Abstract
The lysine residues of rat heme oxygenase-1 (HO-1) were acetylated by acetic anhydride in the absence and presence of NADPH-cytochrome P450 reductase (CPR) or biliverdin reductase (BVR). Nine acetylated peptides were identified by MALDI-TOF mass spectrometry in the tryptic fragments obtained from HO-1 acetylated without the reductases (referred to as the fully acetylated HO-1). The presence of CPR prevented HO-1 from acetylation of lysine residues, Lys-149 and Lys-153, located in the F-helix. The heme degradation activity of the fully acetylated HO-1 in the NADPH/CPR-supported system was significantly reduced, whereas almost no inactivation was detected in HO-1 in the presence of CPR, which prevented acetylation of Lys-149 and Lys-153. On the other hand, the presence of BVR showed no protective effect on the acetylation of HO-1. The interaction of HO-1 with CPR or BVR is discussed based on the acetylation pattern and on molecular modeling.
Keywords: Heme oxygenase, NADPH-cytochrome P450 reductase, biliverdin reductase, MALDI-TOF mass spectrometry, chemical modification, protein-protein interaction
Heme oxygenase (HO, EC 1.14.99.3) is a membrane-bound microsomal enzyme that catalyzes the degradation of heme to biliverdin, carbon monoxide (CO), and free iron [1, 2]. Biliverdin is subsequently converted to bilirubin by a soluble cytosolic enzyme, biliverdin IXα reductase (BVR, EC 1.2.1.24) [3]. The electrons required for HO catalysis in mammals are provided by NADPH-cytochrome P450 reductase (CPR, EC 1.6.2.4), a 78-kDa membrane-bound flavoprotein containing one molecule each of FAD and FMN [4]. For electron transfer, CPR and its redox partners need to associate with each other. Indeed, the molecular surface of HO-1 surrounding the exposed δ-meso edge of heme is positively charged, and can associate, through electrostatic interaction, with the negatively charged surface of CPR [5]. In contrast to cytochrome P450, HO activity can be reconstructed with the soluble forms of HO-1 and CPR, both of which are lacking their membrane binding regions, C-terminal 22 amino acids and N-terminal 57 amino acids, respectively [6].
Thus far, crystal structures of rat and/or human HO-1 in the heme-free [7, 8], heme-bound [9, 10], verdoheme-bound [11], biliverdin-iron chelate-bound [12], and biliverdin-bound forms [13] have been reported. The crystal structures of rat CPR [5] and rat BVR [14, 15] have been also reported. However, less is known about how the HO protein interacts with these two reductases.
Recently, using site-specific mutation and docking modeling, we found that Arg-185 and Lys-149 of rat HO-1 are important in both the HO activity and its association with CPR [16]. In our model, the guanidino group of Arg-185 interacts electrostatically with 2′-phosphate of NADPH bound to CPR. On the other hand, Lys-149 is close to the acidic amino acid clusters near the FMN binding site of CPR; mutation of the acidic clusters causes reduction of the electron transfer rates from CPR to cytochrome P450s [17]. Thus, Arg-185 and Lys-149 appear to interact with CPR in such a way as to orient the redox partners for optimal electron transfer from CPR to the heme of HO-1. The model also suggested that the electrons from CPR are transferred through the distal side of the heme pocket of HO-1 to heme and that Lys-149 is involved in the electron transfer from FMN. Moreover, Wang and Ortiz de Montellano have reported that CPR and BVR bind competitively to human HO-1 and that several amino acid residues involved in binding of these two reductases to human HO-1 are located not only in the distal helices but also in the proximal helices of HO-1 [18]. However, the detailed interacting surfaces have yet to be characterized.
The combination of chemical modification of protein functional groups and mass spectrometry has become a powerful method for investigating molecular interfaces in protein complexes [19]. High sensitivity, accuracy, rapid analysis and low sample consumption are great advantages of mass spectrometry in studies on protein-protein interaction. Recently, the interactions between CPR and cytochrome P450 17α [20] and between human replication protein A and single-stranded DNA [21] have been investigated by a combination of chemical modification and mass spectrometry.
In the present study, employing this approach, we identified two lysine residues crucial for the interaction of rat HO-1 with CPR. Our results clearly show that HO-1-CPR complex formation disturbed the modification of these residues, leading to protection of HO catalytic activity.
