Abstract
Study Objectives:
In order to characterize the genetic mechanisms underlying sleep, we have carried out a large-scale screen in Drosophila to identify short-sleeping mutants. The objectives of this study were as follows: (1) characterize the phenotypes of the shortest-sleeping mutants; (2) examine whether changes in arousal threshold or sleep homeostasis underlie short-sleeping phenotypes; (3) clone a gene affected in one of the shortest sleepers; and (4) investigate whether circadian mutants can be identified using light:dark (L:D) locomotor data obtained for studying sleep behavior.
Design:
Locomotor activity was measured using the Drosophila Activity Monitoring System in a 12:12 L:D cycle.
Setting:
Drosophila research laboratory.
Participants:
Adult flies from the 2nd chromosome Zuker collection, which contain mutations in most of the nonessential genes on the Drosophila 2nd chromosome.
Measurements and Results:
Our analysis of sleep characteristics suggests that daily activity (but not waking activity) correlates with daily sleep time and that defects in sleep maintenance are more common than defects in sleep initiation. Our shortest sleepers have intact or increased sleep rebound following sleep deprivation but show reduced thresholds for arousal. Molecular analysis of one of the short-sleeping lines indicates that it is a novel allele of a dopamine transporter (DAT). Finally, we describe a novel approach for identifying circadian mutants using L:D data.
Conclusions:
Our data suggest that most short-sleeping mutant phenotypes in Drosophila are characterized by an inability to stay asleep, most likely because of a reduced arousal threshold.
Citation:
Wu MN; Koh K; Yue Z; Joiner WJ; Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. SLEEP 2008;31(4):465-472.
Keywords: Drosophila, sleep, circadian, clock, screen, fumin, DAT, Zuker collection
THROUGHOUT THE ANIMAL KINGDOM, ORGANISMS SLEEP FOR HOURS EVERY DAY, EVEN THOUGH DOING SO LEAVES THEM DEFENSELESS AND UNABLE TO feed or mate.1 Sleep clearly performs an important physiological function, but the nature of this function remains unknown. Nonetheless, it is suspected that sleep is necessary for proper functioning of the brain, including learning and memory, synaptic or cellular plasticity, and energy metabolism in the brain.2–5 Our current understanding of sleep and its regulation derives mainly from work in mammalian systems. Electrophysiological and lesion studies have defined anatomical regions and neurotransmitters important for sleep/wake transitions6. Behavioral and electroencephalogram (EEG) studies have suggested that sleep is controlled by circadian and homeostatic processes.7 In addition, sleep regulation is influenced by the level of arousal of an animal, which may be affected by multiple factors, including circadian time, sensory processing, and emotional state.8,9 However, we have a limited understanding of the molecular and genetic pathways underlying the regulation and execution of sleep,10,11 which hinders our ability to gain further insights into the functions of sleep.
Forward genetic analysis can be a powerful way to dissect the molecular pathways underlying a biological process such as sleep. For example, in Drosophila this approach was successful in identifying the core components of a related process—the circadian clock.12–15 It has been shown that Drosophila engage in sleep-like behavior. They have consolidated periods of quiescence at night during which they are less responsive to external stimuli, and, if they are deprived of these periods at night, they will exhibit increased sleep behavior the next morning, a phenomenon known as “sleep rebound.”16–19 Fly sleep is modulated by drugs that affect sleep in humans.17,20,21 In addition, as in humans, sleep in fruit flies changes with age; young flies sleep more and older flies have more fragmented sleep.17,22 Electrophysiological studies show changes during fly sleep, including reduced local field potentials near mushroom bodies, a structure required for learning and memory in the adult Drosophila brain.23 Interestingly, recent data have demonstrated an important role for the mushroom bodies in regulating sleep.24,25
A forward genetic screen for sleep mutants on the X chromosome in Drosophila was recently carried out by Cirelli and colleagues.26,27 This screen resulted in the identification of mini-sleep, a short-sleeping mutant, whose phenotype results from a mutation in the Shaker potassium channel. In addition, it was recently shown that mutations in Hyperkinetic cause a short-sleeping phenotype in Drosophila.28 Hyperkinetic encodes a β subunit that binds Shaker α subunits and regulates their activity. Another short-sleeping fly mutant, fumin, identified serendipitously, carries a mutation in a dopamine transporter (DAT) on the 2nd chromosome.29 Thus, the Drosophila system holds significant promise for the identification of sleep mutants.
In order to perform a forward genetic screen for sleep mutants, we screened the Zuker collection to isolate short-sleeping mutants. The Zuker collection is composed of more than 12,000 mutant lines carrying EMS (ethylmethane sulfonate)-induced mutations.30 This collection has been well characterized and has been used successfully in other screens for adult viable phenotypes.30,31 Although cloning genes affected in EMS mutants is more laborious than from transposon-tagged mutants, using EMS allows for unbiased mutagenesis of the entire genome and the generation of qualitatively distinct alleles. The latter point may be important since sleep is essential32,33 and therefore extreme reduction of sleep may result in lethality. By screening the Zuker collection, we were able to identify several short-sleeping mutants and clone the gene corresponding to one of these mutations. Our analysis of the mutant lines suggests that changes in arousal threshold commonly underlie the phenotypes of short-sleeping mutants.
