Abstract
ClpA, a member of the Clp/Hsp100 family of ATPases, is a molecular chaperone and, in combination with a proteolytic component ClpP, participates in ATP-dependent proteolysis. We investigated the role of ClpA in protein degradation by ClpAP by dissociating the reaction into several discrete steps. In the assembly step, ClpA–ClpP–substrate complexes assemble either by ClpA–substrate complexes interacting with ClpP or by ClpA–ClpP complexes interacting with substrate; ClpP in the absence of ClpA is unable to bind substrates. Assembly requires ATP binding but not hydrolysis. We discovered that ClpA translocates substrates from their binding sites on ClpA to ClpP. The translocation step specifically requires ATP; nonhydrolyzable ATP analogs are ineffective. Only proteins that are degraded by ClpAP are translocated. Characterization of the degradation step showed that substrates can be degraded in a single round of ClpA–ClpP–substrate binding followed by ATP hydrolysis. The products generated are indistinguishable from steady-state products. Taken together, our results suggest that ClpA, through its interaction with both the substrate and ClpP, acts as a gatekeeper, actively translocating specific substrates into the proteolytic chamber of ClpP where degradation occurs. As multicomponent ATP-dependent proteases are widespread in nature and share structural similarities, these findings may provide a general mechanism for regulation of substrate import into the proteolytic chamber.
Keywords: ATP-dependent proteolysis/molecular chaperones
An exciting recent development in cell biology is the emergence of the Clp/Hsp100 proteins as a new family of ATP-dependent molecular chaperones, found in both eukaryotes and prokaryotes (reviewed in refs. 1–3). Clp proteins have roles in many cellular processes including protein reactivation, protein degradation, DNA replication, regulation of gene expression, thermotolerance, inheritance of prion-like factors, and protein translocation through membranes (4–9). For example, Hsp104, a Clp protein in Saccharomyces cerevisiae, disaggregates proteins aggregated by heat shock, reactivates luciferase and RNA-splicing enzymes following heat inactivation, and alters the conformation of a prion-like protein (6–8). Escherichia coli ClpA activates the latent DNA-binding activity of the plasmid P1 replication initiator protein in vitro by remodeling inactive dimers into active monomers (10, 11). ClpA also prevents irreversible heat inactivation of luciferase in vitro (10). ClpX of E. coli acts as a chaperone in bacteriophage Mu transposition and DNA replication by disassembling stable complexes of MuA tetramers and Mu DNA, allowing the initiation of phage DNA replication (12, 13). ClpX, in vitro, also prevents and reverses heat-induced aggregation of bacteriophage λ O protein and activates DNA binding by the TrfA replication initiator of plasmid RK2 by converting inactive dimers to active monomers (14, 15).
Some of the Clp ATPases, including E. coli ClpA, ClpX and HslU (ClpY), are regulatory components of two-component ATP-dependent proteases (16–21). For instance, either ClpA or ClpX can activate degradation by ClpP, a peptidase sharing no homology with the Clp ATPase family. ClpP itself exists as a stable tetradecamer, composed of two stacked heptameric rings (22, 23). Its structure resembles that of the inner core of β subunits of the 20S proteasome of eukaryotes (24, 25) with the 14 active sites located within a roughly spherical chamber formed by the junction of the rings (26). Thus, it appears that cytoplasmic proteins are protected from indiscriminate degradation by isolation of the proteolytic active sites within the interior of ClpP. Axial pores, only large enough to allow access by small polypeptides and unfolded proteins, are seen at either end of ClpP and are presumed to be the sites where substrates enter. The Clp ATPases, including ClpA, ClpB, ClpX, and HslU of E. coli and Hsp104 of S. cerevisiae, undergo self-assembly in the presence of ATP or a nonhydrolyzable ATP analog, forming hexameric or multimeric rings as visualized by electron microscopy (23, 27–30). Complexes of ClpP and ClpA contain a tetradecamer of ClpP flanked at one or both ends by a hexamer of ClpA (23). Similar structures are seen with ClpXP and HslVU complexes (28–30). Thus, Clp ATPase components are situated in an ideal position to regulate the entry of specific substrates into the aqueous core of ClpP.
In many cases the proteins that are substrates for a Clp ATPase chaperone activity are also degraded by the corresponding Clp protease. For example ClpA, but not ClpX, activates RepA; and ClpAP, but not ClpXP degrades RepA (ref. 10; S.W., unpublished observations). Similarly, ClpX disassembles MuA–DNA complexes and disaggregates λ O; and ClpXP degrades MuA and λ O (13, 14, 18, 19). Although the mechanism of substrate selection by Clp ATPases is not well understood, it has recently been discovered that ClpX recognizes the C-terminal sequence of some substrates (31, 32). It has been proposed that the substrate-binding domains of the Clp ATPases are homologous and may be distantly related to PDZ domains, which also mediate C-terminal specific protein–protein interactions, implicating C-terminal recognition by other Clp ATPases (33).