MATERIALS AND METHODS
Enzymes
A soluble form of rat HO-1 lacking the 22-amino acid C-terminal hydrophobic segment (referred to as rat HO-1) was expressed in Escherichia coli and purified as described previously [22]. The heme-rat HO-1 complex was reconstituted with 1.2 equiv. of heme and purified by hydroxyapatite (Bio-Rad) column chromatography as previously described [22]. A recombinant rat liver CPR that lacks the N-terminal 57 hydrophobic amino acids was expressed and purified as described elsewhere [6]. The purification of BVR from rat liver was carried out according to published procedures [3].
Chemical modification and trypsin digestion
Chemical modification was performed according to the method of Nikfarjam et al [20] with slight modifications. Fifty picomoles of heme-rat HO-1 complex were incubated with either CPR or BVR (150 pmol each) in a total volume of 100 μl at pH 8.0 for 1 h on ice. NADP+ (100 pmol) was also included in the incubation mixture; NADPH is the physiological electron donor for CPR [16] and BVR [3], and is known to stabilize the interaction between heme-rat HO-1 complex and CPR [16]. Acetylation of the lysine residues of the heme-rat HO-1 complex was carried out with 2 μl of 15 mM acetic anhydride (Nacalai Tesque, Japan) for 15 min on ice. The molar ratio of heme-rat HO-1 complex to acetic anhydride was 1:600 in the reaction solution containing 20 mM potassium phosphate buffer at pH 8.0. The reaction was terminated by the addition of 20 μ l of 0.1M NH4HCO3. The heme-acetylated rat HO-1 complex was separated from CPR or BVR using Sulfo-Link coupling gel (PIERCE), which covalently immobilized the sulfhydryl-containing reductases. The eluate containing the heme-acetylated rat HO-1 complex was lyophilized and then incubated with 8 M urea in 50 mM Tris-HCl buffer (pH 8.0) for 1hr at 37 °C. After denaturation, trypsin digestion was performed using Tosyl-Phe Chloromethyl Ketone (TPCK)-treated trypsin (Promega) according to the manufacturer’s instruction.
Mass spectrometry and identification of acetylated positions
MALDI-TOF peptide mass fingerprintings were performed using a Bruker Autoflex instrument (Bruker Daltnics) with an α-cyano-4-hydroxyl-trans-cinnamic acid matrix (Sigma). All MALDI-TOF mass spectra were calibrated externally using a standard peptide mixture (angiotensin II (m/z 1046.5), adrenocorticotropic hormone fragment 18-39 (m/z 2465.2), and somatostatin (m/z 3147.5)). Identification of the peptide fragments was carried out with Protein Prospector software (http://prospector.ucsf.edu). The acetylated fragments were identified by an increase in mass number of 42 in the fragments containing lysine residue. An error in mass number of ±0.5 was tolerated in the identification of the modified peptides, if there were no unmodified fragments in this error range.
Measurements of single turnover HO reaction
Single turnover HO reactions of the unmodified and the acetylated HO-1 in complex with heme were monitored by optical absorption changes with a JASCO V-560 UV-visible spectrophotometer at 25 °C. In general, the standard reaction mixture (100 μl) contained 5 μM of rat HO-1 or its acetylated form complexed with heme, 40 nM CPR and 25 μM NADPH (5 molar equivalents to the heme-HO-1 complex) in 0.1 M potassium phosphate buffer (pH 7.4). The reduction rates of ferric heme complexed with unmodified and acetylated HO-1 were measured as reported previously [16]
Computer modeling
The program Hex [23] was used to predict the structure of the complex of rat HO-1 (Protein Data Bank Code 1DVE) and rat BVR (Protein Data Bank Code 1GCU) as described previously [16].
RESULTS
To investigate the effect of acetylation on heme degradation activity of HO-1, we measured absorption spectral changes of the heme-HO-1 complex during the HO reaction in air. The NADPH/CPR-supported single-turnover reaction of the heme-unmodified rat HO-1 complex is shown in Fig. 1A. The heme complexed with rat HO-1 immediately changed to the ferrous oxy-form upon the addition of NADPH, and then was transformed to biliverdin as indicated by the decrease of the Soret band and the increase of absorbance around 670 nm. The heme bound to rat HO-1 was completely degraded to biliverdin within 30 min under these conditions. Fig. 1B shows the NADPH/CPR-supported single-turnover reaction of the heme-acetylated rat HO-1. The spectrum of the heme complexed with acetylated rat HO-1 changed slowly to its oxy-form upon addition of NADPH. Although a decrease in absorbance at 340 nm indicated the consumption of NADPH, no biliverdin formation was observed within 30 min. To further investigate electron transfer from CPR to HO-1, we compared the rate of reduction of the ferric heme bound to the unmodified HO-1 with that of the acetylated form. In the NADPH/CPR-supported system, the rate of reduction of the heme complexed with acetylated HO-1 was decreased to 25% of that of the unmodified HO-1 (data not shown). On the other hand, it should be noted that the heme bound to the HO-1 acetylated in the presence of CPR was converted into biliverdin (Fig. 1C). This indicates that some acetylated lysine residues in HO-1 are involved in the interaction with CPR and that these residues were protected from acetylation by association with CPR.