METHODS
Fly Stocks
Fly stocks were raised at 23°C on standard cornmeal-molasses medium. Zuker collection lines were obtained from Andrea Lougheed, Robert Hardy, and Charles Zuker (Howard Hughes Medical Institute, University of California, San Diego).30 The Zuker collection mutant lines were generated in a cn bw background. The fumin mutants were obtained from K. Kume (Kumamoto University, Japan).29 For recombination mapping experiments with Z2-1744, stocks # 156 al b pr c px sp flies and #214 al b pr c px sp/CyO from the Bloomington Stock Center were used (Bloomington, Indiana).
Behavioral Assays
Drosophila Activity Monitoring System devices (DAMS) from Trikinetics (Waltham, MA) were used to collect locomotor activity and sleep data in 1 minute bins. After loading into 5 × 65 mm glass tubes using CO2 anesthesia, flies were allowed to recover and adapt for approximately 1.5 days. Baseline sleep was then recorded for 2 days, and data for the 2 days were averaged. Flies were 5-10 days old at the start of testing and 5% sucrose was used as their food source during testing. For measurements of sleep, flies were subjected to a 12:12 L:D cycle in a well-humidified incubator (Thermo Scientific) at 25°C. As described previously, sleep was measured using the definition of locomotor inactivity lasting a minimum of 5 minutes17 and analyzed using custom-designed MATLAB (Mathworks, Natick, MA) based software.34 For the primary screen, 4 male flies were tested for each mutant line, and short-sleeping lines were kept if their sleep was reduced ≥2.5 SDs from the population mean. During retesting of 8-16 males and female flies, males were considered short-sleeping if their sleep was ≥2.5 SDs less than the control genetic background cn bw, and females were considered short-sleeping if their sleep was ≥2 SDs less than cn bw. For statistical analyses, either Microsoft Excel or Statistica (StatSoft Inc, Tulsa, OK) was used. Sleep deprivation was performed with the flies in the DAMS monitors, by using a custom-built mechanical deprivator housed in an incubator. The deprivator was set to give 2-sec pulses at random intervals averaging 20 seconds with a computer controlled power source (Trikinetics). Deprivation was performed from ZT18-24, and sleep lost during this period was calculated relative to unstimulated controls. Sleep rebound was calculated by measuring the difference in the amount of sleep from ZT0-6 for deprived animals relative to control non-deprived animals. To correct for differences in the absolute amount of sleep lost, sleep rebound was also expressed as a percent of the amount of sleep lost during deprivation. To measure arousal threshold, a qualitative assay was performed, using increasing stimulus intensities at ZT16 (mild stimulus), ZT18 (moderate stimulus), and ZT20 (severe stimulus). In the dark, tubes in DAMS monitors were gently scraped once with a thin wooden stick (mild stimulus), scraped strongly once (moderate stimulus), or scraped strongly 6 times (severe stimulus). Data were averaged from 3 separate experiments. Flies inactive for 5 minutes prior to the stimulus were considered asleep, and flies displaying at least 1 count of activity within the 2 minutes after the stimulus were considered aroused. To control for the increased chance of spontaneous awakenings in short-sleeping mutants, the percentage of aroused flies was corrected using the following formula: (% aroused−% spontaneously awake)/(100%−% spontaneously awake). The percentage of spontaneous awakenings was counted the day prior to arousal experiments at corresponding circadian times.
Using activity records from the primary screen, determination of the presence of morning and evening anticipation in a 12:12 L:D cycle was determined by visual inspection. Lines that had poor anticipation were retained for circadian analysis. For these subsequent studies, locomotor rhythms were studied in constant darkness (D:D), and data analyzed using Clocklab (Actimetrics Software, Wilmette, IL).
Molecular Biology
Genomic DNA was prepared by homogenizing a single fly in SB buffer (10 mM Tris pH 8.2, 1 mM EDTA, pH 8.0, and 25 mM NaCl) with Proteinase K (200 μg/mL), incubated at 37°C for 30 minutes and then at 80°C for 10 minutes. PCR was performed using HotStarTaq kit (Qiagen). Primers were designed using Omiga (Oxford Molecular) software. Sequencing was performed by the University of Pennsylvania DNA Sequencing Facility.