In this study we explored the role of ClpA in proteolysis by ClpAP in vitro. We found that ClpA–ClpP–substrate degradation complexes assemble by either ClpA–substrate complexes binding ClpP or by ClpA–ClpP complexes binding substrates. We discovered that in an ATP-dependent reaction, ClpA translocates substrates from their binding sites on ClpA to ClpP and that substrates can be degraded to the final small polypeptide products following a single round of substrate-binding to ClpAP.
MATERIALS AND METHODS
Proteins.
P1 RepA (34), ClpA (35), and ClpP [and ClpP(S111A), an inactive mutant (36)] (35) were purified as described. To label proteins in vitro, RepA (150 μg), ClpA (400 μg) and α-casein (200 μg) were separately added to a 2-fold molar excess of succinimidyl [2,3-3H]propionate in 200 μl of 20 mM potassium phosphate, pH 7.5/100 mM KCl/0.5 mM DTT. Mixtures were incubated with gentle mixing for 15 min at 23°C followed by 1 hr at 4°C. Unincorporated label was removed by Sephadex G-25 column chromatography in 20 mM Tris⋅HCl, pH 7.5/100 mM KCl/5 mM DTT/0.1 mM EDTA/0.005% Triton X-100 followed by dialysis against the same buffer without Triton. Both RepA and ClpA retained greater than 90% of their initial specific activity after this procedure as measured in the RepA activation reaction. Solubility in trichloroacetic acid (TCA) was 0.05% for the [3H]RepA and 3% for the [3H]α-casein using the degradation conditions described in the figure legends. In vivo labeling of RepA with [14C]leucine has been previously described (11). ClpP was chemically inactivated by treatment with 100 μM succinyl-Leu-Tyr-chloromethyl ketone (Bachem), which reacts with the catalytic His residue of serine proteases (37). After treatment, excess reagent was removed by gel filtration on Sephadex G25. Throughout, proteins are expressed as moles of RepA dimers, ClpA hexamers, ClpP tetradecamers, and α-casein monomers.
Immunoprecipitation of ClpP and Associated Proteins.
Reaction mixtures containing [3H]RepA or [3H]α-casein, ClpA, and active-site mutant ClpP or chemically inactivated ClpP (in the amounts indicated in the legends) were incubated for 20 min at 23°C in 20 μl of buffer A [20 mM Tris·HCl, pH 7.5/100 mM KCl/5 mM DTT/5 mM magnesium acetate/0.1 mM EDTA/10% glycerol (vol/vol)] containing 0.25 mM adenosine 5′-O-(3-thiotriphosphate) [ATP(γ-S)] and 0.5 mg/ml BSA. Reaction volumes were adjusted to 100 μl with buffer A, and 0.5 mM ATP was added. The samples were incubated at 23°C for 10 min followed by the addition of 100 μl of buffer A containing 2 M NaCl. Samples were incubated an additional 10 min at 23°C and then diluted to 1 ml with 20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/0.05% Tween 20 (buffer B). Rabbit anti-ClpP serum (1:1000) was added, and samples were rotated at 20 rpm for 1 hr at 4°C. Then, 50 μl of a slurry of Protein A Sepharose CL-4B (Pharmacia) in buffer B was added, and the mixtures were rotated for 45 min at 4°C. Protein A Sepharose was collected by centrifugation at 10,000 × g for 20 sec and was washed three times, each time with 1 ml of 20 mM Tris⋅HCl, pH 7.5/500 mM NaCl/0.05% Tween 20/5 mM EDTA and rotated for 10 min at 4°C. Coprecipitated proteins were recovered in 10% SDS (wt/vol) and quantitated by measuring recovered radioactivity. In the absence of ClpA and ClpP, 0.04 pmol of [3H]RepA and 0.02 pmol of [3H]α-casein nonspecifically associated with the immunoprecipitates; the numbers reported account for this nonspecific association.
RESULTS
Assembly of Proteolytic Complexes.
In an effort to dissect the pathway of degradation by ClpAP, we studied the order of assembly of proteolytic complexes. Although there is no evidence to suggest that ClpP recognizes substrates in the absence of ClpA, previous work showed that both ClpA–ClpP complexes (38) and ClpA–RepA complexes could be isolated (10, 11). We wanted to know whether both of these complexes could be intermediates in the degradation pathway.
