Abstract
The eEF1A1 and eEF1A2 isoforms of translation elongation factor 1A have 98% similarity and perform the same protein synthesis function catalyzing codon-dependent binding of aminoacyl-tRNA to 80S ribosome. However, the isoforms apparently play different non-canonical roles in apoptosis and cancer development which are awaiting further investigations. We hypothesize that the difference in non-translational functions could be caused, in particular, by differential ability of the isoforms to be involved in phosphotyrosine-mediated signalling.
The ability of eEF1A1 and eEF1A2 to interact with SH2 and SH3 domains of different signalling molecules in vitro was compared. Indeed, contrary to eEF1A1, eEF1A2 was able to interact with SH2 domains of Grb2, RasGAP, Shc and C-terminal part of Shp2 as well as with SH3 domains of Crk, Fgr, Fyn and phospholipase C-gamma1.
Interestingly, the interaction of both isoforms with Shp2 in vivo was found using stable cell lines expressing eEF1A1-His or eEF1A2-His. The formation of a complex between endogenous eEF1A and Shp2 was also shown. Importantly, a higher level of tyrosine phosphorylation of eEF1A2 as compared to eEF1A1 was demonstrated in several independent experiments and its importance for interaction of eEF1A2 with Shp2 in vitro was revealed.
Thus, despite the fact that both isoforms of eEF1A could be involved in the phosphotyrosine-mediated processes, eEF1A2 apparently has greater potential to participate in such signalling pathways. Since tyrosine kinases/phosphatases play a prominent role in human cancerogenesis, our observations may gave a basis for recently found oncogenicity of the eEF1A2 isoform.
Keywords: eEF1A1, eEF1A2, SH2 domain, SH3 domain, Shp2
1. Introduction
Translation elongation factor 1A (eEF1A) is one of the core members of the elongation machinery providing high efficiency and processivity of the protein synthesis process in the eukaryotic cell (Negrutskii & El'skaya, 1998). eEF1A forms a ternary complex with aminoacyl-tRNA and GTP and delivers the correct aminoacyl-tRNA to the A site of mRNA programmed ribosome in the GTP hydrolysis-dependent mode. The GDP-bound form of eEF1A has been shown to interact with deacylated tRNA (Petrushenko et al., 1997) and can transport it to aminoacyl-tRNA synthetase as the tRNA recycling arm of a tRNA channelling cycle (Petrushenko, Shalak, Budkevich, Negruskii, & El'skaya, 2002).
It is believed that the translation function of eEF1A might be combined with its involvement in cytoskeletal network maintenance and various signalling pathways (Lamberti et al., 2004). eEF1A has been shown to interact with phospholipase C-gamma1 (PLC-gamma1) (Chang et al., 2002; Kim et al., 1999) and non-receptor tyrosine kinase Txk of the Tec family (Maruyama, Nara, Yoshikawa, & Suzuki, 2007). Importantly, Txk can form a complex with poly(ADP-ribose) polymerase 1 and eEF1A to influence interferon-gamma gene transcription in Th1 cells. Recently Lau, Castelli, Lin, and Macaulay (2006) identified eEF1A as a novel binding partner for Akt2 rho-associated kinase. In most cases it is not known whether interactions of eEF1A with protein kinase might lead to phosphorylation of eEF1A. Neither has the role of eEF1A in signal transduction pathways been identified.
In mammalian cells, two tissue- and development-specific isoforms, eEF1A1 and eEF1A2, are present. The expression of the 98% similar isoforms is mutually exclusive (Kahns et al., 1998). Interestingly, eEF1A2, which is present normally only in muscles and neurons, was recently associated with tumor development in tissues that normally express only eEF1A1 (Amiri et al., 2007; Anand et al., 2002). How eEF1A2 isoform is involved in malignant transformation is not as yet understood. Since tyrosine kinases are prominent players in cancer development, direct comparison of the ability of eEF1A1 and eEF1A2 to be involved in phosphoTyr-specific signalling processes could help to interpret cancer-related properties of eEF1A2.
In this study, we compared for the first time the interactions of eEF1A1 and eEF1A2 with SH2 and SH3 domains of various signalling molecules. Contrary to eEF1A1, eEF1A2 was able to interact with SH2 domains of Grb2, RasGAP, Shc and Shp2 as well as with SH3 domains of Crk, Fgr, Fyn and PLC-gamma1. Both eEF1A1 and eEF1A2 formed complexes with SH2 domain of PLC-gamma1. In vivo interaction between endogenous eEF1A and tyrosine phosphatase Shp2 was confirmed in HEK293 cell line and tyrosine phosphorylation of eEF1A1 and eEF1A2 was demonstrated by Western blots with anti-phosphotyrosine-specific antibodies. Importantly, the level of phosphorylation of eEF1A2 was higher than eEF1A1 when isolated from tissues, and also when overexpressed in cells. Furthermore, the level of phosphorylation has been shown to be crucial for in vitro complex formation of eEF1A2 and SH2 domain of Shp2.