RESULTS
Screening the Zuker Collection for Short-Sleeping Mutants
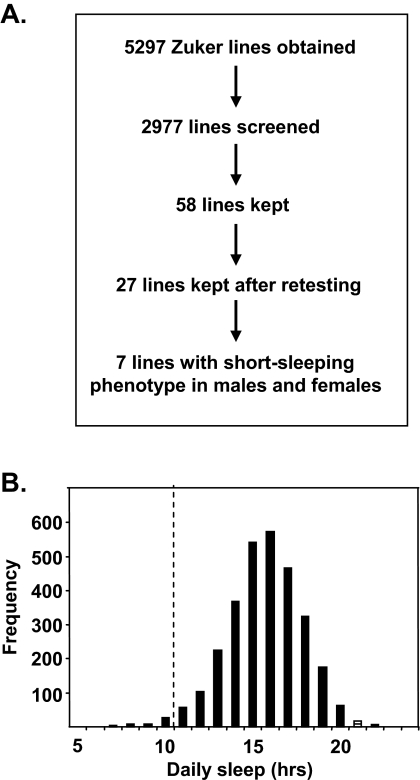
In order to identify novel short-sleeping mutants in Drosophila, we chose to screen the Zuker collection.30 The overall screen schematic is shown in Figure 1A. We obtained 5,297 mutant lines affecting genes on the 2nd chromosome from Zuker and colleagues. Of these, we were able to collect 4 appropriately aged males from each of 2,977 lines; the remaining lines were apparently semi-lethal. In the primary screen, we retained lines whose baseline sleep in a 12:12 L:D cycle was 2.5 SDs lower than the population mean. Fifty-eight of the 2,977 lines (1.9%) met this criterion (Figure 1B). These lines were then retested with 8-16 males and 8-16 females each, and after retesting, 27 out of the 58 lines retained the short-sleeping phenotype for males. Interestingly, 20 out of the 27 lines failed to meet our criteria for short sleep in both males and females (see Methods and Supplemental Figure 1). This sexual dimorphism of the sleep phenotype in Drosophila was also noted by Cirelli and colleagues in their screen for short-sleeping mutants.27 Histograms of nighttime sleep duration, waking activity, bout number, and nighttime bout duration show that the distribution of nighttime bout duration in particular shows a large right-sided skew (Supplemental Figure 2). Nighttime bout duration was analyzed because daytime sleep bouts are typically shorter and may reflect brief naps rather than consolidated sleep.19,35 The marked asymmetry of the distribution of nighttime bout duration emphasizes the importance of applying nonparametric statistical methods to studying nighttime bout duration.
Figure 1.
Overall screen schematic. A. Flow-chart demonstrating the number of 2nd chromosome mutants selected at each stage of screening for short-sleeping mutants. B. Histogram showing daily sleep for all mutant lines tested (group means). The dashed line represents the 2.5 SD cutoff below which mutants were selected for retesting.
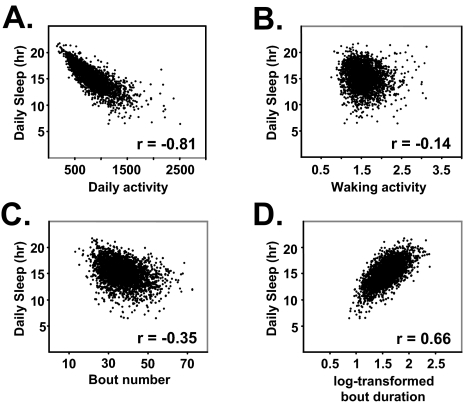
The screening of thousands of mutants provides the opportunity to study the relationship among different sleep characteristics. We examined how daily sleep in Drosophila covaries with two different measures of locomotor activity—total daily activity and waking activity (the amount of activity per waking minute) (Figures 2A and 2B). The amount of daily sleep is strongly inversely correlated with total daily activity (r = −0.81), suggesting that flies that walk more tend to sleep less. In contrast, daily sleep time is poorly correlated with waking activity (r = −0.14). Waking activity has been proposed to be a measure of hyperactivity in flies; that is, flies that have high waking activities are considered hyperactive.35 The lack of correlation between waking activity and daily sleep implies that hyperactivity can be seen in both flies that are short sleepers and flies that are long sleepers. Further, these data suggest that waking activity, but not total daily activity, is a useful independent measure of hyperactivity for sleep mutants. Next, we asked whether daily sleep correlates with bout number or nighttime sleep bout duration. As shown in Figures 2C and 2D, the amount of daily sleep was strongly correlated with bout duration and weakly inversely correlated with bout number. While nighttime sleep and daytime sleep were similarly correlated with daily activity and waking activity (Supplemental Figure 3), bout number was better inversely correlated with nighttime sleep than daytime sleep. Overall, these results suggest that short-sleeping flies tend to have an increased number of short sleep bouts at night.
Figure 2.
Correlations between sleep time and measures of activity and sleep bouts. A. Correlation between daily sleep and daily locomotor activity for all lines. B. Correlation between daily sleep and waking activity (activity/time awake). C. Correlation between daily sleep and bout number. D. Correlation between daily sleep and log-transformed nighttime bout duration in h. The logarithm of the bout duration is used here because of the large right sided skew for bout duration. Pearson correlation coefficient (r) is shown for all panels, and P < 0.0001 for A–D.