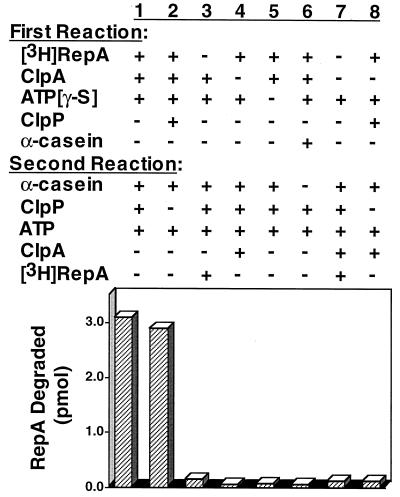
To determine whether ClpA–substrate complexes are able to present substrates to ClpP, we incubated a labeled substrate, RepA, with ClpA and ATP[γ-S] to generate stable ClpA–RepA complexes. This was followed by the addition of a large excess of an unlabeled substrate, α-casein, to compete with free RepA for binding to ClpA. Last, ATP (which exchanges with ATP[γ-S] bound to ClpA with a half-time of less than 1 min [S.W., unpublished results]) and ClpP were added. If the ClpA–RepA complex could interact with ClpP without first releasing RepA, then the bound RepA would be preferentially degraded even in the presence of an excess of competing substrate. We found by using this approach that RepA was degraded (Fig. 1, column 1), and the amount degraded (15%) was similar to that when all three proteins were present in the first reaction (Fig. 1, column 2). In control experiments, when RepA, ClpA, or ATP[γ-S] was omitted from the first reaction and added after α-casein to the second reaction, degradation of RepA was inhibited (Fig. 1, columns 3–5, respectively). Similarly, when casein was added in the first reaction (Fig. 1, column 6) or when ClpA and RepA were both omitted from the first reaction and added in the second (Fig. 1, column 7), degradation of RepA was inhibited. When RepA and ClpP were incubated together in the first reaction and ClpA was added in the second, RepA degradation was inhibited by the competing α-casein, suggesting that ClpP does not bind RepA in the absence of ClpA (Fig. 1, column 8). These results show that ClpA with bound substrate can interact with ClpP, thereby preferentially presenting the bound substrate for degradation by ClpP in the presence of a large excess of a competing substrate.
Figure 1.
α-Casein trap experiment to determine whether ClpP recognizes ClpA–RepA complexes. [3H]RepA–ClpA complexes were formed by incubating 28 pmol of ClpA with 18 pmol of [3H]RepA in 40 μl of buffer A containing 0.5 mM ATP[γ-S] for 10 min at 23°C. Reaction volumes were adjusted with 80 μl of buffer A containing 100 μM α-casein, 9 pmol of ClpP, and 2 mM ATP. After a 5-min incubation at 23°C, TCA was added to 20% concentration, and RepA degradation was quantitated by measuring radioactivity in the acid-soluble fractions. Control experiments with added or omitted components in the two steps are as indicated.
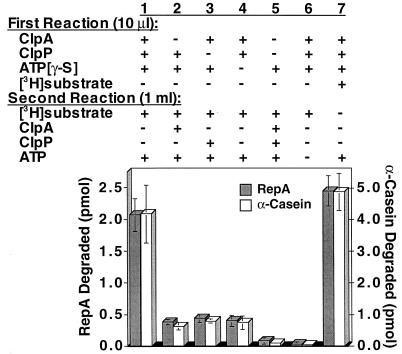
Next we asked whether preassembled ClpA–ClpP complexes are able to directly interact with and degrade substrates. We first incubated ClpA, ClpP, and ATP[γ-S] to generate stable ClpA–ClpP complexes that dissociate very slowly (38) and then diluted the reaction 1:100. Finally ATP and labeled substrate, either RepA or α-casein, were added. Because we used dilution conditions in which ClpA and ClpP assemble poorly, we expected to observe degradation only if preformed ClpA–ClpP complexes bind substrates and not if preformed ClpA–ClpP complexes must dissociate and reassemble after ClpA binds the substrate. We found that the substrates were degraded when ClpA, ClpP, and ATP[γ-S] were present in the first reaction (Fig. 2, column 1); the amount of degradation seen was similar to that observed when the substrate was present in the first reaction (Fig. 2, column 7). In control experiments, when ClpA, ClpP, ATP[γ-S] or both ClpA and ClpP were omitted from the first reaction and added after dilution, degradation was less than 20% of the control, showing that ClpA and ClpP assembled poorly in dilute conditions of the second reaction (Fig. 2, columns 2–5). When ATP was omitted, insignificant degradation was seen (Fig. 2, column 6). Thus, assembled ClpAP is able to recognize substrates. Taken together these results show that the interactions between substrate–ClpA complexes and ClpP and between ClpA–ClpP complexes and substrate need not be ordered in the degradation pathway.
Figure 2.