2. Materials and methods
2.1. Cell lines and plasmid constructs
HEK293 cells were purchased from the American Type Culture Collection (Manassas, VA) and grown according to their instructions. To produce eEF1A1 and eEF1A2 stable cell lines pcDNA 3.1 eEF1A1 and eEF1A2 with the C-terminal His-tags were linearized by MunI and transfected in HEK293 cells using ExGene500 (Fermentas) according to manufacturer recommendations. G418 was used to select stable cell lines.
2.2. Bioinformatics
ScanSite Program was used for bioinformatic identification of possible SH2 and SH3 domains binding sites within eEF1A1 and eEF1A2. ScanSite employs the matrix of selectivity values for amino acids at each position relative to an orienting residue as determined by the oriented peptide library technique (Obenauer, Cantley, & Yaffe, 2003).
2.3. Purification of the eEF1A1 and eEF1A2 isoforms
eEF1A1 was purified from rabbit liver using a combination of the gel filtration, anion exchange, cation exchange and hydroxyapatite chromatographies as described previously (Budkevich et al., 2002). eEF1A2 was isolated from rabbit muscles using the same procedure except the gel-filtration step was omitted in some cases. To confirm the biological activity of the isolated isoforms, the GDP/[3H]GDP exchange was carried out as in (Carvalho, Carvalho, & Merrick, 1984).
2.4. Pull down assay
Purified GST-SH2 and GST-SH3 domains of various signalling molecules were kindly provided by Prof. Ivan Gout (London, UK) (Gout et al., 1993). In a pull down assay, 1.5 μg of GST, GST-SH2 or GST-SH3 domains fusion protein was incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) for 2 h at 4 °C. The beads were then washed to remove unbound proteins and incubated at 4 °C for 3 h with 1.5 μg of purified eEF1A1 or eEF1A2. Non-specific interactions were removed by extensive washing and bound proteins eluted by boiling in Laemmli sample buffer. Eluted proteins were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and detected by immunoblotting with specific antibodies.
2.5. Phosphatase treatment
Calf intestine alkaline phosphatase (CIAP) (Fermentas) has been used to dephosphorylate eEF1A1 and eEF1A2. The proteins were precipitated from the lysates of stable cell lines on NiNTA and extensively washed with 1× reaction buffer for CIAP (10 mM Tris–HCl (pH 7.5), 10 mM MgCl2), then 10 U of CIAP was added and reaction mix was incubated at 37 °C for 1 h. Thereafter the beads were washed two times with 1× reaction buffer and the proteins bound were eluted by boiling in Laemmli sample buffer.
For dephosphorylation of total protein fraction HEK293 cells overexpressing eEF1A1 or eEF1A2 were extracted with 1× CIAP reaction buffer. To facilitate lysis the cells were passed through a 20-gauge needle 20 times and whole cell extracts were centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant was incubated with 10 U of CIAP at 37 °C for 1 h, in control samples sodium orthovanadate was added to final concentration 1 mM.
2.6. Immunoprecipitation
HEK293 cells were treated under indicated conditions, then washed with ice-cold phosphate-buffered saline (PBS) and extracted with the lysis buffer containing 10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 1% Triton X-100, 1 mM sodium orthovanadate and a mixture of protease inhibitors (Roche Applied Science). Whole cell extracts were centrifuged at 10,000 × g for 15 min at 4 °C. Endogenous protein complexes were immunoprecipitated from 0.5 mg of total cell lysates with corresponding antibodies (1 μg/probe) and immobilized on protein A-Sepharose beads (Amersham Biosciences). Immune complexes were washed three times with the lysis buffer and used for immunoblotting. As a loading control Western blotting (WB) of total cell lysate with anti-β-actin has been used. The effective serum starvation (24 h in medium without serum) and stimulation (10% FBS, 30 min) has been confirmed in WB of total cell lysates with antibodies against phosphorylated residues S240/S244 in ribosomal protein S6, which is phosphorylated by S6K1/2 (effector kinases in PI3K/mTOR pathway) in response to growth factors stimulation.
2.7. Immunoblot analysis
Proteins were separated by SDS-PAGE, transferred onto polyvinylidene difluoride membrane, and incubated for 1 h with blocking solution (2% BSA, Tris-buffered saline, 0.1% Tween-40). After blocking the membranes were probed 2 h at 4 °C with anti-GST, anti-eEF1A (Upstate Biotechnology), anti-His tag (Cell Signalling), anti-Shp2 (Cell Signalling) or anti-pTyr 4G10 (Upstate Biotechnology) antibodies. After extensive washing with the solution of Tris-buffered saline and 0.1% Tween-40, the membranes were incubated with secondary horseradish peroxidase-conjugated antibody for 1 h at room temperature. The antigen–antibody complexes were detected using the ECL system (Millipore). When immunoblots had to be re-probed, the membranes were initially stripped and re-blocked prior to incubation with another type of primary antibody.