Phenotypic Characterization of the Short-Sleeping Mutants
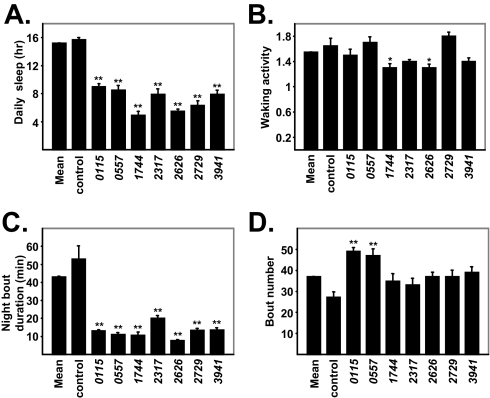
After screening and retesting, we identified 7 lines in the 2nd chromosome Zuker collection as short-sleeping mutant lines (Z2-0115, Z2-0557, Z2-1744, Z2-2317, Z2-2626, Z2-2729, and Z2-3941). The daily sleep of these lines is depicted in Figure 3A, which shows that each of these 7 lines has a significant reduction in daily sleep compared to the population mean and background controls, with the most severe reductions occurring in Z2-1744, Z2-2626, and Z2-2729. Both nighttime and daytime sleep are significantly reduced in these 7 mutants, compared to controls (Supplemental Figures 4 and 5). We next examined waking activity in these 7 mutants and found that Z2-1744 and Z2-2626 have mildly reduced waking activities compared to the population mean and background controls. The other short-sleepers have waking activities that are similar to the screen mean or background controls (Figure 3B). The 24-h profiles of locomotor activity for the short-sleeping mutants are shown in Supplemental Figure 6.
Figure 3.
Sleep characteristics of the short-sleeping mutants. A. Daily sleep is shown for the screen mean, background control cn bw (n=27), Z2-0115 (n=44), Z2-0557 (n=15), Z2-1744 (n=24), Z2-2317 (n=25), Z2-2626 (n=24), Z2-2729 (n=12), and Z2-3941 (n=23). B. Waking activity (activity/minutes awake) of the short-sleeping mutant lines. C. Sleep bout duration at night for the short-sleeping mutant lines. D. Sleep bout number for the short-sleeping mutant lines. * denotes P < 0.05 and ** denotes P < 0.01 using Mann-Whitney U test and is shown only if significant compared to both population mean and background controls. Data for male flies is shown. Error bars represent SEM.
In order to characterize the sleep architecture in these short-sleepers, we analyzed nighttime sleep bout duration and bout number. Figure 3C shows that all of the mutants have defects in sleep maintenance, as evidenced by a significant reduction in sleep bout duration during the night, compared to controls. In terms of bout number, Z2-0115 and Z2-0557 mutants appear to exhibit a compensatory increase in the number of daily sleep bouts, compared to both the population mean and background controls (Figure 3D). However, none of the mutant lines have a significant reduction in the number of sleep bouts, compared to controls, suggesting that these short-sleeping mutants do not have defects in sleep initiation. Overall, the sleep architecture of the short-sleeping mutants suggests that their phenotypes are driven in large part by their inability to stay asleep during the night.
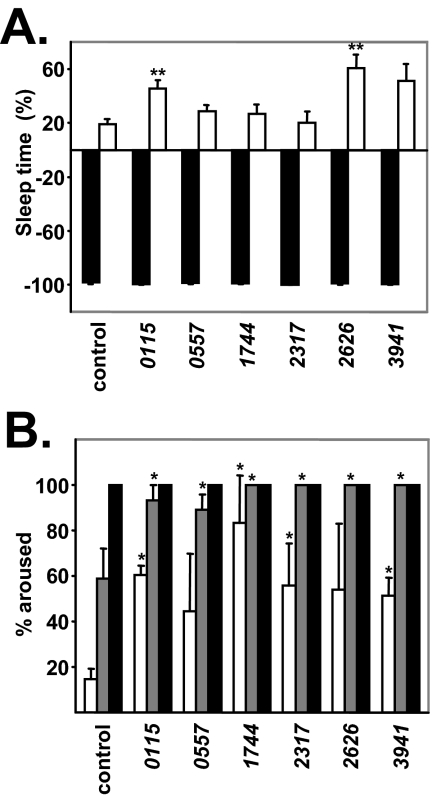
We next asked whether changes in homeostatic drive or arousal threshold underlie the sleep maintenance phenotypes observed in our short-sleepers. During the course of our analysis, one of the lines, Z2-2729, appeared to accumulate a semi-lethal modifier and was therefore not included in further assays. This phenomenon has been described for a number of mutant lines from the Zuker collection.30 In order to study homeostatic drive following sleep deprivation, we performed mechanical deprivation of these lines for 6 h from ZT18-24 and measured sleep rebound for 6 h following lights on (from ZT0-6). Figure 4A shows the percentage of sleep lost during deprivation compared to unstimulated controls and the percentage of sleep gained during recovery relative to amount of sleep lost. In addition, the total amount of sleep lost during deprivation or gained during recovery (in minutes) is shown in Supplemental Figure 7. All of the short-sleeping mutant lines demonstrate sleep rebound after mechanical deprivation, similar to or greater than controls. Two lines (Z2-0115 and Z2-2626) exhibit significantly increased rebound, suggesting that these lines are more sensitive to sleep loss.
