Abstract
The chromatin-associated protein DEK was first identified as a fusion protein in patients with a subtype of acute myelogenous leukemia. It has since become associated with diverse human ailments ranging from cancers to autoimmune diseases. Despite much research effort, the biochemical basis for these clinical connections has yet to be explained. We have identified a structural domain in the C-terminal region of DEK [DEK(309–375)]. DEK(309–375) implies clinical importance because it can reverse the characteristic abnormal DNA-mutagen sensitivity in fibroblasts from ataxia-telangiectasia (A-T) patients. We determined the solution structure of DEK(309–375) by nuclear magnetic resonance spectroscopy, and found it to be structurally homologous to the E2F/DP transcription factor family. On the basis of this homology, we tested whether DEK(309–375) could bind DNA and identified the DNA-interacting surface. DEK presents a hydrophobic surface on the side opposite the DNA-interacting surface. The structure of the C-terminal region of DEK provides insights into the protein function of DEK.
Keywords: NMR structure, oncogene product, ataxia-telangiectasia, acute myeloid leukemia, chromatin-associated protein, chromosomal translocation
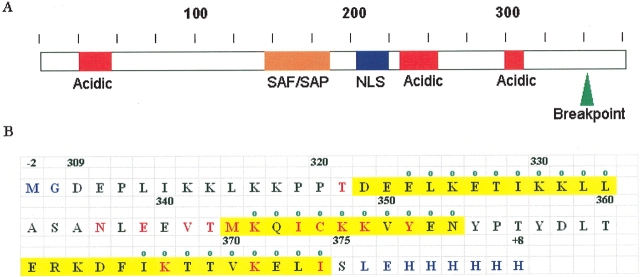
The 375-residue DEK protein is abundant in the nucleus and is associated with chromatin. DEK was first discovered in patients with a subtype of AML as a fusion product in which its C-terminal 26 amino acids are replaced by the N-terminal two-thirds of the nucleoporin CAN (break point is shown in Fig. 1A ▶; von Lindern et al. 1992). Although DEK’s role in AML has remained elusive, DEK has been linked in other ways to malignancy. A C-terminal fragment of DEK containing the last 65 residues is known to partially reverse the DNA-mutagen-sensitive phenotypes that characterize cells from A-T patients (Meyn et al. 1993). In addition, increased levels of DEK mRNA are reported in four neoplastic conditions: hepatocellular carcinoma, glioblastoma, melanoma, and AML (Kondoh et al. 1999; Grottke et al. 2000; Kroes et al. 2000; Larramendy 2002). Primary sequence analysis of DEK reveals acidic regions, a putative nuclear localization signal, and a nuclear scaffold attachment protein motif (SAF/SAP motif; Fig. 1A ▶).
Figure 1.
DEK(309–375) forms a structural domain. (A) Characterization of DEK amino acid sequence. DEK has a nuclear scaffold attachment protein motif, namely, SAF/SAP motif, which extends from residue 149 to 183 (orange); a putative nuclear localization signal that extends from residue 205 to 221 (blue); and three acidic regions including residues 30–49, 228–254, and 300–310 (red). The breakpoint of the chromosomal translocation is residue 350, shown in green, which produces DEK-CAN fusion protein. (B) The amino acid sequence of the DEK(309–375) construct used in these studies is provided with the experimentally determined helices shaded in yellow. Residues shown in red experienced chemical shift perturbations on mixing with the E2F/DP consensus sequence. Residues with a green circle are protected from exchanging with water in the deuterium exchange experiment. Residues with nonnative amino acids are colored blue.
DEK has been reported to interact with DNA in a sequence-dependent manner in two different biological processes. First, it was shown that DEK can bind to class II MHC gene promoters including the DQA1 Y-box (Adams et al. 2003). Thus, DEK could alter class II MHC gene regulation, and may alter or enhance the reactivity of antibodies. Interestingly, DEK has been identified as an antigen for several autoimmune diseases including JRA (Szer et al. 1991, 1994; Sierakowska et al. 1993), systemic lupus erythematosus, sarcoidosis, and systemic sclerosis (Dong et al. 2000). Second, DEK can modulate the transcription of human immunodeficiency virus type 2 (HIV-2) by binding to its peri-ets DNA sequence (Markovitz et al. 1990, 1992; Hannibal et al. 1994; Fu and Markovitz 1996). The importance of the peri-ets site is highlighted by the longer latency period of HIV-2 compared with that of HIV-1, which lacks this site (Markovitz et al. 1992; Perkins et al. 1993).