Fig. 1. Absorption spectral changes of the heme-rat HO-1 complex during the NADPH/CPR-supported single-turnover reaction.
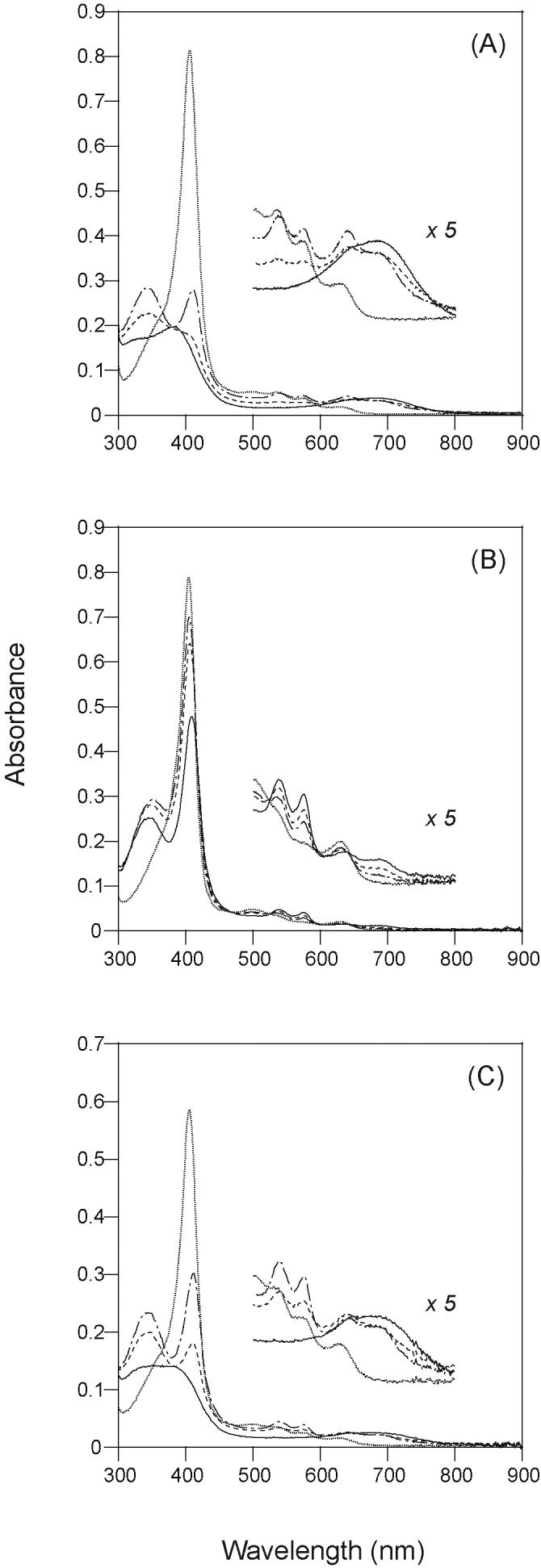
The spectra were recorded before (dotted line; ········) and after the addition of NADPH; 5 (dashed and dotted line; -·-·-), 10 (dashed line; ------) and 30 (solid line; —) min. (A) Unmodified HO-1, (B) HO-1 acetylated in the absence of CPR, (C) HO-1 acetylated in the presence of CPR. All reaction mixtures contained 5 μM heme-HO-1 complex, 40 nM CPR and 25 μM NADPH in 0.1M potassium phosphate buffer (pH 7.4).