3. Results
3.1. Existence of SH2/SH3 domain binding sites in the eEF1A1 and eEF1A2 molecules are predicted by bioinformatic analysis
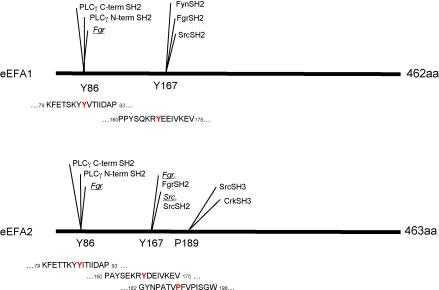
First we examined the possibility if the 98% similar eEF1A1 and eEF1A2 might have distinct binding motifs for signalling molecules. A low stringency Scansite (Obenauer et al., 2003) search revealed potential SH2/SH3 domain containing protein partners and predicted the existence of several phosphorylation sites for tyrosine kinases in both isoforms (Fig. 1). However, the SH3 recognition motif specific for Src kinase and adaptor protein Crk was found only in the eEF1A2 molecule. This is an important indication of the possibility of differential involvement of eEF1A2 in the formation of signalling complexes.
Fig. 1.
Schematic representation of potential SH2 and SH3 interaction sites of eEF1A1 and eEF1A2. Predicted sites were identified by ScanSite search of eEF1A1 and eEF1A2 peptide sequences: core amino acids are indicated. Y: tyrosine kinase phosphorylation site; SH2: SH2 domain binding site; SH3: SH3 domain binding site.
3.2. eEF1A1 and eEF1A2 interact in vitro with SH2/SH3 domains of different proteins
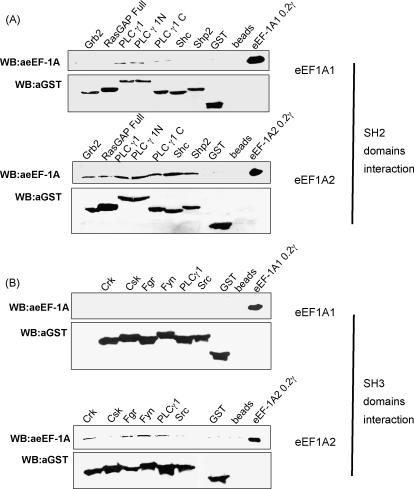
The ability of eEF1A1 and eEF1A2 to form complexes with SH2 and SH3 domains of different signalling molecules was directly analysed in pull down experiments. The preparative amounts of native eEF1A1 and eEF1A2 were purified from rabbit tissues as described in Section 2. Importantly, the rabbit and human proteins are 100% identical. SH2 domains of signalling proteins Grb2, RasGAP, PLC-gamma1, Shc, Shp2 and SH3 domains of Crk, Csk, Fgr, Fyn, Src were expressed as recombinant proteins fused with GST, purified from bacteria, immobilized on glutathione sepharose and mixed with eEF1A1 or eEF1A2. Complexes of eEF1A1 and eEF1A2 with SH2 and SH3 domains of different proteins were detected (Fig. 2A and B). Both eEF1A1 and eEF1A2 interacted with SH2 domain of PLC-gamma1, while eEF1A2 exclusively formed complexes with SH2 domains of Grb2, RasGAP, Shc, Shp2 and SH3 domains of Crk, Fgr, Fyn, PLC-gamma1.
Fig. 2.
Interaction of eEF1A1 and eEF1A2 with different GST-SH3 and GST-SH2 domain fusion proteins. GST-SH2 (A) or GST-SH3 (B) fusion proteins were coupled to glutathione-Sepharose beads and incubated with purified eEF1A1 (upper panels in A and B) or eEF1A2 (lower panels in A and B). The beads were washed extensively and the proteins were eluted by boiling in Laemmli sample buffer. Eluted proteins were resolved by SDS-PAGE and visualized by immunoblotting with anti-eEF1A antibodies (0.2 μg of purified eEF1A1 or eEF1A2 were loaded as controls). Amount of the GST-SH2 (A) or GST-SH3 (B) fusion proteins was controlled by immunoblotting with anti-GST antibodies. As a control, non-specific binding of eEF1A1 (upper panels in A and B) or eEF1A2 (lower panels in A and B) to GST alone or to beads alone has been analysed.
It should be noted that principal possibility of the interaction of PLC-gamma1 and eEF1A was shown earlier. The interactions of SH2 and SH3 domains (Kim et al., 1999) or PH domain (Chang et al., 2002) of PLC-gamma1 with eEF1A were reported. Our results confirmed the interaction of eEF1A with SH2 and SH3 domains of PLC-gamma1 and were used as internal positive control of interaction with known eEF1A binding partner.