Figure 4.
Homeostatic measurements and arousal thresholds of the short-sleeping mutants. A. Sleep rebound following mechanical deprivation. Sleep time lost during deprivation (black bars) is expressed as a percentage relative to amount of sleep time for unshaken controls during ZT18-24. The white bars represent sleep time gained during recovery compared to unshaken controls, expressed as a percentage of sleep time lost during deprivation. Z2-0115 (n=23), Z2-0557 (n=30), Z2-1744 (n=29), Z2-2317 (n=19), Z2-2626 (n=22), and Z2-3941 (n=28) are compared to control background line cn bw (n=19). B. Arousal threshold for control cn bw (n=30 for mild stimuli, n=26 for moderate stimuli, and n=29 for strong stimuli) and short-sleeping lines Z2-0115 (n=23; n=15; n=19), Z2-0557 (n=34; n=26; n=38), Z2-1744 (n=13; n=7; n=14), Z2-2317 (n=16; n=16; n=14), Z2-2626 (n=11; n=5; n=5), Z2-3941 (n=16; n=10; n=7). Responses to a mild stimulus (white bars), moderate stimulus (gray bars), and strong stimulus (black bars) are shown. Data is for male flies, and error is expressed as SEM. * denotes P < 0.05 and ** denotes p < 0.01 using Mann-Whitney U test compared versus control.
We examined whether arousal thresholds were altered in these short-sleeping mutants. Figure 4B demonstrates that all of the short-sleeping mutants have reduced arousal thresholds, compared to the control background (cn bw). At a mild stimulus intensity, controls were aroused ~15% of the time, compared to 44%-85% for short-sleeping mutants. At a moderate stimulus intensity, controls were aroused ~51% of the time, whereas mutants were aroused 88%-100% of the time. Together, these data suggest that changes in arousal threshold commonly underlie the defects in sleep maintenance observed in short-sleeping mutants.
Identification of Z2-1744 as a Novel DAT Allele
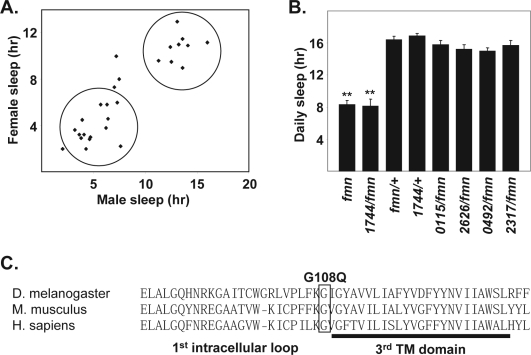
Given that Z2-1744 was one of the shortest sleeping lines, we decided to map this mutant first. Rough recombination mapping was performed using a multiply marked line (al bw c px sp). Analysis of the sleep phenotypes of the resulting recombinants back-crossed to the mutant chromosome demonstrated segregation into wild-type and mutant groups (Figure 5A). These mapping experiments suggested that Z2-1744 mapped near and to the right of curved which is located at cytological location 52D5. Currently, the only known Drosophila short-sleeping mutant on the 2nd chromosome is fumin, which affects a dopamine transporter. fumin is located at 53C7, leading us to test if Z2-1744 fails to complement fumin. As shown in Figure 5B, Z2-1744 fails to complement fumin, whereas a number of other short-sleeping mutants complement fumin. We next sequenced the DAT open reading frame in Z2-1744 mutants, using Z2-0115 mutants as a background control. We found a single base pair mutation causing a glycine (GGA) to a glutamine (GAA) change at amino acid 108. This amino acid is located at the putative junction of the 1st intracellular loop and the 3rd transmembrane domain and is conserved between fly, mouse, and human dopamine transporters (Figure 5C). The identification of a DAT mutant in our screen for short-sleepers reinforces the importance of monoaminergic, and specifically dopaminergic, signaling in sleep regulation.
Figure 5.
Z2-1744 is a novel DAT allele. A. Daily sleep is shown for female sleep versus male sleep from independent lines generated via meiotic recombination between Z2-1744 and a mapping line. This data suggests that the sleep phenotype segregates into two discrete populations—mutant and wild-type. B. Daily sleep for male flies (mean ± SEM) is shown—fmn (n=16), Z2-1744/fmn (n=14), fmn/+ (n=26), Z2-1744/+ (n=30), Z2-0115/fmn (n=16), Z2-2626/fmn (n=16), Z2-0492/fmn (n=8), Z2-2317/fmn (n=8). ** denotes P < 0.01 (Mann-Whitney U test) for fmn and Z2-1744/fmn compared to other fmn heterozygous combinations. C. Amino acid sequence of part of Drosophila DAT aligned with mouse DAT and human DAT. The region depicted includes the 1st intracellular loop and the 3rd transmembrane domain. The point mutation identified in Z2-1744 affects glycine 108, changing it to a glutamine, and this glycine is conserved between fly, mouse, and human.