In addition to these sequence-specific DNA-binding functions, the chromatin association of DEK has been linked to several important biological processes. DEK is capable of changing the topology of chromatin and is suggested to participate in chromatin remodeling (Alexiadis et al. 2000). It is also reported to substantially reduce the replication efficiency of chromatin but not of naked DNA templates (Alexiadis et al. 2000). The chromatin association of DEK is also important for maintaining the latency of Kaposi’s sarcoma associated herpesvirus (KSHV). DEK forms a complex with the viral protein LANA (Kaposi’s sarcoma herpes viral latency-associated nuclear antigen) and this interaction allows the viral DNA of KSHV to be tethered to the chromosome of host cells (Krithivas et al. 2002).
Here we describe the structure of the C-terminal region of DEK that extends amino acids 309–375 [DEK(309–375)]. Structural similarities between DEK(309–375) and the DNA-binding domain of DP2, which is a member of the winged-helix DNA-binding proteins, is discussed. Using NMR chemical shift perturbation analysis, we have identified a surface of DEK(309–375) that can interact with DNA.
Results and Discussion
The C-terminal region of DEK defines a structural domain
We produced DEK fragments that contain residues 195–375 [DEK(195–375)] or 309–375 [DEK(309–375)] in order to define a structural domain in the C-terminal region of DEK. Comparison of HSQC spectra of DEK(309–375) and DEK(195–375) indicated that the residues N-terminal to DEK(309–375) have amide proton and nitrogen chemical shifts characteristic of a random coil (data not shown). We therefore used DEK(309–375) to determine the structure of the C-terminal region of DEK. The cDNA of DEK(309–375) was cloned into the pET28 Escherichia coli expression vector (Novagen) in frame with a C-terminal histidine tag. Residues that were introduced for cloning and ease of purification are highlighted in Figure 1B ▶. Sedimentation velocity analysis of DEK(309–275) indicates that this fragment is in a monomeric association state at 0.7 mg/mL concentration in 50 mM sodium phosphate (pH 7.0), 5mM DTT, and 100 mM KCl (data not shown).
The structure of DEK(309–375) is provided in Figure 2A ▶. The structured portion of this construct includes residues 320–375 and consists of three α-helices that span residues 322–333, 343–353, and 361–374. The two additional residues at the N terminus (MG) are unstructured, whereas the introduced LE residues in the C-terminal region form a helical turn. However, the amide protons of these residues are not protected from exchange with water in a deuterium exchange experiment (protected residues are shown in Fig. 1B ▶).
Figure 2.
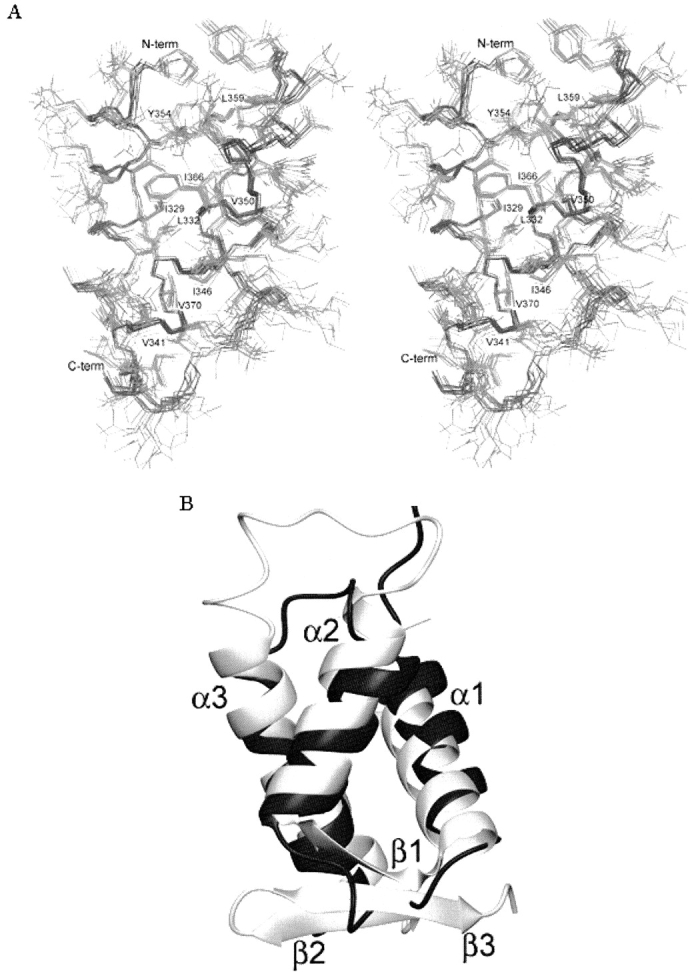
(A) The stereoview of the three-dimensional structure of DEK(309–375) reveals a hydrophobic core. The backbone atoms of the 10 lowest energy structures are superimposed in this figure. This figure was prepared using MOLMOL (Koradi et al. 1996). (B) The structure of DEK(309–375) closely resembles that of DP2. The α-helices of DEK(309–375; black) is superimposed onto the α-helices of the DNA-binding domain of DP2 (gray). DEK(309–375) lacks the β sheet present in DP2. This figure was prepared using MOLMOL (Koradi et al. 1996).