Contrary to eEF1A1, eEF1A2 was found to interact with SH2/SH3 domains of several adaptor proteins. Grb2 and Shc provide a critical link between cell surface growth factor receptors and the Ras and Myc signalling pathways via interactions with cytoplasmic Tyr kinases, G-proteins and other cellular Tyr-phosphorylated proteins. Interestingly, Shc is constitutively activated (tyrosine-phosphorylated) in most breast cancer cells where non-typical expression of eEF1A2 was reported (Frackelton et al., 2006; Tomlinson et al., 2005).
Identification of RasGAP protein as a direct partner of G-protein eEF1A2 may be interesting in two ways. On the one hand, RasGAP may play GTPase activating role for eEF1A2, because the initial rate of GTP hydrolysis by the intrinsic eEF1A2 GTPase is slightly lower when compared with eEF1A1 (Kahns et al., 1998 and our unpublished observation). On the other hand, eEF1A2 may influence RasGAP activity and consequently modulate Ras.
eEF1A2 interacts also with SH3 domain of Crk which is involved in MAPK signalling, actin cytoskeleton reorganization and insulin receptor signalling (Feller, 2001). It should be noted that eEF1A is involved in actin cytoskeleton regulation as well (Gross & Kinzy, 2005; Shiina, Gotoh, Kubomura, Iwamatsu, & Nishida, 1994).
Fgr and Fyn are Tyr kinases that participate, in particular, in the MAPK/ERK and insulin receptor signalling pathways involved in the control of initiation of translation (Alper & Bowden, 2005). The interaction of eEF1A2 with SH3 domains of Fgr and Fyn suggests eEF1A2 as a prospective substrate for these enzymes.
Thus, eEF1A2 is capable of forming complexes with SH2 and SH3 domains of several signalling molecules. The potentially increased role of eEF1A2, as compared to eEF1A1, in phosphoTyr-mediated signalling pathways lends support for the putative oncogenic role assigned to eEF1A2 earlier (Amiri et al., 2007; Lamberti et al., 2004; Lee, 2003).
3.3. eEF1A1 and eEF1A2 are in complex with Shp2 in vivo
Verification of the in vitro formed complexes was done in vivo taking as an example interaction of eEF1A with Shp2.
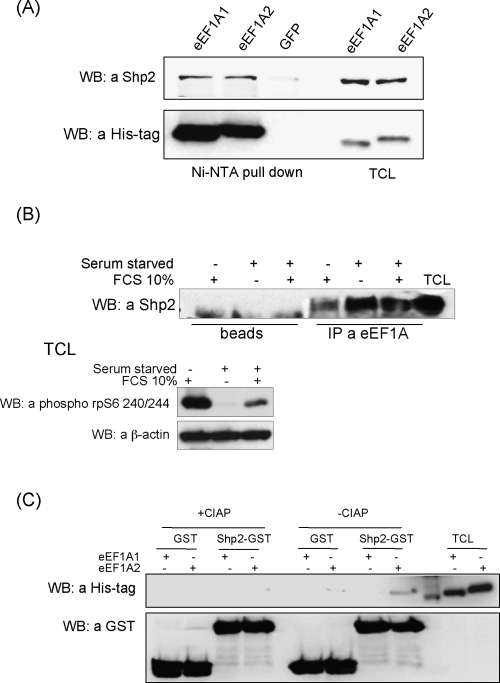
First, HEK 293 stable cell lines overexpressing eEF1A1-His, eEF1A2-His or GFP protein were used to confirm that endogenous Shp2 protein interacts with eEF1A2. Interestingly, specific binding of endogenous Shp2 to both isoforms of eEF1A was detected (Fig. 3A). Since only the C-terminal SH2 domain of Shp2 was used as a bait in the experiments in vitro, we suggest eEF1A1 and eEF1A2 bind to different domains of Shp2.
Fig. 3.
eEF1A1 and eEF1A2 form complexes with Shp2. (A) Complexes of His-tagged eEF1A1/eEF1A2 and Shp2 were precipitated from HEK293 stable cell lines using NiNTA sepharose. NiNTA sepharose beads were washed extensively and complexes were eluted by boiling in Laemmli sample buffer. Eluted proteins were resolved by SDS-PAGE and visualized by immunoblotting with anti-Shp2 (upper panel) or anti-His (lower panel) antibodies. NiNTA sepharose beads incubated with lysate of GFP overexpressing stable cell line were used as a control of non-specific binding of Shp2. TCL: total cell lysate. (B) Complexes were immunoprecipitated with anti-eEF1A antibodies from HEK293 cells treated under different conditions (growing, serum starved and serum starved followed by 30 min 10% serum stimulation). Protein A sepharose beads were washed extensively and the complexes were eluted by boiling in Laemmli sample buffer. Eluted proteins were resolved by SDS-PAGE and visualized by immunoblotting with anti-Shp2 antibodies. Protein A sepharose beads alone incubated with the same cell lysates were used as a control of Shp2 non-specific binding. Total cell lysates (TCL) of HEK293 cells were immunobloted with phosphospecific antibodies to phosphorylated Ser240/Thr244 in ribosomal protein S6 and β-actin to control the efficiency of treatment conditions and total protein amount used for immunoprecipitation in each sample, respectively. (C) GST or Shp2 GST-SH2 fusion proteins were coupled with glutathione-sepharose beads and incubated with phosphatase treated (10 U CIAP per 350 μg of total cell lysate) or untreated total cell lysates of stable cell lines overexpressing His-tagged eEF1A1 or eEF1A2 proteins. The beads were washed extensively and the proteins were eluted by boiling in Laemmli sample buffer. Eluted proteins were resolved by SDS-PAGE and visualized by immunoblotting with anti-His antibodies (upper panel). Amount of GST or Shp2 GST-SH2 proteins in each probe was controlled by immunoblotting with anti-GST antibodies (lower panel).