A Novel Approach to Identifying Circadian Mutants in Drosophila
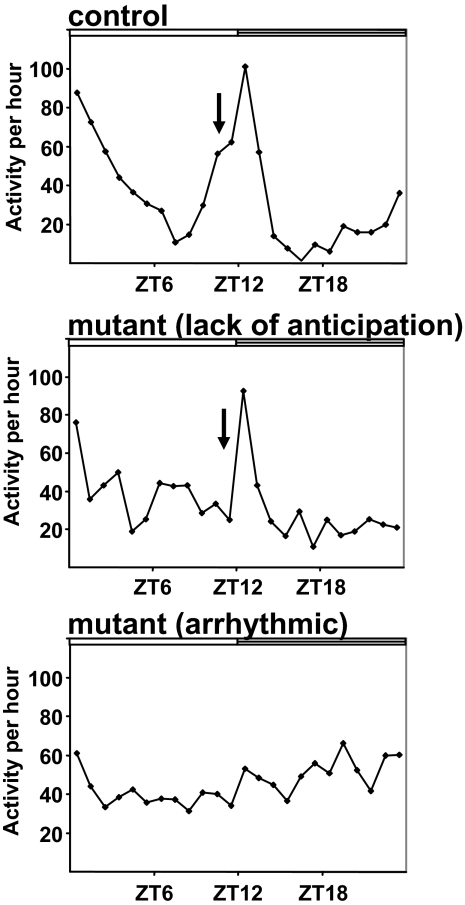
Finally, we asked whether we could use our L:D data-set to also identify novel circadian mutants. We examined the activity records of the roughly 3,000 lines from the 2nd chromosome Zuker collection and an additional 73 lines from the 3rd chromosome Zuker collection. Since the presence of an L:D cycle can mask the rhythm phenotypes of certain Drosophila circadian mutants,36 we examined the locomotor activity records for the presence of morning and evening anticipation which denotes endogenous clock activity. Flies with an intact circadian clock increase their locomotor activity in anticipation of light:dark transitions.37 We found a number of lines where this anticipation was absent. In addition, some lines appeared arrhythmic even in L:D (Figure 6). In total, we identified 21 lines that appeared arrhythmic in L:D or had absent morning and evening anticipation. We then examined their rhythm phenotypes in D:D. Of these 21 lines, we found that 6 lines had abnormal circadian rhythms in D:D. 1 mutant had a long period phenotype and the other 5 lines were arrhythmic. Through complementation and sequencing analyses, we determined that Z2-3326 is a novel allele of timeless (tim) and Z3-0084 is an allele of Clock (Clk) (Table 1). The L:D activity patterns for these novel tim and Clk alleles are shown in Supplemental Figure 8. The other mutants are currently undergoing further characterization and will be described elsewhere. These data demonstrate that circadian mutants can be identified with L:D data by examining morning and evening anticipation and thus provide a novel approach for identifying such mutants.
Figure 6.
Lack of anticipation as a means for identifying circadian mutants using L:D activity data. Representative activity profiles from primary screen data are shown for a control Zuker mutant line, for a mutant line lacking evening anticipation, and for a mutant line which appears arrhythmic in L:D. ZT=zeitgeber time. Light bar represents lights on, dark bar represents lights off. Arrows point to evening anticipation, which is impaired in the mutants.
Table 1.
Circadian Mutants Identified Using L:D Activity Data
| Genotype | %AR (n) | period |
|---|---|---|
| + | 0 (20) | 23.6 ± 0.05 |
| ClkZ3-84 | 100 (38) | n.a. |
| +/Clkjrk | 44 (25) | 24.2 ± 0.48 |
| ClkZ3-84/Clkjrk | 100 (22) | n.a. |
| timZ2-3326 | 77.4 (31)* | 29 ± 1 |
| +/tim01 | 0 (22) | 23.6 ± 0.05 |
| timZ2-3326/tim01 | 16.7 (18) | 28.7 ± 0.28 |
Circadian analysis of novel alleles of Clock and timeless. The molecular lesions for the novel alleles are as follows: ClkZ3-84 (Q684Stop) and timZ2-3326 (N214Y, N348T). % arrhythmic (%AR), number of male flies (n), and period length (±SEM) are shown for control (+) flies, the two novel alleles individually, as well as in combination with previously identified Clkjrk and tim01 alleles.13,14 Note that the Clkjrk mutation has been shown to have a dominant effect on circadian rhythms. *The majority of these timeless flies were weakly rhythmic and appeared to have long periods in the first 2–3 days of D:D.