The three helices of DEK(309–375) are tightly arranged around a well-packed core of hydrophobic residues including L325, I329, L332, V341, I346, V350, Y354, L359, I366, and V370 (Fig. 2A ▶). The overall structure of DEK(309–375) is well defined, as the RMSD for all heavy atoms of residues 320–375 is 1.29 Å (Table 1).
Table 1.
Structural statistics for the NMR conformers
| NOE distance restraints | Average RMSD | ||
| All | 1456 | From distance restraints | 0.36 ± 0.0001 |
| Intraresidue | 802 | From dihedral restraints | 1.03 ± 0.1 |
| Interresidue | 654 | From idealized geometry | |
| Sequential (|i - j| = 1) | 293 | bonds (Å) | 0.0039 ± 0.0001 |
| Medium | 231 | angles (°) | 0.67 ± 0.009 |
| I, i + 2 | 82 | impropers (°) | 0.57 ± 0.015 |
| I, i + 3 | 124 | Of atomic coordinates between 10 structures: (2° structure) | |
| I, i + 4 | 25 | backbone (Å) | 0.539 |
| Long | 130 | all heavy atoms (Å) | 1.29 |
| Hydrogen bonds | 33 | Ramachandran plot appearance | |
| Dihedral angle restraints | Most favored regions (%) | 84.9 | |
| Additionally allowed regions (%) | 15.1 | ||
| φ | 46 | Generously allowed region s(%) | 0.0 |
| ψ | 46 | Disallowed regions (%) | 0.0 |
The first 11 amino acids of DEK(309–375) are mobile in solution as revealed through 15N relaxation experiments (supplemental data). These residues have weak NOEs in NOESY spectra and fast deuterium exchange rates, which confirms their flexibility. In contrast, the deuterium exchange rates of amide protons located within the three helices, including residues 324–333, 344–353, and 366–374, indicate that these residues are protected from exchange with water (Fig. 1B ▶).
The structure of DEK(309–375) is related to the winged helix DNA-binding motif
We tested whether DEK(309–375) shares structural homology with other protein structures by using the program DALI (Holm and Park 2000). The DALI search revealed 10 protein structures with a Z-score higher than 3.0. Inspection of these structures indicated that the DNA-binding domain of the DP2 protein complexed with its heterodimeric partner E2F4 shares significant similarity with DEK(309–375), with a Z-score of 3.7. Residues 319–375 of DEK(309–375) superimpose onto the helical region of the DNA-binding domain of DP2(68–133) with an RMSD of 2.3Å (Fig. 2B ▶). The overall fold of the DNA-binding domain of DP2 is very similar to that of DEK(309–375). Both proteins contain three tightly packed, twisted α-helices (Fig. 2B ▶). In both structures, the middle helix is the shortest and the C-terminal helix is the longest. The DNA-binding domain of DP2 contains an additional β-sheet consisting of a short strand preceding α2 and two strands C-terminal to the α-helices (Fig. 2B ▶).
Several protein families have winged helix motifs, including the hepatocyte nuclear factor-3 (HNF-3) family (Costa et al. 1989; Sheng et al. 2002), Drosophila homeotic forkhead proteins (Weigel et al. 1989), forkhead box family (Anderson et al. 1998; Biggs et al. 2001; Weigelt et al. 2001; Carlsson and Mahlapuu 2002), E2F/DP family (Slansky and Farnham 1996), Genesis (Sutton et al. 1996), and regulatory factor X 1(RFX1; Siegrist et al. 1993). With the exception of Drosophila homeotic forkhead protein family, these protein families are transcription factors. Forkhead transcription factors including FOX proteins, HNF-3, genesis, and RFX1 bind double-strand DNA (dsDNA) as monomers, whereas E2F/DP binds dsDNA as a heterodimer.