Dilution experiment to determine whether assembled ClpAP recognizes substrates. ClpAP complexes were formed by incubating 0.8 pmol each of ClpA and ClpP in 10 μl of buffer A containing 1 mM ATP[γ-S] for 10 min at 23°C. Complexes were diluted to 1 ml with buffer A containing 0.005% Triton X-100. Then 90 pmol of [3H]RepA or 70 pmol of [3H]α-casein and 1 mM ATP were added, and the mixtures were incubated for 5 min at 23°C. TCA was added to 20% and radioactivity in the acid-soluble fractions was measured. Control experiments were performed with added or omitted components as indicated. Results are means (±SEM) of three independent experiments.
Knowing that ClpA could form complexes either with substrate or with ClpP, we looked directly for complexes of all three proteins. We incubated [3H]ClpA, ClpP, and [14C]RepA with ATP[γ-S], and then isolated complexes by ultrafiltration. We found that RepA was retained on the filters only when ClpA, ClpP, and ATP[γ-S] were present in the incubation mixture (Table 1). Free ClpA hexamers, RepA dimers, ClpP tetradecamers, and RepA–ClpA complexes were not retained. ClpA–ClpP complexes were retained on the filters in an ATP[γ-S]-dependent manner and could be detected when labeled ClpA was used and RepA was omitted (Table 1). Both RepA–ClpA–ClpP and ClpA–ClpP complexes were dissociated by treatment with 1 M NaCl (Table 1).
Table 1.
Isolation of RepA–ClpA–ClpP and ClpA–ClpP complexes
| Experiment | Additions | Amount retained, pmol
|
|
|---|---|---|---|
| RepA | ClpA | ||
| 1 | RepA, ClpA, ClpP, ATP[γ-S] | 6.3 ± 0.2 | 8.3 ± 0.3 |
| 2 | RepA, ClpA, ClpP | <0.5 | <0.5 |
| 3 | RepA, ClpA, ATP[γ-S] | <0.5 | <0.5 |
| 4 | RepA, ClpP, ATP[γ-S] | <0.5 | — |
| 5 | RepA, ClpA, ClpP, ATP[γ-S] followed by treatment with 1 M NaCl | <0.5 | <0.5 |
| 6 | ClpA, ClpP, ATP[γ-S] | — | 6.0 ± 0.2 |
| 7 | ClpA, ClpP | — | <0.5 |
| 8 | ClpA, ATP[γ-S] | — | <0.5 |
| 9 | ClpA, ClpP, ATP[γ-S], followed by treatment with 1 M NaCl | — | <0.5 |
Complexes were formed by incubating 28 pmol of [3H]ClpA, 9 pmol ClpP, and 18 pmol of [14C]RepA in 20 μl of buffer A containing 1 mM ATP[γ-S] for 20 min at 23°C. Reaction volumes were adjusted to 100 μl with buffer A lacking Mg2+ and containing 0.1 mg/ml BSA and 0.005% Triton X-100. Samples were centrifuged at 2040 × g for 6 min through Ultrafree-MC 300,000 NMWL filters (Millipore). The amount of [14C]RepA and [3H]ClpA was quantitated by measuring radioactivity in the retentates and filtrates. In the presence of ATP[γ-S], 0.9 pmol of ClpA was nonspecifically bound and has been subtracted from all ClpA values. Similarly, 1.5 pmol of RepA was nonspecifically bound and has been subtracted from all RepA values. ClpP alone was not retained as demonstrated by the recovery of 95% of ClpP in the filtrate as detected by Bio-Rad Protein assay. Results are means (±SEM) of three to seven independent experiments.
Translocation of Substrates from ClpA to ClpP.
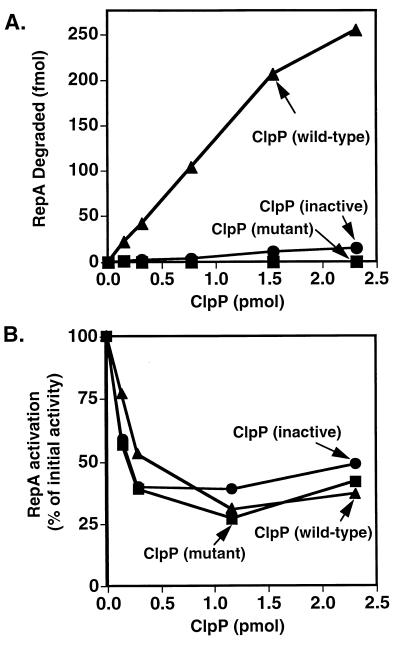
The translocation of substrates from their binding sites on ClpA into the chamber of ClpP is one of the possible energy-dependent roles of ClpA in degradation. Our first indication that ClpA may, in fact, have a translocation role came from experiments with two inactive forms of ClpP. One inactive form had a mutation that changed the active-site serine to alanine, and the other inactive form was made by treating wild-type ClpP with succinyl-Leu-Tyr-chloromethyl ketone, a reagent that inactivates ClpP by reacting with His-136 in the active site (M.R.M. and H. Y. Yong, unpublished data). Surprisingly, these proteolysis-deficient forms of ClpP (Fig. 3A) inhibited the ClpA-dependent activation of RepA (Fig. 3B). The wild-type protein caused a decrease in RepA activation as expected because RepA was degraded. The inhibition by the inactive ClpP proteins was never more than 75%, for unknown reasons. This inhibition may be expected if RepA is actively translocated by ClpA to ClpP and sequestered by the inactive ClpP.