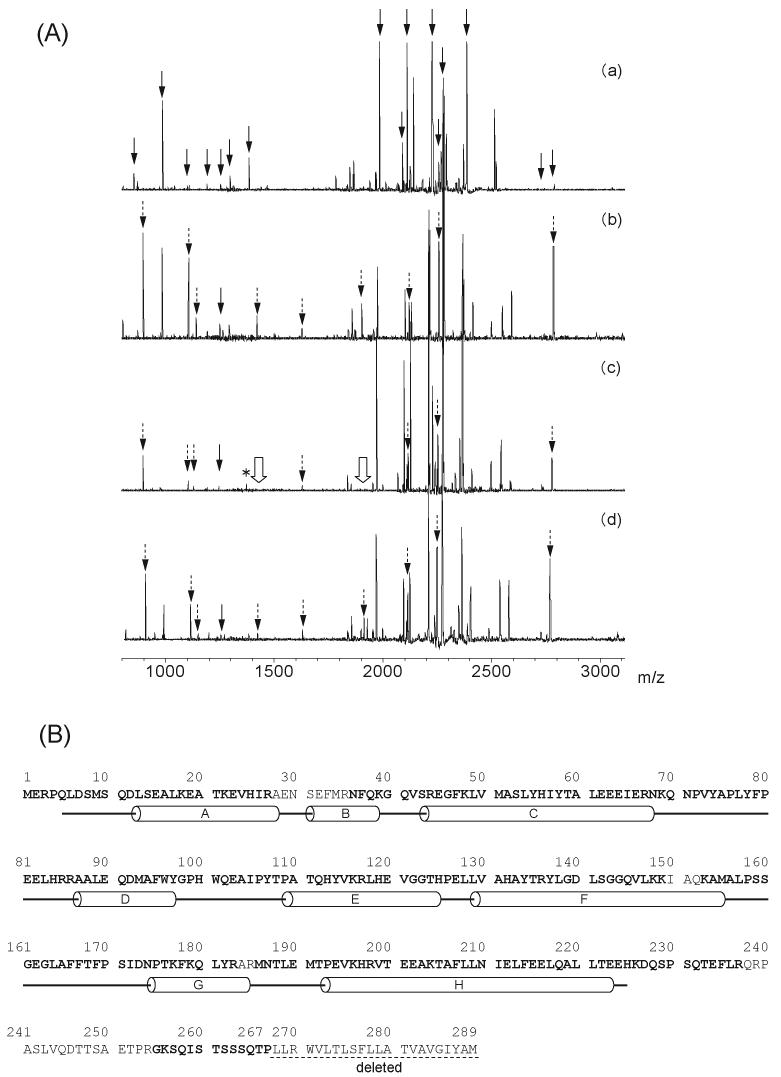
With the heme-unmodified rat HO-1 complex, 16 tryptic peptide fragments derived from HO-1 protein were clearly detected in the mass spectra in the region of m/z 800-3000 (indicated by solid arrows in Fig. 2A-a). Identified peptides in the mass spectra from heme-unmodified rat HO-1 complex covered about 89 % of the sequence of the parent protein (indicated by bold characters in Fig. 2B). A similar degree of sequence coverage was found for heme-acetylated rat HO-1. Nine acetylated peptides could be detected in the MALDI-TOF mass spectrum of the tryptic peptides of heme-acetylated rat HO-1 (indicated by dashed arrows in Fig. 2A-b). The identified peptides and the acetylated positions are summarized in Table 1.
Fig. 2. Mass spectrometric analysis of the heme-rat HO-1 complex.
(A) MALDI-TOF mass spectra of the tryptic peptides from rat HO-1: (a) unmodified, (b) acetylated in the absence of the reductases, (c) acetylated in the presence of CPR, (d) acetylated in the presence of BVR. The 16 solid arrows in (a) indicate the positions of the peptide fragment derived from the unmodified HO-1. The dashed arrows, 9 in (b) and (d) and 7 in (c) indicate the peaks of the acetylated peptides. The single solid-line arrows at m/z 1249 in (b), (c) and (d) indicate the Tyr-137-Lys-148 peptide without acetylation. In (c), the peak at m/z 1377 (asterisk) indicates the Tyr-137-Lys-149 peptide without acetylation and the 2 hollow arrows indicate the peaks, m/z 1419 and 1902, which were not detected in HO-1 acetylated in the presence of CPR. (B) Sequence and secondary structure of rat HO-1. Identified amino acid sequences in the mass spectra from heme-unmodified rat HO-1 complex are indicated by bold characters. The α-helices are shown as cylinders.
Table 1.