The topology of the winged helix motif consists of three α helices (α1, α2, and α3), three β strands (β1, β2, and β 3), and two loops (“wings,” W1 and W2), arranged as α1–β1–α2–α3–β2-W1–β3-W2. E2F and DP proteins lack the W2 wing. Many winged helix proteins, including HNF-3, E2F/DP, and genesis, bind the major groove of dsDNA by using polar residues within α3 (Clark et al. 1993; Jin et al. 1999; Zheng et al. 1999). The DP and E2F protein families comprise closely related transcription factors and form E2F/DP heterodimers to bind their target DNA sequences (for reviews, see Slansky and Farnham 1996; Helin 1998). These proteins bind dsDNA using an RRxYD motif, where x is an aliphatic residue (Slansky and Farnham 1996; Helin 1998). In DP2, α3 is situated in the DNA major groove and contains an RRVYD sequence with each arginine forming hydrogen bonds to guanine bases of sense- and antisense-strands (Zheng et al. 1999). Interestingly, DEK(309–375) contains a similar amino acid sequence in its α2; KKVYE. We therefore tested whether DEK can bind the E2F/DP consensus DNA sequence through contacts with α2.
DEK(309–375) interacts with dsDNA through α2
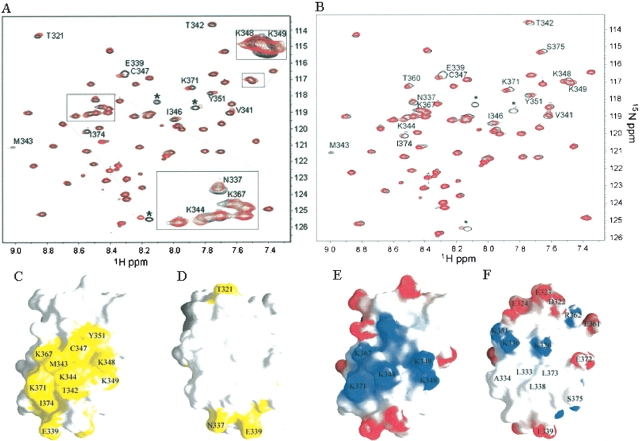
We monitored the chemical shift perturbation of the amide nitrogen and proton atoms of DEK(309–375) in [1H-15N] HSQC spectra on addition of the E2F/DP consensus DNA sequence (ds5′-TTTCGCGCG-3′; Fig. 3A ▶). Although DEK(309–375) appears to interact with the E2F/DP consensus sequence, the binding is weak, as the chemical shift perturbations were not saturated at a 1:4 molar ratio, and further chemical shift changes and line broadening are observed at a 1:8 molar ratio (data not shown). The residues that experienced chemical shift perturbations include those in the KKVYE sequence (K348, K349, Y351) as well as those in surrounding residues within α2 (M343, K344, I346, C347; colored red in Fig. 1B ▶). Residues within the loop between α1 and α2 (N337, E339, V341) and two lysines of α3 (K367, K371) also experienced chemical shift perturbations (colored red in Fig. 1B ▶). All of the residues that experienced chemical shift perturbations on addition of the dsDNA were mapped onto a surface representation of DEK(309–375; Fig. 3C,D ▶). It is clear that the surface shown in Figure 3C ▶ is the DNA-interacting surface of DEK(309–375) and that the surface shown in Figure 3D ▶ does not interact with dsDNA. The electrostatic properties of these surfaces are illustrated in Figure 3, E and F ▶, respectively. The DNA-interacting surface has a positively charged region that includes the five lysines that experienced chemical shift perturbation on addition of the dsDNA. In contrast, the opposite side has exposed hydrophobic residues, including L333 and A334 of α1, L338 of the loop between α1 and α2, and L373 from α3.
Figure 3.
DEK(309–375) interacts with dsDNA. (A,B) Superimposed 1H-15N HSQC NMR spectra of DEK(309–375) alone (black) and with DNA (red) containing 4 molar equivalents of the E2F/DP DNA consensus sequence (ds5′-TTTCGCGCG-3′; A) and a mutant E2F/DP sequence (ds5′-CCCATATAT-3′; B). The residues within the linker and C-terminal His tag are labeled with asterisks. (C,D) The residues that experience chemical shift perturbations caused by adding the E2F/DP consensus DNA sequence (ds5′-TTTCGC-GCG-3′) are colored yellow. (E,F) The electrostatic properties of surfaces for the surfaces shown in C and D, respectively. Surfaces D and F are rotated 180° from relative to C and E, respectively. These figures were made using GRASP (Nicholls et al. 1991).