Figure 3.
Inhibition of ClpA-dependent RepA activation by proteolysis-deficient forms of ClpP. (A) [3H]RepA (0.9 pmol) and ClpA (0.8 pmol) were incubated in 20 μl of buffer A containing 0.5 mM ATP[γ-S] and increasing amounts of wild-type ▴, mutant ■, or chemically inactivated • ClpP. After a 15-min incubation at 23°C, the mixtures were diluted 1:2 with buffer A containing 2 mM ATP and 5 μM α-casein. After a 5-min incubation at 23°C, TCA was added, and protein degradation was determined by measuring radioactivity in the acid-soluble fractions. (B) RepA (0.9 pmol) and ClpA (0.8 pmol) were incubated in 10 μl of buffer A containing 1 mM ATP[γ-S] with increasing amounts of mutant ClpP ■, chemically inactive ClpP •, or wild-type ClpP ▴ for 15 min at 23°C. Two microliters was then diluted 1:100 into buffer A containing 0.005% Triton X-100, 1 mM ATP, and 4 μM α-casein, and incubated for 5 min at 23°C. RepA activation was quantitated by measuring radioactivity retained on nitrocellulose filters following a 5-min incubation at 23°C with 10 fmol of [3H]oriP1 plasmid DNA and 0.5 μg of calf thymus DNA (11). Values correspond to the percent of activation relative to the control without ClpP.
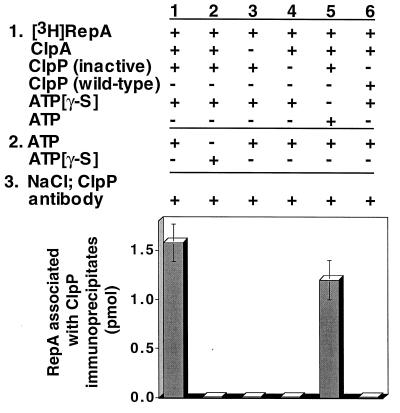
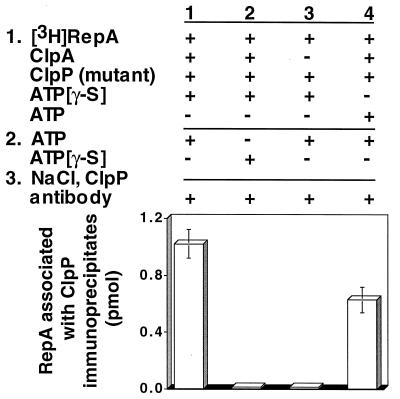
To search directly for translocation of substrates to ClpP, we designed the following experiment. First, ClpA was incubated with radiolabeled substrate, ATP[γ-S], and chemically inactivated ClpP to allow the assembly of substrate–ClpA–ClpP complexes. Next, ATP was added in excess of ATP[γ-S], in expectation that the translocation of substrate may be the step that specifically required ATP. After a short incubation, 1 M NaCl was added to dissociate ClpA and ClpP. Lastly, ClpP was immunoprecipitated, and associated radiolabeled substrate was measured. We discovered that the substrate, RepA, coprecipitated with the inactivated ClpP (Fig. 4, column 1). After correcting for the recovery of ClpP, the immunoprecipitates contained about 0.6 mol of RepA2 per mol of ClpP14. When ATP was omitted, RepA was not detectable in the immunoprecipitates, demonstrating that translocation required ATP (Fig. 4, column 2). Similarly, ClpA was required for the transfer reaction (Fig. 4, column 3). The control in which ClpP was omitted showed that ClpP was necessary for the immunoprecipitation of RepA (Fig. 4, column 4). When ATP was present during the first incubation as well as the second, RepA was found with immunoprecipitated ClpP, although the amount was slightly less than when ATP[γ-S] was present in the assembly step (Fig. 4, column 5). As expected, when wild-type ClpP was used, RepA was not associated with ClpP immunoprecipitates because RepA was degraded and the degradation products were released from ClpP (Fig. 4, column 6). When the active-site ClpP mutant protein was used, the results were similar to those obtained with the chemically inactivated ClpP (Fig. 5).
Figure 4.