DISCUSSION
Forward Genetic Screening for Short-Sleeping Mutants Using the Zuker Collection
In order to identify short-sleeping mutants in Drosophila, we have screened the 2nd chromosome mutants of the Zuker collection, a collection of EMS-mutagenized stocks. The generation of baseline sleep data for nearly 3,000 mutant lines allows us to examine the variability and correlations of different sleep characteristics. Among several commonly measured sleep variables, we find that bout duration shows a particularly large skew, which should be taken into consideration in the analysis and interpretation of bout duration data. While daily sleep is strongly inversely related to daily locomotor activity, daily sleep is poorly correlated with waking activity. These data support the idea that waking activity can be used to determine if a short-sleeping mutant is “hyperactive,” i.e., that the mutant shows excessive locomotor activity while awake. Daily sleep is better correlated with sleep bout duration than sleep bout number, suggesting that changes in bout duration and not bout number drive the differences in sleep time. A similar finding has also been described for wild-type flies.19
A Novel Approach for Identifying Circadian Mutants
We used our L:D locomotor activity data-set to identify circadian mutants by considering not only behavioral arrhythmicity, but also examining morning and evening anticipation. Most genetic screens for circadian mutants using locomotor rhythms involve using a D:D schedule. These screens typically require at least 5-7 days of recording in order to generate reliable circadian data. In the case of some large scale-screens, such as performed here for sleep phenotypes, it may be preferable to screen for a shorter period of time.
We find that, as expected, some circadian mutants can be identified by behavioral arrhythmicity even in L:D. For instance, clock mutants and cyc mutants are poorly rhythmic even in L:D.14,15. However, we also find that examination of morning and evening anticipation can be a useful tool for finding circadian mutants in L:D, as demonstrated by of our identification of an allele of timeless in this manner. Finally, an additional advantage of an L:D screen is that a single assay may be employed for isolation of both circadian and sleep mutants.
Phenotypic Characterization of the Short-Sleeping Mutants Reveals Aspects of Sleep Regulation in Drosophila
After screening nearly 3,000 lines, we identified 7 short-sleeping mutants. A short-sleeping phenotype could be caused by reduced sleep bout duration (defective sleep maintenance), reduced sleep bout number (defective sleep initiation), or both. Our analysis of the phenotypes of these mutants suggests that impairment of sleep maintenance rather than sleep initiation is a more common mechanism by which sleep is perturbed in fruit flies. We find that in our short-sleeping mutants, nighttime bout duration is severely reduced, compared to controls, suggesting that these mutants have difficulty with sleep maintenance, or staying asleep. Along these lines, studies using inbred strains of mice also reveal significant differences in slow wave sleep fragmentation across strains.38 In contrast, none of our mutants have a significant reduction in sleep bout number compared to controls, suggesting that mutations causing reduced sleep initiation are uncommon.
In order to identify possible mechanisms underlying the reduced and fragmented sleep seen in our short-sleepers, we characterized the responses of these mutants to sleep deprivation and arousing stimuli during sleep. We find that, compared to controls, all of our short-sleepers exhibit at least as much sleep rebound following deprivation, suggesting that homeostatic response to sleep deprivation is intact in the mutants. However, it remains possible that these short-sleeping mutants have impaired homeostatic regulation of baseline sleep, which may contribute to their short-sleep phenotype. Along these lines, most of our short-sleepers do not exhibit a marked compensatory increase in sleep bout number, despite having reduced and fragmented baseline sleep.
In contrast, we find that all of our short-sleeping mutants have reduced arousal thresholds, although we cannot exclude the possibility that this phenotype is secondary to an inability of our short sleepers to achieve deeper stages of sleep through longer uninterrupted sleep bouts. Interestingly, hyperarousal is proposed to be an important mechanism underlying problems with sleep initiation and sleep maintenance for patients with insomnia.39 The finding that all of our short-sleeping mutants have reduced arousal thresholds and severely reduced sleep bout duration suggests that, at least in flies, hyperarousal leads to sleep fragmentation.
Finally, it is worth considering why we did not identify any short-sleeping mutants with impaired sleep initiation or significantly reduced sleep rebound after deprivation. In addition to the possibility that these are simply rare phenotypes, it is also possible that these two phenotypes are linked. In other words, short-sleeping mutants that specifically have a reduction in sleep initiation but not sleep maintenance may represent mutants that lack sleep drive; despite having reduced sleep, such mutants would tend not to initiate sleep bouts because they do not feel sleepy. In surveying the Drosophila genome for mutations causing reduced sleep, our data suggest that there are multiple pathways that affect arousal threshold but few pathways that directly impact homeostatic sleep drive (unless these also affect viability).
Identification of a Novel DAT Allele
We characterized the genetic lesion in the Z2-1744 mutant, as this mutant was one of the shortest sleepers. By recombination mapping, complementation analysis, and sequencing analysis, we find that Z2-1744 is a novel allele of DAT. The point mutation in Z2-1744 is qualitatively different than the single pre-existing mutation, which results from a transposon insertion causing aberrant splicing and putative truncation of the DAT protein. The identification of another DAT allele is important, because there is only one previously characterized short-sleeping dopamine transporter mutant in Drosophila and because no deficiency is described that uncovers this region. Thus, loss-of-function analyses of DAT are currently limited to a single allele, which makes genetic and phenotypic studies more difficult. In our arousal assay, Z2-1744 appears to be the most arousable of our short-sleeping mutants, consistent with prior observations of reduced arousal threshold in fumin mutants.29 Our isolation of a DAT mutant from an unbiased screen of short-sleepers emphasizes the importance of arousal, and in particular monoaminergic tone, in regulation of sleep behavior.