The DNA-interacting specificity of DEK(309–375) was tested using dsDNA containing a mutated E2F/DP sequence, the HIV-2 peri-ets, or the DQA1 Y-box sequence as well as an RNA oligomer. We generated a mutated E2F/DP sequence (ds5′-CCCATATAT-3′) by replacing all of the AT and GC pairs with GC and AT pairs, respectively. Addition of the mutant E2F/DP sequence caused chemical shift perturbations in the [1H-15N] HSQC spectrum of DEK(309–375) that were similar to those observed by adding the E2F/ DP consensus DNA sequence (Fig. 3B ▶). In addition, DEK(309–375) showed similar chemical shift perturbations on adding dsDNA containing the HIV-2 peri-ets sequence (ds5′-GCTATACTTGGTCAGG-3′) and the DQA1 Y-box sequence (ds5′-CTAATTGGCC-3′; data not shown). Furthermore, addition of an RNA (5′-UUGAUUGGUUGUCAA-3′) or single-strand DNA (5′-GCTATACTTGGTCAGG-3′) showed weaker but similar chemical shift perturbations (data not shown). These results indicate that although DEK(309–375) interacts with dsDNA, DEK(309–375) does not specifically recognize the E2F/DP consensus, the HIV-2 peri-ets, or the DQA1 Y-box sequence. It should be noted that additional residues, including T360 and S375, showed small but comparable chemical shift perturbations when the mutant DNA was added (Fig. 3B ▶). It is possible that the third helix has more interaction with a DNA that contains a specific sequence for DEK(309–375).
In this work, we reveal that the C-terminal region of DEK [DEK(309–375)] forms a putative dsDNA-binding domain. This domain is structurally related to the winged-helix motif (Gajiwala and Burley 2000). Despite the structural similarity with winged helix proteins, the DNA-interaction mechanism of DEK(309–375) appears different from that of winged helix proteins, as it uses the second helix to interact with DNA, whereas other winged helix proteins use the third helix to interact with DNA. It is possible that DEK(309–375) requires another protein to form a stable protein–DNA complex. Further experiments are required to reveal the functional significance of this C-terminal region’s DNA-binding properties.
Materials and methods
Protein expression and purification
The 67-amino-acid C-terminal fragment of DEK [DEK(309–375)] was cloned into the pET28a expression vector in frame with a C-terminal histidine tag (Novagen; C & P Biotech Corp.). Primers containing the NcoI and XhoI sites were used for PCR to produce 5′ and 3′ cloning sites, respectively. The plasmid for expression of the 180-amino-acid C-terminal fragment of DEK [DEK(195–375)] was constructed in the same manner. For preparation of isotope labeled protein, cells were grown in M9 minimal media containing 15 NH4Cl, and/or 13C glucose at 37°C until OD600 0.6 and induced with 0.4 mM IPTG for 2 h before harvesting. Cells were lysed using Y-PER (Pierce), applied to Ni-NTA (Qiagen) Sepharose for 1 h, washed with 100 mL of 50 mM sodium phosphate buffer (pH 8.0), and eluted with 20 mL of the sodium phosphate buffer containing 250 mM imidazole. After dialyzing against 50-mM sodium phosphate buffer (pH 6.5) with 100 mM KCl to remove imidazole, the protein was concentrated and applied to a G-75 column (Amersham Pharmacia Biotech). The protein concentration was measured at 280 nm using an extinction coefficient of 3840 M−1 cm−1. The final yield of soluble protein was ~30 mg/L of bacterial culture.
Sedimentation velocity analysis
The association status of DEK(309–375) was analyzed by sedimentation velocity measurements performed at the National Analytical Ultracentrifugation Facility (University of Connecticut Bio-technology Center). The sample solutions were prepared at three protein concentrations, including 0.1, 0.3, and 0.7 mg/mL, and subjected to a sedimentation velocity run, using the 8-hole rotor at 20°C and 50,000 rpm. The data for each loading concentration were analyzed by the program Sedfit2 (version 8.7) using the model of a single, noninteracting, discrete species.