Identification of the positions of the lysine residues of HO-1 acetylated in the presence or absence of reductases
| Observed mass (m/z) | Peptide | Peptide sequence | Acetylated lysine | Without reductasea | With CPRa | With BVRa |
|---|---|---|---|---|---|---|
| 2119.891 | 1-18 | MERPQLDSMSQDLSEALK | 18 | yes | yes | yes |
| 1124.671 | 19-27 | EATKEVHIR | 22 | yes | yes | yes |
| 1105.643 | 36-44 | NFQKGQVSR | 39 | yes | yes | yes |
| 2783.232 | 45-67 | EGFKLVMASLYHIYTALEEEIER | 48 | yes | yes | yes |
| 2257.072 | 68-85 | NKQNPVYAPLYFPEELHR | 69 | yes | yes | yes |
| 1249.676 | 137-148 | YLGDLSGGQVLK | none | yes | yes | yes |
| 1377.802 | 137-149 | YLGDLSGGQVLKK | none | no | yes | no |
| 1419.823 | 137-149 | YLGDLSGGQVLKK | 149 | yes | no | yes |
| 1902.010 | 137-153 | YLGDLSGGQVLKKIAQK | 149 and 153 | yes | no | yes |
| 896.596 | 178-183 | FKQLYR | 179 | yes | yes | yes |
| 1627.821 | 186-198 | MNTLEMTPEVKHR | 196 | yes | yes | yes |
Yes and no indicate whether or not the corresponding peptide (the 3rd column in the same line) was detected.
Without reductases, two acetylated peptides, Tyr-137-Lys-149 with one acetylated lysine residue and Tyr-137-Lys-153 with two acetylated lysine residues were observed at m/z 1419 and 1902, respectively (Fig. 2A-b). However, when CPR was present during the acetylation reaction, these acetylated peptides were not produced (indicated by the hollow arrows in Fig. 2A-c); instead, non-acetylated Tyr-137-Lys-149 peptide was detected at m/z 1377 (indicated by an asterisk in Fig. 2A-c). These findings indicate that acetylation at Lys-149 and Lys-153 was prevented during the acetylation of heme-rat HO-1 complex in the presence of CPR. Although we failed to detect the non-acetylated Tyr-137-Lys-153 peptide (m/z 1818), this was probably due to further digestion at Lys-149 of this peptide. Indeed, non-acetylated Tyr-137-Lys-153 peptide was not observed in the peptide fragments of heme-unmodified HO-1 (Fig. 2A-a). It should be noted that the peak at m/z 1249, corresponding to non-acetylated Tyr-137-Lys-148 peptide, was observed irrespective of the presence or absence of CPR (Fig. 2A-b, c). These findings are consistent with the rat HO-1 crystal structure [9] that shows that Lys-149 and Lys-153 are fully exposed on the protein surface, whereas Lys-148 is buried inside the protein, so that it is not amenable to acetylation. On the other hand, the mass spectrum of heme-rat HO-1 complex acetylated in the presence of BVR was similar to that of heme-rat HO-1 complex acetylated in the absence of reductase (Fig. 2A-d and Table 1).
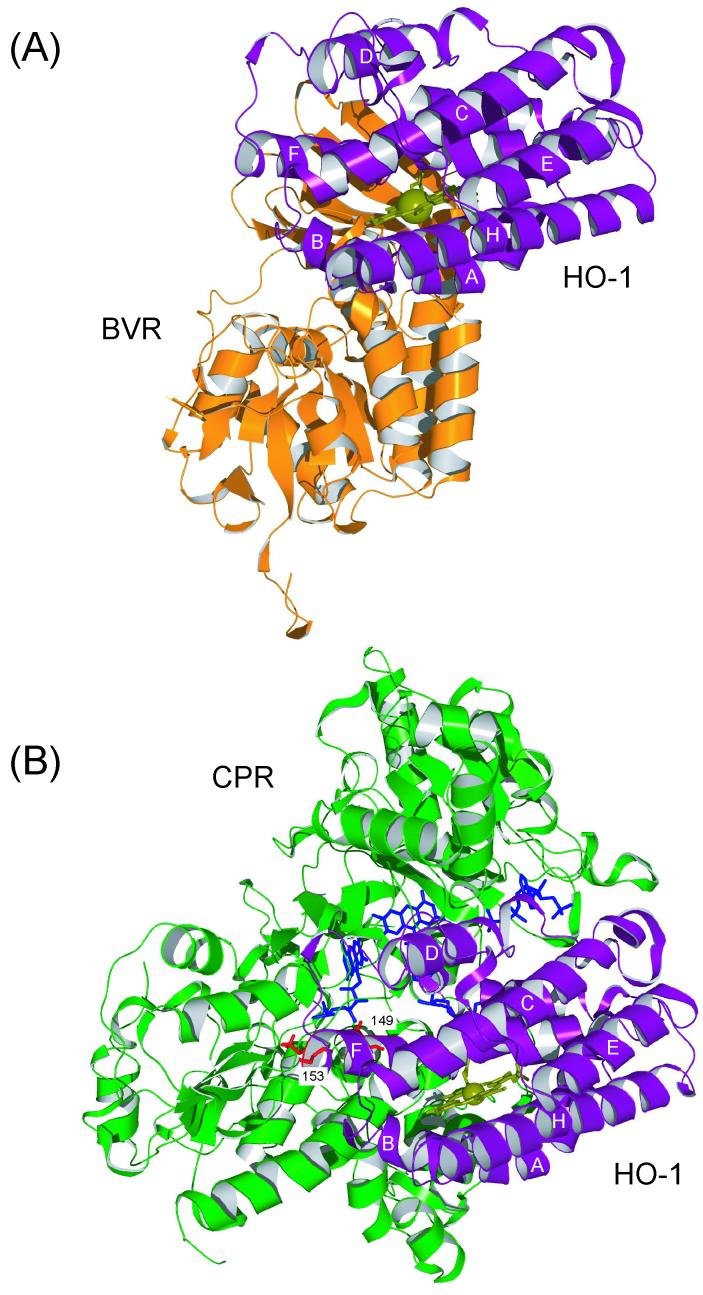