Also, a complex of endogenous eEF1A and Shp2 was immunoprecipitated from HEK293 cells using anti-eEF1A antibodies (Fig. 3B). The complex eEF1A/Shp2 was detected in growing cells. Interestingly, complex formation was more effective in cells synchronized by growth factors starvation and less efficient in cells stimulated with serum, indicating the regulatory nature of this interaction (Fig. 3B). Importantly, eEF1A2 was shown earlier to accumulate in G0 rather than in G1 and S phase cells (Ann et al., 1991). However, since anti-eEF1A monoclonal antibodies are specific both for eEF1A1 and eEF1A2, and the presence of both isoforms in embryonic cells is possible, it is difficult to determine precisely which isoform of eEF1A interacts with Shp2 in vivo.
3.4. eEF1A2 and Shp2 interaction is sensitive to dephosphorylation
To test if the interaction of eEF1A2 with C-terminal SH2 domain of phosphatase Shp2 is dependent on phosphorylation status of eEF1A, equal amounts of total cell lysates of the stable cell lines expressing eEF1A1 or eEF1A2 were treated with calf intestine alkaline phosphatase, or, in control samples, incubated with phosphatase inhibitor (sodium orthovanadate). Specific interaction of SH2-Shp2 and eEF1A2 was detected only in the samples not treated with CIAP indicating that phosphorylation of eEF1A2 is a necessary condition for eEF1A2/Shp2 complex formation (Fig. 3C).
3.5. eEF1A1 and eEF1A2 are phosphorylated at Tyr in vivo
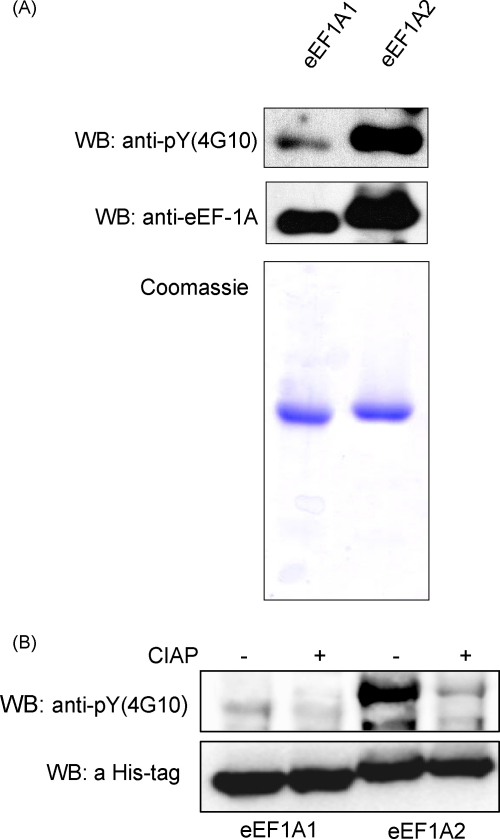
The interaction of eEF1A with SH2 domains of different proteins in vitro as well as the eEF1A–Shp2 complexes formation in vivo indicate the possibility of tyrosine phosphorylation in the eEF1A molecules. To test if native eEF1A1 and eEF1A2 contain such modifications we probed the isolated eEF1A proteins with monoclonal antibodies specific to phosphorylated tyrosines (4G10). As a result, tyrosine phosphorylation of both isoforms was detected (Fig. 4A). Moreover, the phosphorylation of eEF1A1 and eEF1A2 at tyrosine residues was confirmed using the His-tagged eEF1A isoforms precipitated on Ni-NTA sepharose (Fig. 4B). The eEF1A1 and eEF1A2 samples dephosphorylated with calf intestine alcaline phosphatase were used to control specificity of 4G10 antibodies. Notably, eEF1A2 was characterized by greater phosphorylation level as compared to eEF1A1 in both experiments.