NMR spectroscopy and collection of structure restraints
NMR spectra were recorded at 20°C, using 1 mM DEK(309–375) in 50 mM sodium phosphate (pH 6.5), 100 mM KCl, and 95% H2O/5% 2H2O or 100% 2H2O at 1H NMR frequencies of 600 and 800 MHz on Varian Unity INOVA NMR spectrometers equipped with triple resonance probes and pulsed field gradients. Sequential resonance assignments were obtained from standard three-dimensional CBCANH (Grzesiek and Bax 1992b), CBCA(CO)NH (Grzesiek and Bax 1992a), total correlated spectroscopy (TOCSY)-15N-HSQC (Marion et al. 1989a) and HCCHTOCSY experiments (Bax et al. 1990). NOE restraints were obtained from 1H-1H two-dimensional NOESY and 13C and 15N edited three-dimensional NOESY experiments (Solomon 1955; Fesik et al. 1989; Marion et al. 1989a,b) with 150 msec mixing times and total recording times of 24, 92, and 87 h, respectively. Slowly exchanging protons were identified after exchanging the H2O buffer to a 2H2O buffer. Dihedral angle restraints were calculated from chemical shifts using the program TALOS (Cornilescu et al. 1999). The protection of amide protons from exchange with water was observed by the deuterium exchange experiment. DEK(309–375) is lyophilized in 50 mM NaPO4, and 100 mM KCl, 100% H2O and dissolved in 100% 2H2O. 1H-15N HSQC spectra were recorded every 5 min after being dissolved in 2H2O.
Structure calculations
Spectra were processed using the NMRPipe software (Delaglio et al. 1995) on SGI workstations. Cross-peaks in the NOESY spectra were assigned and integrated using the program XEASY (Bartels et al. 1995). The NMR structure was calculated using a simulated annealing protocol in the program XPLOR (Brunger 1993). Eighty random conformers were annealed in 60,000 steps using Cartesian dynamics. All simulated annealed conformers were energy-minimized using the refinement protocol in the XPLOR (Brunger 1993). Table 1 shows an overview of the restraints used and structural statistics. The backbone dihedral angles were analyzed by Ramachandran plot using PROCHECK-NMR (Laskowski et al. 1996). No cis-peptide bonds were observed and no residues were found with a positive φ angle. No residue was found in the disallowed regions. Secondary structure elements and RMSD values were calculated using the program MOLMOL (Koradi et al. 1996), which was also used to generate Figure 2 ▶.
Mapping DNA-interacting surfaces of DEK(309–375)
DNA-interacting surfaces of DEK(309–375) were studied using chemical shift perturbation analysis of the amide nitrogen and proton signals in 1H-15N HSQC spectra on addition of DNA. We used chemically synthesized dsDNA that has the consensus E2F/ DP binding site (ds5′-TTTCGCGCG-3′), a nonspecific sequence (ds5′-CCCATATAT-3′), the HIV-2 peri-ets sequence (ds5′-GCT ATACTTGGTCAGG-3′), and the DQA1 Y-box sequence (ds5′-CTAATTGGCC-3′). Each dsDNA was titrated into 15N-labeled DEK(309–375) at molar ratios of 1:1, 1:2, 1:4, and 1:8. Perturbed chemical shifts were followed through the DNA titration in order to assign signals at each molar ratio. Reference spectra were taken at the same protein concentration in the same buffer conditions. In addition, the chemical shift perturbation on addition of an RNA (5′-UUGAUUGGUUGUCAA-3′) or a single-strand DNA (5′-GCTATACTTGGTCAGG-3′) was observed.
Data bank accession codes
The coordinates of the 20 energy-refined XPLOR conformers of DEK(309–375) have been deposited in the PDB with the accession code 1Q1V. The NMR chemical shifts will be deposited in the BioMagResBank (BMRB).
Electronic supplemental material
Supplemental material includes NMR relaxation data of 15N amide nitrogens.
Acknowledgments
NMR instrumentation was provided with funds from the NSF (BIR-961477), the University of Minnesota Medical School, and the Minnesota Medical Foundation. We thank Basic Science Computer Laboratory for providing us with SGI workstations. We thank Drs. David Markovitz, Vlodek Szer, and Claudia Gruss for helpful discussions. We thank the National Analytical Ultracentrifugation Facility for conducting the sedimentation velocity analysis. This work is partially supported by a grant from Grant-in-Aid of Research, Artistry, and Scholarship from the Graduate School of University of Minnesota to H.M. M.D. was partially supported by the National Institute of Health predoctoral training grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
DEK(309–375), deletion of the N-terminal 308 amino acids of the human DEK protein
AML, acute myelogenous leukemia
A-T, ataxia telangiectasia
JRA, juvenile rheumatoid arthritis
HSQC, hetero-nuclear single quantum coherence
NOE, nuclear Overhauser effect
RMSD, root mean square deviation.