Computational modeling was attempted to identify the surface contact regions in the complex of HO-1 and BVR. The X-ray structure of rat HO-1 (Protein Data Bank Code 1DVE) and rat BVR (Protein Data Bank Code 1GCU) were used as input to the program Hex [23]. The lowest energy solution was selected from the 50 candidate docking solutions (Fig. 3A). In this model, the distance between the heme bound to HO-1 and the substrate binding site of BVR is about 20 Å. The docking surface of HO-1 consists of the A-, B-, F- and G-helices. This is similar to that seen in the docking model of the complex of HO-1 and CPR (Fig. 3B) as previously reported [16]. Interestingly, although lysine residues at positions 18, 22, 39, 149, 177 and 179 in rat HO-1 are located within 5 Å from BVR, they are not close to any acidic residues of BVR. On the other hand, Lys-149 and Lys-153 in rat HO-1 are surrounded by two acidic clusters of CPR, Asp-207-Asp-208-Asp-209 and Glu-213-Glu-214-Asp-215 (Fig. 3B, ref 16). Thus Lys-149 and Lys-153 of HO-1 are crucial for association with CPR, whereas these lysine residues of HO-1 are not very important in the interaction with BVR.
Fig. 3. Putative docking model of rat HO-1 (PDB 1DVE) with rat BVR (1GCU) (A) and with CPR (1AMO) (B; ref 16) calculated by the program Hex.
HO-1 is represented by a ribbon (purple). Heme is shown as ball-and-sticks (olive). The side chains of Lys-149 and Lys-153 in F helix of HO-1 are represented in stick form (red). BVR and CPR are shown as orange and green ribbons, respectively. The cofactors of CPR are in stick form (blue).
DISCUSSION
Using site-specific mutation, we have recently found that mutation of Lys-149 and Lys-153 to Ala, i.e. K149A and K149A/K153A, caused a 7- and 9-fold decrease, respectively, in the affinity of rat HO-1 for CPR [16]. Moreover, we and Yoshida’s group reported that replacement of Lys-149 and Lys-153 with Ala reduced the HO activity as well as the reduction rate of the ferric heme bound to rat HO-1 [16, 24]. On the other hand, Wang and Ortiz de Montellano, using fluorescence resonance energy transfer, reported that the mutation of Lys-149 in human HO-1 had no significant effect on binding to CPR [18]. Thus there has been some disagreement regarding the role of Lys-149 of HO-1.
In this work, we employed a mass spectrometric protein fingerprinting method [19-21] to search for lysine residues in rat HO-1 that are critical for binding to CPR and BVR. This method is based on the principle that, when particular amino acid residues are located at the interacting site with other proteins, these residues may be specifically protected from chemical modification [20, 21]. When CPR was present, Lys-149 and Lys-153 in F-helix of rat HO-1 actually escaped acetylation (Fig. 2A-c and Table 1), suggesting that these residues were masked by CPR. Thus we could confirm the direct involvement of Lys-149 and Lys-153 in the association with CPR. This was also supported by computer modeling of the complex of rat HO-1 and rat CPR, wherein Lys-149 and Lys-153 of rat HO-1 are close to the acidic amino acid clusters near the FMN binding site of CPR (Fig. 3B, ref 16).