Conclusions
The analysis of short-sleeping mutants identified in our large-scale screen suggests that sleep maintenance is more easily genetically perturbed than sleep initiation. Further, we suggest that these defects in sleep maintenance are usually caused by changes in arousal threshold, mediated by molecules such as dopamine. The promise of using a genetically tractable system such as Drosophila to study sleep lies in its ability to facilitate the identification of novel genes, molecules, and pathways involved in sleep and its regulation. The characterization of the molecular lesions in the mutants identified in this screen and others will aid us in this endeavor.
ACKNOWLEDGMENTS
We are especially indebted to Andrea Lougheed, Robert Hardy, and Charles Zuker for generously providing the Zuker collection mutants for screening. We thank K. Kume for the fumin mutant. M.N.W. is supported by a Burroughs-Wellcome Foundation Career Award for Medical Scientists. K.K. is supported by NIH 2P01AG017628-06. W.J. is supported by the HHMI. A.S. is an Investigator of the HHMI and is also supported by NIH 2P01AG017628-06.
Footnotes
Disclosure Statement
This was not an industry supported study. The authors have indicated no conflicts of interest.
REFERENCES
- 1.Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 2.Siegel JM. Clues to the functions of mammalian sleep. Nature. 2005;437:1264–71. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker MP, Stickgold R. Sleep, memory, and plasticity. Annu Rev Psychol. 2006;57:139–66. doi: 10.1146/annurev.psych.56.091103.070307. [DOI] [PubMed] [Google Scholar]
- 4.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45:347–60. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- 6.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–63. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 7.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–68. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- 8.Saper CB, Cano G, Scammell TE. Homeostatic, circadian, and emotional regulation of sleep. J Comp Neurol. 2005;493:92–8. doi: 10.1002/cne.20770. [DOI] [PubMed] [Google Scholar]
- 9.Steriade M. Brain electrical activity and sensory processing during waking and sleep states. In: Kryger M, Roth T, Dement WC, editors. Principles and practice of sleep medicine. 4th ed. Philadelphia, PA: Elsevier Saunders; 2005. pp. 101–119. [Google Scholar]
- 10.Taheri S, Zeitzer JM, Mignot E. The role of hypocretins (orexins) in sleep regulation and narcolepsy. Annu Rev Neurosci. 2002;25:283–313. doi: 10.1146/annurev.neuro.25.112701.142826. [DOI] [PubMed] [Google Scholar]
- 11.Tafti M, Maret S, Dauvilliers Y. Genes for normal sleep and sleep disorders. Ann Med. 2005;37:580–9. doi: 10.1080/07853890500372047. [DOI] [PubMed] [Google Scholar]
- 12.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–6. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–6. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 14.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 15.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–14. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 16.Hendricks JC, Finn SM, Panckeri KA, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 17.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 18.Greenspan RJ, Tononi G, Cirelli C, Shaw PJ. Sleep and the fruit fly. Trends Neurosci. 2001;24:142–5. doi: 10.1016/s0166-2236(00)01719-7. [DOI] [PubMed] [Google Scholar]
- 19.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep Homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–39. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 20.Hendricks JC, Kirk D, Panckeri K, Miller MS, Pack AI. Modafinil maintains waking in the fruit fly Drosophila melanogaster. Sleep. 2003;26:139–46. doi: 10.1093/sleep/26.2.139. [DOI] [PubMed] [Google Scholar]
- 21.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–75. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 22.Koh K, Evans JM, Hendricks JC, Sehgal A. From the Cover: A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–7. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol. 2002;12:1934–40. doi: 10.1016/s0960-9822(02)01300-3. [DOI] [PubMed] [Google Scholar]
- 24.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–60. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 25.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–6. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 26.Cirelli C. Searching for sleep mutants of Drosophila melanogaster. Bioessays. 2003;25:940–9. doi: 10.1002/bies.10333. [DOI] [PubMed] [Google Scholar]
- 27.Cirelli C, Bushey D, Hill S, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 28.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27:5384–93. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–84. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167:203–6. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsunoda S, Sierralta J, Sun Y, et al. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–9. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 32.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–4. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 33.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–62. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 35.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 2005;393:759–72. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- 37.Helfrich-Forster C. The locomotor activity rhythm of Drosophila melanogaster is controlled by a dual oscillator system. J Insect Physiol. 2001;47:877–87. [Google Scholar]
- 38.Franken P, Malafosse A, Tafti M. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22:155–69. [PubMed] [Google Scholar]
- 39.Roth T, Roehrs T, Pies R. Insomnia: pathophysiology and implications for treatment. Sleep Med Rev. 2007;11:71–9. doi: 10.1016/j.smrv.2006.06.002. [DOI] [PubMed] [Google Scholar]