Supplemental material: see www.proteinscience.org
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04797104.
References
- Adams, B.S., Cha, H.C., Cleary, J., Haiying, T., Wang, H., Sitwala, K., and Markovitz, D.M. 2003. DEK binding to class II MHC Y-box sequences is gene- and allele-specific. Arthritis Res. Ther. 5 R226–R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiadis, V., Waldmann, T., Andersen, J., Mann, M., Knippers, R., and Gruss, C. 2000. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes & Dev. 14 1308–1312. [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J., Viars, C.S., Czekay, S., Cavenee, W.K., and Arden, K.C. 1998. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics 47 187–199. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Xia, T.-H., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- Bax, A., Clore, G.M., and Gronenborn, A.M. 1990. 1H-1H correlation via isotropic mixing of 13C magnetization: A new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88 425–431. [Google Scholar]
- Biggs 3rd, W.H., Cavenee, W.K., and Arden, K.C. 2001. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome 12 416–425. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. 1993. XPLOR Version 3.1: A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Carlsson, P. and Mahlapuu, M. 2002. Forkhead transcription factors: Key players in development and metabolism. Dev. Biol. 250 1–23. [DOI] [PubMed] [Google Scholar]
- Clark, K.L., Halay, E.D., Lai, E., and Burley, S.K. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364 412–420. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Costa, R.H., Grayson, D.R., and Darnell Jr., J.E. 1989. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and α 1-antitrypsin genes. Mol. Cell. Biol. 9 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Dong, X., Wang, J., Kabir, F.N., Shaw, M., Reed, A.M., Stein, L., Andrade, L.E., Trevisani, V.F., Miller, M.L., Fujii, T., et al. 2000. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 43 85–93. [DOI] [PubMed] [Google Scholar]
- Fesik, S.W., Gampe, R.T.J., Zuiderweg, E.R., Kohlbrenner, W.E., and Weigl, D. 1989. Heteronuclear three-dimensional NMR spectroscopy applied to CMP-KDO synthetase (27.5 kD). Biochem. Biophys. Res. Commun. 159 842–847. [DOI] [PubMed] [Google Scholar]
- Fu, G.K. and Markovitz, D.M. 1996. Purification of the pets factor. A nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J. Biol. Chem. 271 19599–19605. [DOI] [PubMed] [Google Scholar]
- Gajiwala, K.S. and Burley, S.K. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10 110–116. [DOI] [PubMed] [Google Scholar]
- Grottke, C., Mantwill, K., Dietel, M., Schadendorf, D., and Lage, H. 2000. Identification of differentially expressed genes in human melanoma cells with acquired resistance to various antineoplastic drugs. Int. J. Cancer 88 535–546. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax, A. 1992a. Correlating backbone amide and side chain resonances in large proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114 6291–6293. [Google Scholar]
- ———. 1992b. An efficient experiment for sequential backbone assignment of medium sized isotopically enriched proteins. J. Magn. Reson. 99 201–207. [Google Scholar]
- Hannibal, M.C., Markovitz, D.M., and Nabel, G.J. 1994. Multiple cis-acting elements in the human immunodeficiency virus type 2 enhancer mediate the response to T-cell receptor stimulation by antigen in a T-cell hybridoma line. Blood 83 1839–1846. [PubMed] [Google Scholar]
- Helin, K. 1998. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev. 8 28–35. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Park, J. 2000. DaliLite workbench for protein structure comparison. Bioinformatics 16 566–567. [DOI] [PubMed] [Google Scholar]
- Jin, C., Marsden, I., Chen, X., and Liao, X. 1999. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J. Mol. Biol. 289 683–690. [DOI] [PubMed] [Google Scholar]
- Kondoh, N., Wakatsuki, T., Ryo, A., Hada, A., Aihara, T., Horiuchi, S., Goseki, N., Matsubara, O., Takenaka, K., Shichita, M., et al. 1999. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 59 4990–4996. [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Krithivas, A., Fujimuro, M., Weidner, M., Young, D.B., and Hayward, S.D. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76 11596–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes, R.A., Jastrow, A., McLone, M.G., Yamamoto, H., Colley, P., Kersey, D.S., Yong, V.W., Mkrdichian, E., Cerullo, L., Leestma, J., et al. 2000. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 156 191–198. [DOI] [PubMed] [Google Scholar]