Fig. 4.
eEF1A1 and eEF1A2 are phosphorylated at tyrosine residues in vivo. (A) 0.5 μg of purified eEF1A1 and eEF1A2 was immunobloted with anti-phosphotyrosine-specific antibodies (4G10) followed by WB with anti-eEF1A antibodies (upper panel). SDS-PAGE of purified eEF1A1 and eEF1A2 visualized by Coomassie staining, 1 μg of each protein was loaded per line (lower panel). (B) NiNTA sepharose beads with precipitated eEF1A1/eEF1A2 proteins were divided on two samples, the first was treated with 5 U CIAP at 37 °C for 1 h, second was incubated at 37 °C for 1 h in the presence of 1 mM NaOVa. NiNTA sepharose beads were washed with the lysis buffer two times; proteins were eluted by boiling in Laemmli sample buffer and resolved by SDS-PAGE. Phosphorylated eEF1A1 and eEF1A2 were visualized by immunoblotting with anti-phosphotyrosine-specific antibodies (4G10) (upper panel) followed by WB with anti-His tag antibodies (lower panel).
4. Discussion
Besides its function in translation, eEF1A is thought to be involved in several cellular processes, including embryogenesis, senescence, oncogenic transformation, cell proliferation and organization of cytoskeleton (reviewed in Gross & Kinzy, 2005; Lamberti et al., 2004; Lee, 2003). There are two tissue-specific isoforms of eEF1A. eEF1A1 functions in all mammalian tissues except neurons, cardiomyocytes and myotubes, where eEF1A2 is taking role and is expressed. Recently eEF1A2 was also found to express in human pancreatic islets (Ahmed, Forsberg, & Bergsten, 2005). Few attempts to differentiate the specific functions of the isoforms have been carried out so far. In vitro, eEF1A2 has slightly decreased translation functions [our unpublished observation]. Discovery of putative oncogenic role of eEF1A2 in ovarian cancer (Lee, 2003) as well as opposite apoptosis-related functions of the isoforms (Chang & Wang, 2007) indicated a possibility of distinct participation of eEF1A1 and eEF1A2 in cellular signalling processes. The mechanisms of cancer-related functions of eEF1A2 are not known. We suggested that difference of the isoforms could be related to their distinct ability to be involved into phosphoTyr-mediated signalling pathways.
To examine such a possibility we used the panel of cloned SH2 and SH3 domains of various signalling molecules. SH2 domains are known to associate with specific phosphoTyr-containing sites (Schlessinger & Lemmon, 2003). SH3 domains of adaptor proteins can support the association with a number of signalling proteins and upon cell stimulation can engage adaptor proteins with Tyr-phosphorylated cytoplasmic or membrane-attached partners (Li, 2005). As a result, we detected the interaction of eEF1A1 and eEF1A2 with SH2 domain of PLC-gamma1 indicating that both isoforms could contain phosphoTyr residues. In addition, eEF1A2 was found associated with SH2 domains of Grb2, RasGAP, Shc and Shp2 (Fig. 2A). This suggests eEF1A2 plays a more universal role in signal pathways. The finding that eEF1A2 rather than eEF1A1 can interact with SH3 domains of Crk, Fgr and Fyn supports this notion (Fig. 2B).
The association of eEF1A and Shp2 is very appealing in terms of the role Shp2 is playing in the cell-activation of Ras, with subsequent activation of PI3K and MAPK signalling pathways (Mohi & Neel, 2007). In addition, Shp2 regulates actin cytoskeleton reorganization by modulating the activity of small GTP-binding protein Rho (Burks & Agazie, 2006; Harada, Katoh, & Negishi, 2005). It is interesting that eEF1A is an actin-binding protein which can regulate actin bundling as well, but its target substrates have not been identified so far (Murray, Edmonds, Liu, & Condeelis, 1996). Since direct binding of eEF1A to Shp1 was recently found to have some regulatory consequence, namely eEF1A2-dependent increase of Shp1 phosphatase activity (Nandan, Yi, Lopez, Lai, & Reiner, 2002) the examination of possible effect of eEF1A2 on the Shp2 activity is a tempting task for future investigations.
We have observed direct interaction of eEF1A2 with SH2 domain of RasGAP protein. Here we can only speculate about possible involvement of eEF1A2 in the activation of PI3K and MAPK pathways by Ras through RasGAP inactivation (Yarwood, Bouyoucef-Cherchalli, Cullen, & Kupzig, 2006). Otherwise, RasGAP may play a role of GAP for eEF1A2. The last notion could be supported by the observation that the subunits of eEF1B, regular exchange factor for eEF1A1, were not identified as interacting partners for eEF1A2 (Mansilla et al., 2002).
The phosphorylation of native and overexpressed eEF1A1 and eEF1A2 at tyrosine residues was confirmed by WB with phosphotyrosine-specific antibodies (Fig. 4A and B). Several preps of eEF1A1 and eEF1A2 were analysed and eEF1A2 always demonstrated higher phosphorylation level than eEF1A1 (data not shown).