With cytochrome P450s, it has been widely accepted that basic residues on the surface of the proximal side of the heme pocket are involved in binding to the negatively charged region on the CPR surface [25]. In the present study, we found that Lys-18, -22, -39, -48, -69, -148, -179 and -196 seem not to be significantly involved in the interaction with CPR. Indeed we and Yoshida’s group have shown that replacement of Lys-18 and Lys-22 with Ala did not affect HO activity [16, 24]. These results indicate that HO-1 interacts with CPR in a fashion somewhat different from that of cytochrome P450s.
Computer modeling of the complex of rat HO-1 and rat BVR showed Lys-18, -22, -39, -149, -177 and -179 in rat HO-1 are located within 5 Å of the surface of BVR. The presence of BVR during acetylation of HO-1, however, did not prevent acetylation of either of the reactive lysine residues (Fig. 2A-d, 3A and Table 1). An earlier kinetic study reported that the release of biliverdin from human HO-1 was accelerated in the presence of BVR [26]. Moreover, previous studies showed that CPR and BVR compete with each other in binding to HO-1 [16, 18], suggesting that the binding sites on HO-1 for BVR and CPR partially overlap. The docking model clearly indicates the overlapped binding site on the HO-1 for these two reductases (Fig. 3). Interestingly, the surface of HO-1 around the heme is primarily positively charged, and the surface of CPR in the vicinity of the FMN group is predominantly negatively charged. Thus they must electrostatically associate with each other. On the other hand, the surface of BVR at the interface between rat HO-1 and BVR is far less electronegative compared to that of CPR. Although the fluorescence resonance energy transfer study suggested that Lys-18, -22, -179, -183 and -185 in human HO-1 contribute to the binding to BVR, the effects of alanine substitution of these residues on the binding affinity for BVR is less marked compared to that of CPR [18], suggesting that the interaction mechanism of BVR with HO-1 might be somewhat different from that of CPR with HO-1
ACKNOWLEDGEMENTS
This work was supported in part by Grants-in-Aid for Young Scientists 18770121 (to Y. H.) and 19750150 (to H. Sato) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Grants-in-Aid for Scientific Research 18590278 (to M. N.) and 18550153 (to H. Sakamoto) from the Japan Society for the Promotion of Science, by a grant GM 080575 from the National Institute of Health (to G. P.), and by grant from the Ishibashi Foundation for the Promotion of Science (to H. Sato).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Tenhunen R, Marver HS, Schmid R. Microsomal heme oxygenase. Characterization of the enzyme. J. Biol. Chem. 1969;244:6388–6394. [PubMed] [Google Scholar]
- [2].Yoshida T, Noguchi M, Kikuchi G. Oxygenated form of heme. heme oxygenase complex and requirement for second electron to initiate heme degradation from the oxygenated complex. J. Biol. Chem. 1980;255:4418–4420. [PubMed] [Google Scholar]
- [3].Noguchi M, Yoshida T, Kikuchi G. Purification and properties of biliverdin reductases from pig spleen and rat liver. J Biochem. (Tokyo) 1979;86:833–848. doi: 10.1093/oxfordjournals.jbchem.a132615. [DOI] [PubMed] [Google Scholar]
- [4].Strobel HW, Hodgson AV, Shen S. In: Cytochrome P450. Ortiz de Montellano, editor. Plenum Press; New York: 1995. pp. 225–244. [Google Scholar]
- [5].Wang M, Robert DL, Paschke R, Shea TM, Masters BSS. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. USA. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hayashi S, Omata Y, Sakamoto H, Hara T, Noguchi M. Purification and characterization of a soluble form of rat liver NADPH-cytochrome P-450 reductase highly expressed in Escherichia coli. Protein Expr. Purif. 2003;29:1–7. doi: 10.1016/s1046-5928(03)00023-8. [DOI] [PubMed] [Google Scholar]
- [7].Sugishima M, Sakamoto H, Kakuta Y, Omata Y, Hayashi S, Noguchi M, Fukuyama K. Crystal structure of rat apo-heme oxygenase-1 (HO-1): mechanism of heme binding in HO-1 inferred from structural comparison of the apo and heme complex forms. Biochemistry. 2002;41:7293–7300. doi: 10.1021/bi025662a. [DOI] [PubMed] [Google Scholar]