- Larramendy, M.L., Niini, T., Elonen, E., Nagy, B., Ollila, J., Vihinen, M., and Knuutila, S. 2002. Overexpression of translocation-associated fusion genes of FGFRI, MYC, NPMI, and DEK, but absence of the translocations in acute myeloid leukemia. A microarray analysis. Haematologica 87 569–577. [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Marion, D., Driscoll, P.C., Kay, L.E., Wingfield, P.T., Bax, A., Gronenborn, A.M., and Clore, G.M. 1989a. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Applied to interleukin 1β. Biochemistry 28 6150–6156. [DOI] [PubMed] [Google Scholar]
- Marion, D., Kay, L.E., Sparks, S.W., Torchia, D.A., and Bax, A. 1989b. Three-dimensional heteronuclear NMR of 15N-labeled proteins. J. Am. Chem. Soc. 111 1515–1517. [Google Scholar]
- Markovitz, D.M., Hannibal, M., Perez, V.L., Gauntt, C., Folks, T.M., and Nabel, G.J. 1990. Differential regulation of human immunodeficiency viruses (HIVs): A specific regulatory element in HIV-2 responds to stimulation of the T-cell antigen receptor. Proc. Natl. Acad. Sci. 87 9098–9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovitz, D.M., Smith, M.J., Hilfinger, J., Hannibal, M.C., Petryniak, B., and Nabel, G.J. 1992. Activation of the human immunodeficiency virus type 2 enhancer is dependent on purine box and kappa B regulatory elements. J. Virol. 66 5479–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyn, M.S., Lu-Kuo, J.M., and Herzing, L.B. 1993. Expression cloning of multiple human cDNAs that complement the phenotypic defects of ataxia-telangiectasia group D fibroblasts. Am. J. Hum. Genet. 53 1206–1216. [PMC free article] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Perkins, N.D., Edwards, N.L., Duckett, C.S., Agranoff, A.B., Schmid, R.M., and Nabel, G.J. 1993. A cooperative interaction between NF-κ B and Sp1 is required for HIV-1 enhancer activation. EMBO J. 12 3551–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, W., Rance, M., and Liao, X. 2002. Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence. Biochemistry 41 3286–3293. [DOI] [PubMed] [Google Scholar]
- Siegrist, C.A., Durand, B., Emery, P., David, E., Hearing, P., Mach, B., and Reith, W. 1993. RFX1 is identical to enhancer factor C and functions as a transactivator of the hepatitis B virus enhancer. Mol. Cell. Biol. 13 6375–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowska, H., Williams, K.R., Szer, I.S., and Szer, W. 1993. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin. Exp. Immunol. 94 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slansky, J.E. and Farnham, P.J. 1996. Introduction to the E2F family: Protein structure and gene regulation. Curr. Top. Microbiol. Immunol. 208 1–30. [DOI] [PubMed] [Google Scholar]
- Solomon, I. 1955. Relaxation processes in a system of two spins. Physiol. Rev. 99 559–565. [Google Scholar]
- Sutton, J., Costa, R., Klug, M., Field, L., Xu, D., Largaespada, D.A., Fletcher, C.F., Jenkins, N.A., Copeland, N.G., Klemsz, M., et al. 1996. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J. Biol. Chem. 271 23126–23133. [DOI] [PubMed] [Google Scholar]
- Szer, W., Sierakowska, H., and Szer, I.S. 1991. Antinuclear antibody profile in juvenile rheumatoid arthritis. J. Rheumatol. 18 401–408. [PubMed] [Google Scholar]
- Szer, I.S., Sierakowska, H., and Szer, W. 1994. A novel autoantibody to the putative oncoprotein DEK in pauciarticular onset juvenile rheumatoid arthritis. J. Rheumatol. 21 2136–2142. [PubMed] [Google Scholar]
- von Lindern, M., Fornerod, M., Soekarman, N., van Baal, S., Jaegle, M., Hagemeijer, A., Bootsma, D., and Grosveld, G. 1992. Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Baillieres Clin. Haematol. 5 857–879. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Jurgens, G., Kuttner, F., Seifert, E., and Jackle, H. 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57 645–658. [DOI] [PubMed] [Google Scholar]
- Weigelt, J., Climent, I., Dahlman-Wright, K., and Wikstrom, M. 2001. Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4). Biochemistry 40 5861–5869. [DOI] [PubMed] [Google Scholar]
- Zheng, N., Fraenkel, E., Pabo, C.O., and Pavletich, N.P. 1999. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes & Dev. 13 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]