Translocation of RepA from ClpA to inactive ClpP. [3H]RepA (34 pmol), ClpA (28 pmol), and chemically inactivated ClpP (11 pmol) were incubated first with ATP[γ-S] and then with ATP. Samples were then treated with 1 M NaCl and ClpP antibody. Coimmunoprecipitated RepA was quantitated by measuring the amount of radioactivity in the recovered immunoprecipitates. Control experiments were performed as indicated. Results are means (±SEM) of two to five independent experiments.
Figure 5.
Translocation of RepA from ClpA to mutant ClpP. Reactions were identical to those described in Fig. 4 except that the inactive mutant form of ClpP was used. Control experiments are as indicated. Results are means (±SEM) of two to five independent experiments.
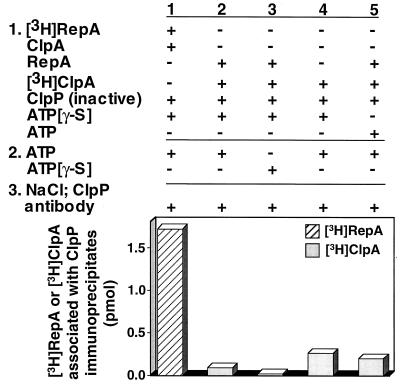
It was important to show that treatment with 1 M NaCl removed most of the ClpA from the immunoprecipitates. The possibility existed that ClpA was associated with ClpP, and RepA was immunoprecipitated because it was associated with ClpA. To quantitate the amount of ClpA in the immunoprecipitates, we repeated the set of experiments using radiolabeled ClpA and unlabeled RepA (Fig. 6). We found that about 1/20 as much ClpA as RepA coimmunoprecipitated with ClpP, not enough to explain the association of the substrate with ClpP by an indirect association with ClpA. Similarly low amounts of ClpA coimmunoprecipitated with ClpP when ATP was omitted. A significant but small amount of ClpA associated with the ClpP immunoprecipitates when RepA was omitted or when ATP was included throughout the experiment. Very likely this is because ClpA is itself a substrate for degradation by ClpAP (10).
Figure 6.
Translocation of RepA from [3H]ClpA to inactivated ClpP. Experimental conditions were identical to those described in Fig. 4. In experiment 1 [3H]RepA and unlabeled ClpA were used, whereas in experiments 2–5 [3H]ClpA and unlabeled RepA were used. Control experiments with added or omitted components are as indicated. In the absence of added ClpP and RepA, 0.02 pmol of [3H]ClpA associated with the immunoprecipitates and has been subtracted.
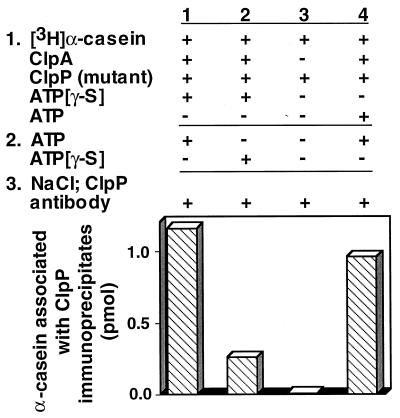
We next showed that ClpA specifically translocated ClpAP substrates. We used the protocol described above but substituted [3H]α-casein for [3H]RepA. We found that the labeled α-casein coimmunoprecipitated with ClpP (Fig. 7). There was no detectable association of α-casein with the ClpP immunoprecipitates when ClpA was omitted. When ATP was omitted, 1/5 the amount of α-casein coimmunoprecipitated, consistent with the observation that a low level of α-casein is degraded in the absence of ATP (Fig. 8, column 7). We next performed competition experiments in which we added other proteins, either substrates or nonsubstrates, to the first reaction with [3H]RepA, ClpA, ATP[γ-S], and inactive ClpP (Table 2). As expected, excess unlabeled RepA or α-casein inhibited the coimmunoprecipitation of labeled RepA with ClpP. When the competing substrate was added after the ATP promoted step, transfer of [3H]RepA was no longer inhibited. Two other proteins that are not degraded by ClpAP, ovalbumin and lysozyme, did not significantly affect the amount of labeled RepA associated with ClpP immunoprecipitates. Thus, these experiments show that specific ClpAP substrates are translocated to ClpP in a reaction requiring ClpA and ATP. It is not known whether the reaction is driven by ATP binding or by ATP hydrolysis.
Figure 7.
Translocation of α-casein from ClpA to mutant ClpP. Experiments were carried out as described in Fig. 4 except that inactive mutant ClpP was substituted for chemically inactivated ClpP and 50 pmol of [3H]α-casein was substituted for [3H]RepA.
Figure 8.