Recently, phosphorylation of Tyr 29 and Tyr 141 of eEF1A was found during an extensive search for all proteins containing phosphotyrosine in human cancer cells (Rush et al., 2005). However, since both identified tryptic peptides contained phosphoTyr29 or phosphoTyr141 (which are identical in eEF1A1 and eEF1A2) and the overall search was limited by the peptides containing phosphotyrosine, the unambiguous identification of the phosphorylated isoform in that case was hardly possible.
Thus, we have identified several binding partners of eEF1A2 previously unknown. Novel interaction of eEF1A with tyrosine phosphatase Shp2 has been confirmed also in vivo. Potentially augmented ability of eEF1A2 as compared to eEF1A1 to be involved in key signal transduction complexes could be linked to the putative oncogenic role of the former found recently in ovarian cancer (Anand et al., 2002). The definite role of eEF1A2 in activation of cell proliferation awaits further investigations.
Acknowledgments
We thank Michal Dadlez and Nadeem Shaikh for critical reading of the manuscript and helpful suggestions. The work on this project was supported by grants from the Fund for Fundamental Research of MON, Ukraine, INTAS and Wellcome Trust (Grant 074742/Z04/Z). Ganna Panasyuk was supported by Royal Society International Incoming Short Visit Grant. The authors thank Prof. I. Gout for providing GST-SH2 and GST-SH3 purified protein domains, Dr. C.R. Knudsen for pcDNAeEF1A1-His and pcDNAeEF1A2-His constructs and O.V. Novosylna for eEF1A1 and eEF1A2 preparations.
References
- Ahmed M., Forsberg J., Bergsten P. Protein profiling of human pancreatic islets by two-dimentional gel electrophoresis and mass-spectrometry. J. Proteome Res. 2005;4(3):931–940. doi: 10.1021/pr050024a. [DOI] [PubMed] [Google Scholar]
- Alper O., Bowden E.T. Novel insights into c-Src. Curr. Pharm. Des. 2005;11(9):1119–1130. doi: 10.2174/1381612053507576. [DOI] [PubMed] [Google Scholar]
- Amiri A., Noei F., Jeganathan S., Kulkarni G., Pinke D.E., Lee J.M. eEF1A2 activates Akt and stimulates Akt-dependent actin remodeling, invasion and migration. Oncogene. 2007;26(21):3027–3040. doi: 10.1038/sj.onc.1210101. [DOI] [PubMed] [Google Scholar]
- Anand N., Murthy S., Amann G., Wernick M., Porter L.A., Cukier I.H. Protein elongation factor eEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 2002;31(3):301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- Ann D.K., Moutsatsos I.K., Nakamura T., Lin H.H., Mao P.L., Lee M.J. Isolation and characterization of the rat chromosomal gene for a polypeptide (pS1) antigenically related to statin. J. Biol. Chem. 1991;266(16):10429–10437. [PubMed] [Google Scholar]
- Budkevich T.V., Timchenko A.A., Tiktopulo E.I., Negrutskii B.S., Shalak V.F., Petrushenko Z.M. Extended conformation of mammalian translation elongation factor 1A in solution. Biochemistry. 2002;41:15342–15349. doi: 10.1021/bi026495h. [DOI] [PubMed] [Google Scholar]
- Burks J., Agazie Y.M. Modulation of alpha-catenin Tyr phosphorylation by SHP2 positively effects cell transformation induced by the constitutively active FGFR3. Oncogene. 2006;25(54):7166–7179. doi: 10.1038/sj.onc.1209728. [DOI] [PubMed] [Google Scholar]
- Carvalho M.D., Carvalho J.F., Merrick W.C. Biological characterization of various forms of elongation factor from rabbit reticulocytes. Arch. Biochem. Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- Chang J.S., Seok H., Kwon T.K., Min D.S., Ahn B.H., Lee Y.H. Interaction of elongation factor-1alpha and pleckstrin homology domain of phospholipase C-gamma 1 with activating its activity. J. Biol. Chem. 2002;277(22):19697–19702. doi: 10.1074/jbc.M111206200. [DOI] [PubMed] [Google Scholar]
- Chang R., Wang E. Mouse translation elongation factor eEF1A-2 interacts with Prdx-1 to protect cells against apoptotic death induced by oxidative stress. J. Cell. Biochem. 2007;100:267–278. doi: 10.1002/jcb.20969. [DOI] [PubMed] [Google Scholar]
- Feller S.M. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20(44):6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Frackelton A.R., Jr., Lu L., Davol P.A., Bagdasaryan R., Hafer L.J., Sgroi D.C. p66 Shc and tyrosine-phosphorylated Shc in primary breast tumors identify patients likely to relapse despite tamoxifen therapy. Breast Cancer Res. 2006;8(6):R73. doi: 10.1186/bcr1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I., Dhand R., Hiles I.D., Fry M.J., Panayotou G., Das P. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- Gross S.R., Kinzy T.G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 2005;12(9):772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- Harada A., Katoh H., Negishi M. Direct interaction of Rnd1 with FRS2 beta regulates Rnd1-induced down-regulation of RhoA activity and is involved in fibroblast growth factor-induced neurite outgrowth in PC12 cells. J. Biol. Chem. 2005;280(18):18418–18424. doi: 10.1074/jbc.M411356200. [DOI] [PubMed] [Google Scholar]