- [8].Lad L, Schuller DJ, Shimizu H, Friedman J, Li H, Ortiz de Montellano PR, Poulos TL. Comparison of the heme-free and -bound crystal structures of human heme oxygenase-1. J. Biol. Chem. 2003;278:7834–7843. doi: 10.1074/jbc.M211450200. [DOI] [PubMed] [Google Scholar]
- [9].Sugishima M, Omata Y, Kakuta Y, Sakamoto H, Noguchi M, Fukuyama K. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- [10].Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat. Struct. Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- [11].Lad L, Ortiz de Montellano PR, Poulos TL. Crystal structures of ferrous and ferrous-NO forms of verdoheme in a complex with human heme oxygenase-1: catalytic implications for heme cleavage. J. Inorg. Biochem. 2004;98:1686–1695. doi: 10.1016/j.jinorgbio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- [12].Sugishima M, Sakamoto H, Higashimoto Y, Noguchi M, Fukuyama K. Crystal structure of rat heme oxygenase-1 in complex with biliverdin-iron chelate. Conformational change of the distal helix during the heme cleavage reaction. J. Biol. Chem. 2003;278:32352–32358. doi: 10.1074/jbc.M303682200. [DOI] [PubMed] [Google Scholar]
- [13].Lad L, Friedman J, Li H, Bhaskar B, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1 in a complex with biliverdin. Biochemistry. 2004;43:3793–3801. doi: 10.1021/bi035451l. [DOI] [PubMed] [Google Scholar]
- [14].Kikuchi A, Park SY, Miyatake H, Sun D, Sato M, Yoshida T, Shiro Y. Crystal structure of rat biliverdin reductase. Nat Struct Biol. 2001;8:221–225. doi: 10.1038/84955. [DOI] [PubMed] [Google Scholar]
- [15].Whitby FG, Philips JD, Hill CP, McCoubrey W, Maines MD. Crystal structure of a biliverdin IXα reductase enzyme-cofactor complex. J Mol Biol. 2002;319:1199–1210. doi: 10.1016/S0022-2836(02)00383-2. [DOI] [PubMed] [Google Scholar]
- [16].Higashimoto Y, Sakamoto H, Hayashi S, Sugishima M, Fukuyama K, Palmer G, Noguchi M. Involvement of NADPH in the interaction between heme oxygenase-1 and cytochrome P450 reductase. J. Biol. Chem. 2005;280:729–737. doi: 10.1074/jbc.M406203200. [DOI] [PubMed] [Google Scholar]
- [17].Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J. Biol. Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- [18].Wang J, Ortiz de Montellano PR. The binding sites on human heme oxygenase-1 for cytochrome P450 reductase and biliverdin reductase. J. Biol. Chem. 2003;278:20069–20076. doi: 10.1074/jbc.M300989200. [DOI] [PubMed] [Google Scholar]
- [19].Akashi S, Shinobu M, Terada T, Ito Y, Yokoyama S, Takio K. Characterization of the structural difference between active and inactive forms of the Ras protein by chemical modification followed by mass spectrometric peptide mapping. Anal. Biochem. 1997;248:15–25. doi: 10.1006/abio.1997.2122. [DOI] [PubMed] [Google Scholar]
- [20].Nikfarjam L, Izumi S, Yamazaki T, Kominami S. The interaction of cytochrome P450 17α with NADPH-cytochrome P450 reductase, investigated using chemical modification and MALDI-TOF mass spectrometry. Biochim. Biophys. Acta. 2006;1764:1126–1131. doi: 10.1016/j.bbapap.2006.04.003. [DOI] [PubMed] [Google Scholar]
- [21].Shell SM, Hess S, Kvaratskhelia M, Zou Y. Mass spectrometric identification of lysines involved in the interaction of human replication protein a with single-stranded DNA. Biochemistry. 2005;44:971–978. doi: 10.1021/bi048208a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Omata Y, Asada S, Sakamoto H, Fukuyama K, Noguchi M. Crystallization and preliminary X-ray diffraction studies on the water soluble form of rat heme oxygenase-1 in complex with heme. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1017–1019. doi: 10.1107/s0907444998003448. [DOI] [PubMed] [Google Scholar]
- [23].Ritchie DW, Kemp GJ. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- [24].Zhou H, Shun D, Zhang X, Sasahara M, Sato M, Chu GC, Yoshida T. Basic amino acid residues of heme oxygenase important for electron transfer between NADPH-cytochrome P450 reductase and heme oxygenase as revealed by alanine-scanning mutagenesis. Yamagata Med. J. 2000;18:25–36. [Google Scholar]
- [25].Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- [26].Liu Y, Ortiz de Montellano PR. Reaction intermediates and single turnover rate constants for the oxidation of heme by human heme oxygenase-1. J. Biol. Chem. 2000;275:5297–5307. doi: 10.1074/jbc.275.8.5297. [DOI] [PubMed] [Google Scholar]