). In this variability scoring scheme, a fully conserved residue is assigned a value of 1, and the maximum variability is 20. The only exposed, fully conserved residue in the structure is Asp-35, present in the center of the groove.
The conservation data presented in Fig. 4 is based on a multiple sequence alignment of the 42 highest scoring sequences to the PLZF BTB domain and may mask patterns seen in more restricted subsets. However, essentially the same result was seen when the subset of the 10 most similar human sequences was used (data not shown). This does not prove that other conserved surface features are not found in specific subsets of BTB domains, but it does support the existence of a conserved surface common to most, if not all BTB proteins.
As expected in the constricted space of this groove, there is a well defined and regular array of water molecules in this pocket. This is a general feature of binding sites in proteins (30). The groove is ≈8 Å wide and 20 Å long and is of a sufficient size to accomodate a 5–6 amino acid peptide. We suggest that the binding groove is a potential interaction site for mSin3A, histone deacetylase 1, SMRT, or N-CoR. It also is possible that this is a binding site for other, as yet unidentified, ligands. The residues of the α3/β4 loop have temperature factors in the range of 40–60 Å2, which are among the highest in the entire structure. Thus, the walls of the groove may be able to adjust their conformation to accommodate a putative ligand. The overall shape and symmetry of the BTB dimer is similar to that of the HIV protease dimer (31), and both share a groove that crosses the narrow dimension of the molecule with a pair of diad-related aspartate residues exposed at the floor. In the protease, these are the active site residues. There are, however, significant differences, in that the BTB domain lacks the conserved Asp-Thr-Gly signature sequence of aspartate proteases, and the details of the orientation of the aspartates in the PLZF structure are not the same as in the protease. In addition, the PLZF BTB domain is mostly α-helical, whereas aspartate proteases are built from a β-sheet core. It remains to be seen whether the BTB groove is a simple binding site or whether it has enzymatic activity.
DISCUSSION
The hydrophobic nature of the dimer interface, the large interface accessible surface area, and the extensive shape complimentarity (as measured by the low gap volume index of 1.44 Å) (32) are all typical of an obligate homodimer. This is consistent with the coupled two-state unfolding transition from a folded dimer to two unfolded monomers observed by differential scanning calorimetry and equilibrium denaturation (22). Protein domains known to form combinatorial hetero-oligomers generally associate either through smaller hydrophobic surfaces, as in the leucine zipper family (33), or through polar interfaces as in the SMAD protein family (34). BTB domains have been shown by in vitro coexpression to form specific homo- and hetero-oligomers through self-association (3), and further work will be necessary to establish the importance of associations between different BTB domains in vivo.
The dimerization of transcription factors has implications for the binding to the target DNA site, and to date, no physiologically relevant DNA-binding site(s) has been reported for PLZF (35). However, GAGA factor binds to high density GA repeats with a large number of potential binding sites in close proximity, Tramtrack binds two monomer sites separated by 24 base pairs in the fushi tarazu promoter (36), and ZF5 binds two sites separated by 16 base pairs in the c-myc promoter (37). In general, the DNA-binding domains of BTB domain transcription factors are not rigidly associated with the BTB domain dimer region, and it is unlikely that the twofold symmetry of the dimer extends through the entire protein–DNA complex. In the case of PLZF, the BTB domain and the zinc-finger regions are separated by residues 127–406, which includes both an acidic region and a proline-rich segment with only weak sequence similarity with other known proteins. Because of the expected flexibility in this region, bidentate binding to putative biological DNA-binding sites is not likely to induce DNA bending or looping unless the two sites are very widely separated. This also explains the observation that the BTB domain can function as an autonomous transrepression module, and maintains its activities when placed in a variety of DNA-binding domain contexts. The presumed flexibility in the relative positions of the DNA-binding domains also may be important for binding to nucleosomal DNA.