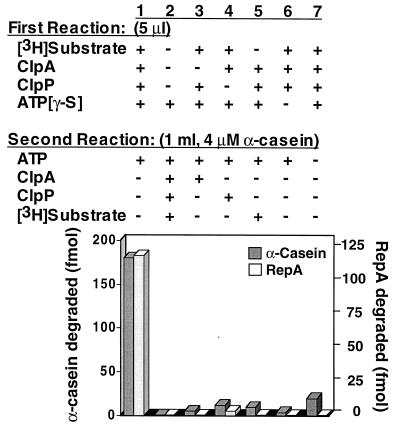
Dilution experiment in the presence of an α-casein trap to determine whether ClpAP can degrade substrates in a single cycle of substrate binding and product release. Substrate–ClpA–ClpP complexes were formed by incubating 0.8 pmol each of ClpA and ClpP with 0.9 pmol of [3H]RepA or [3H]α-casein in 5 μl of buffer A containing 1 mM ATP[γ-S] for 10 min at 23°C. Complexes were diluted to 1 ml with buffer A containing 4 μM α-casein, 0.005% Triton X-100, and 1 mM ATP, and incubated an additional 10 min at 23°C. Reaction products were precipitated with 20% TCA, and radioactivity in the acid-soluble fractions was measured. Control experiments are as shown.
Table 2.
Specificity of ATP-dependent substrate translocation from ClpA to chemically inactivated ClpP
| Experiment | Additions to first reaction | RepA associated with ClpP immunoprecipitates, pmol |
|---|---|---|
| 1 | Complete: [3H]RepA, ClpA, ClpP (inactivated), ATP[γ-S] | 1.09 ± 0.11 |
| 2 | Complete plus 20-fold molar excess of RepA to [3H]RepA | 0.05 ± 0.03 |
| 3 | Complete plus 20-fold molar excess of casein to [3H]RepA | 0.17 ± 0.01 |
| 4 | Complete plus 20-fold molar excess of lysozyme to [3H]RepA | 1.09 ± 0.24 |
| 5 | Complete plus 20-fold molar excess of ovalbumin to [3H]RepA | 0.87 ± 0.01 |
| 6 | Complete, with a 20-fold molar excess of RepA to [3H]RepA added after incubation with ATP | 1.14 ± 0.22 |
Translocation experiments were identical to those described in Fig. 4, but for experiments 2–5 a 20-fold molar excess of unlabeled RepA, α-casein, lysozyme, or ovalbumin was added during the first incubation step as indicated. In experiment 6, a 20-fold molar excess of unlabeled RepA was added after the ATP incubation step. Results are means (±SEM) of two independent experiments.
After the ClpA–ClpP–RepA/α-Casein Complex Is Formed the Substrate Is Degraded to Completion, and This Occurs Without the Release and Rebinding of Substrate by ClpA.
Because ClpAP makes many proteolytic cleavages in its substrates, producing acid-soluble small polypeptides (35, 39), we wanted to know whether complete degradation requires multiple rounds of substrate binding and release. To address this question we incubated a labeled substrate, α-casein, in one experiment and RepA in another, with ClpA, ClpP, and ATP[γ-S] to generate stable complexes. Reaction mixtures were then diluted to prevent further assembly of ClpAP, and a large excess of unlabeled α-casein was added to inhibit rebinding of ClpAP to the labeled substrate. We then added ATP, incubated the mixture for a short time, and analyzed the labeled products. With these conditions, ClpAP degraded a significant amount of the labeled substrate (13–20%) to acid-soluble products (Fig. 8, column 1). Control experiments showed that there was undetectable degradation when the entire reaction was carried out in the presence of excess unlabeled α-casein in the dilute conditions of the second reaction (Fig. 8, column 2). Similarly, when ClpA, ClpP, or labeled substrate was omitted from the first reaction and added to the second, there was very little degradation of the labeled substrate (Fig. 8, columns 3–5). There was only a small amount of substrate degraded when ATP[γ-S] was omitted from the first reaction or when ATP was omitted from the second reaction (Fig. 8, columns 6 and 7). When reaction mixtures from the experiment in column 1 with RepA were analyzed by reverse-phase column chromatography after the second incubation, the labeled products generated were indistinguishable from those generated in a steady-state experiment. Partial degradation products were not detected. This set of experiments shows that ClpAP degrades both RepA and α-casein to small polypeptide products in a single cycle of substrate binding to ClpAP and product release.