- Kahns S., Lund A., Kristensen P., Knudsen C.R., Clark B.F.C., Cavallius J. The elongation factor 1 A-2 isoform from rabbit: Cloning of the cDNA and characterization of the protein. Nucl. Acids Res. 1998;26(8):1884–1890. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Si F., Kim S.J., Hong S.B., Hwang J.I., Lee H.J. The SH2–SH2–SH3 domain of phospholipase C-gamma 1 directly binds to translational elongation factor-1alpha. Mol. Cells. 1999;9(6):631–637. [PubMed] [Google Scholar]
- Lamberti A., Caraglia M., Longo O., Marra M., Abbruzzese A., Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: Review article. Amino Acids. 2004;26(4):443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- Lau J., Castelli L.A., Lin E.C., Macaulay S.L. Identification of elongation factor 1alpha as a potential associated binding partner for Akt2. Mol. Cell Biochem. 2006;286(1–2):17–22. doi: 10.1007/s11010-005-9006-5. [DOI] [PubMed] [Google Scholar]
- Lee J.M. The role of protein elongation factor eEF1A2 in ovarian cancer. Reprod. Biol. Endocrinol. 2003;1:69. doi: 10.1186/1477-7827-1-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.S. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 2005;390(Pt 3):641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla F., Friis I., Jadidi M., Nielsen K.M., Clark B.F., Knudsen C.R. Mapping the human translation elongation factor eEF1H complex using the yeast two-hybrid system. Biochem. J. 2002;365(Pt 3):669–676. doi: 10.1042/BJ20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Nara K., Yoshikawa H., Suzuki N. Txk, a member of the non-receptor tyrosine kinase of the Tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1alpha and regulates interferon-gamma gene transcription in Th1 cells. Clin. Exp. Immunol. 2007;147(1):164–175. doi: 10.1111/j.1365-2249.2006.03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohi M.G., Neel B.G. The role of Shp2 (PTPN11) in cancer. Curr. Opin. Genet. Dev. 2007;17(1):23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Murray J.W., Edmonds B.T., Liu G., Condeelis J. Bundling of actin filaments by elongation factor 1 alpha inhibits polymerization at filament ends. J. Cell. Biol. 1996;135(5):1309–1321. doi: 10.1083/jcb.135.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan D., Yi T., Lopez M., Lai C., Reiner N.E. Leishmania EF-1alpha activates the Src homology 2 domain containing tyrosine phosphatase SHP-1 leading to macrophage deactivation. J. Biol. Chem. 2002;277(51):50190–50197. doi: 10.1074/jbc.M209210200. [DOI] [PubMed] [Google Scholar]
- Negrutskii B., El'skaya A. Eukaryotic translation elongation factor 1α: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Progr. Nucl. Acid Res. Mol. Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- Obenauer J.C., Cantley L.C., Yaffe M.B. Scansite 2.0: Proteome-wide prediction of cell signalling interactions using short sequence motifs. Nucl. Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko Z.M., Ladohin A.S., Budkevich T.V., Shalak V.F., Negrutskii B.S., El'skaya A.V. Evidence for the formation of an unusual ternary complex of rabbit liver EF-1a with GDP and deacylated tRNA. FEBS Lett. 1997;407:13–17. doi: 10.1016/s0014-5793(97)00242-1. [DOI] [PubMed] [Google Scholar]
- Petrushenko Z.M., Shalak V.F., Budkevich T.V., Negruskii B.S., El'skaya A.V. Novel complexes of mammalian translation elongation factor eEF1A*GDP with uncharged tRNA and aminoacyl-tRNA synthetase. Implications for tRNA channeling. Eur. J. Biochem. 2002;269:4811–4818. doi: 10.1046/j.1432-1033.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- Rush J., Moritz A., Lee K.A., Guo A., Goss V.L., Spek E.J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Schlessinger J., Lemmon M.A. SH2 and PTB domains in tyrosine kinase signalling. Sci. STKE. 2003;191:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- Shiina N., Gotoh Y., Kubomura N., Iwamatsu A., Nishida E. Microtubule severing by elongation factor 1 alpha. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- Tomlinson V.A., Newbery H.J., Wray N.R., Jackson J., Larionov A., Miller W.R. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer. 2005;5:113. doi: 10.1186/1471-2407-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood S., Bouyoucef-Cherchalli D., Cullen P.J., Kupzig S. The GAP1 family of GTPase-activating proteins: Spatial and temporal regulators of small GTPase signalling. Biochem. Soc. Trans. 2006;34(Pt 5):846–850. doi: 10.1042/BST0340846. [DOI] [PubMed] [Google Scholar]