At the molecular level, the BTB domain has two roles: it can self-associate to form a dimer, and it can interact with other non-BTB domain proteins. Because the conserved groove in the BTB domain is at the dimer interface, we predict that dimer formation is necessary for the proposed interactions within this groove. We do not rule out the possibility that that the domain contains other protein interaction sites, but these sites would not have the striking sequence conservation seen in the central groove. The PLZF BTB domain has been shown to interact directly with each of mSin3A, histone deacetylase 1, SMRT, and N-CoR (12–14). Clearly, these cannot bind simultaneously to the same site on the domain, but they may bind to nonoverlapping regions of the dimer. We expect that additional ligands for this and other BTB domains remain to be discovered, and the structure presented here provides a framework with which to study these interactions.
Acknowledgments
We thank J.-L. Couderc, F. Laski, J. Licht, C. Richardson, D. Rose, and S. Zollman for helpful discussions; I. Klitsie, D. Kuntz, and V. Ahn for help with cloning and protein expression; and D. Thiel and members of the MacCHESS staff for assistance in data collection. CHESS is supported by the National Science Foundation under award DMR-9311772, and the Macromolecular Diffraction at CHESS is supported by award RR-01646 from the National Institutes of Health. C.K.E. is a postdoctoral fellow of the Deutsche Forschungsgemeinschaft. This work has been supported by grants from the Medical Research Foundation of Canada (MRCC) and the National Cancer Institute of Canada (NCIC).
ABBREVIATION
- PLZF
promyelocytic leukemia zinc finger
- 3D
three-dimensional
- CHESS
Cornell High Energy Synchrotron Source
- RA
retinoic acid
- RARα
RA receptor α
- SMRT
silencing mediator for retinoid and thyroid hormone receptors
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1BUO).
References
- 1. Godt D, Couderc J L, Cramton SE, Laski F A. Development (Cambridge, UK) 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- 2.Zollman S, Godt D, Privé G G, Couderc J L, Laski F A. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell V J, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 4.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 5.Koonin E V, Senkevich T G, Cherenos V I. Trends Biochem Sci. 1992;17:213–214. doi: 10.1016/0968-0004(92)90379-n. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Doolittle R. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 7.Klug A, Schwabe J. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 8.Chen Z, Brand N J, Chen A, Chen S J, Tong J H, Wang Z Y, Waxman S, Zelent A. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S, et al. Proc Natl Acad Sci USA. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong H J, Wang Z Y, Licht J, Waxman S, Chomienne C, et al. Proc Natl Acad Sci USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 12.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 13.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 15.Dhordain P, Albagli O, Ansieau S, Koken M H, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J P, Leprince D. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 16.Wilkins R C, Lis J T. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szebenyi D M E, Arvai A, Ealick S, La Iuppa J M, Nielsen C. J Synchrotron Rad. 1997;4:128–135. doi: 10.1107/S0909049597000204. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 19.Fortelle, E. D. & Brigogne, G. (1997) Methods Enzymol. 276. [DOI] [PubMed]
- 20.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47. [DOI] [PubMed]
- 21.Murshudov G, Vagin A, Dodson E. Acta Crystallogr D. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Lopez-Guisa J M, Ninan N, Weiner E J, Rauscher F J, III, Marmorstein R. J Biol Chem. 1997;272:27324–27329. doi: 10.1074/jbc.272.43.27324. [DOI] [PubMed] [Google Scholar]
- 23.Rice D W, Eisenberg D. J Mol Biol. 1997;267:1026–1038. doi: 10.1006/jmbi.1997.0924. [DOI] [PubMed] [Google Scholar]
- 24.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 27.Bennett M J, Choe S, Eisenberg D. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard S J, Argos P. Protein Sci. 1994;3:2194–2206. doi: 10.1002/pro.5560031205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krem M M, Di Cera E. Proteins. 1998;30:34–42. doi: 10.1002/(sici)1097-0134(19980101)30:1<34::aid-prot3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 31.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana B K, Baldwin E, Weber I T, Selk L M, Clawson L, Schneider J, Kent S B. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alber T. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Hata A, Lo R S, Massague J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 35.Li J Y, English M A, Ball H J, Yeyati P L, Waxman S, Licht J D. J Biol Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 36.Brown J L, Sonoda S, Ueda H, Scott M P, Wu C. EMBO J. 1991;10:665–674. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Numoto M, Niwa O, Kaplan J, Wong K K, Merrell K, Kamiya K, Yanagihara K, Calame K. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]