DISCUSSION
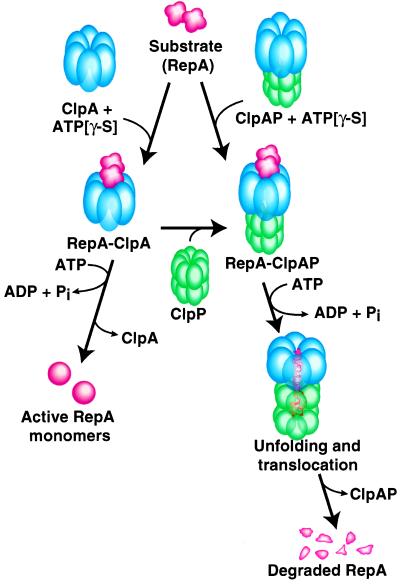
Fig. 9 summarizes our working model of the pathway of degradation by ClpAP. The ClpA hexamers can interact with the substrate, forming stable substrate–ClpA complexes in a reaction that requires ATP binding. For RepA–ClpA complexes about one RepA dimer is bound per ClpA hexamer (11). The substrate–ClpA complexes are able to interact with ClpP, generating stable substrate–ClpAP complexes. Alternatively, assembled ClpAP complexes (23) can interact with the substrate. Following assembly of the degradation complex, the substrate is translocated from its ClpA recognition/binding site to ClpP. Once a substrate is in the ClpP chamber, peptide-bond cleavage occurs, and presumably the peptide products passively diffuse out. For RepA and α-casein, ATP-dependent degradation of the substrate to small polypeptides is accomplished in one cycle of binding to ClpAP. It will be interesting to know whether substrates larger than RepA (32 kDa) and α-casein (24 kDa) will require multiple rounds of binding for complete degradation.
Figure 9.
Working model of the pathway of degradation by ClpAP. See text for discussion.
Fig. 9 also summarizes the alternate fate of substrates in the absence of ClpP: protein remodeling by ClpA. For RepA, inactive dimers are remodeled into active monomers. The observation that RepA monomerized by ClpA or by DnaJ/DnaK is not degraded by ClpP without further participation of ClpA and ATP (ref. 10; S.W. and J.H., unpublished observation) suggests that protein remodeling by ClpA is not sufficient to convert the substrate into a form that passively diffuses into the ClpP chamber. Thus, our current model is that the chaperone activity of ClpA unfolds the substrate and, in the presence of ClpP, translocates the substrate to ClpP.
In summary, we have presented direct evidence that ClpA actively translocates specific substrates to ClpP. Our results suggest that ClpA regulates degradation by translocating those proteins it specifically recognizes into the ClpP chamber through an otherwise inaccessible channel. In eukaryotes, a likely function of one or more of the ATPase components of the proteasome is to regulate substrate entry into the proteolytic chamber. One significant difference is that in eukaryotes the specificity of degradation is determined by the ubiquitin conjugation machinery and not by the ATPases associated with the proteasome.
Acknowledgments
We thank Susan Gottesman and Keith McKenney for their many helpful discussions.
ABBREVIATIONS
- ATP[γ-S]
adenosine 5′-O-(3-thiotriphosphate)
- TCA
trichloroacetic acid
References
- 1. Gottesman S, Wickner S, Maurizi M. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 2.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 3.Suzuki C K, Rep M, Maarten van Dijl J, Suda K, Grivell L A, Schatz G. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 4.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1376. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 5.Mhammedi-Alaoui A, Pato M, Gama M J, Toussaint A. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 6.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 7.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 8.Vogel J L, Parsell D A, Lindquist S. Curr Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 9.Schweder T, Lee K H, Lomovskaya O, Matin A. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruklitis R, Welty D J, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 13.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 14.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konieczny I, Helinski D R. Proc Natl Acad Sci USA. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark W P, Maurizi M R. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 17.Hwang B J, Woo K M, Goldberg A L, Chung C H. J Biol Chem. 1988;263:8727–8734. [PubMed] [Google Scholar]
- 18.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 19.Wojtkowiak D, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 20.Rohrwild M, Coux O, Huang H-C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missiakas D, Schwager F, Betton J M, Georgopoulos C, Raina S. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 22.Flanagan J M, Wall J S, Capel M S, Schneider D K, Shanklin J. Biochemistry. 1995;34:10910–10917. doi: 10.1021/bi00034a025. [DOI] [PubMed] [Google Scholar]
- 23.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 24.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 25.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 27.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 28.Kessel M, Wu W, Gottesman S, Kocsis E, Steven A C, Maurizi M R. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 29.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 30.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 31.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Tousaint A. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 32.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 33.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 34.Wickner S H. Proc Natl Acad Sci USA. 1990;87:2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurizi M R, Thompson M W, Singh S K, Kim S H. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 36.Maurizi M R, Clark W P, Kim S-H, Gottesman S. J Biol Chem. 1990;265:12546–12552. [PubMed] [Google Scholar]
- 37.Stevenson K J, Smillie L B. J Mol Biol. 1965;12:937–941. doi: 10.1016/s0022-2836(65)80342-4. [DOI] [PubMed] [Google Scholar]
- 38.Maurizi M R. Biochem Soc Trans. 1991;19:719–723. doi: 10.1042/bst0190719. [DOI] [PubMed] [Google Scholar]
- 39.Thompson M W, Singh S K, Maurizi M R. J Biol Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]