Abstract
We report a detailed classification of disulfide patterns to further understand the role of disulfides in protein structure and function. The classification is applied to a unique searchable database of disulfide patterns derived from the SwissProt and Pfam databases. The disulfide database contains seven times the number of publicly available disulfide annotations. Each disulfide pattern in the database captures the topology and cysteine spacing of a protein domain. We have clustered the domains by their disulfide patterns and visualized the results using a novel representation termed the “classification wheel.” The classification is applied to 40,620 protein domains with 2–10 disulfides. The effectiveness of the classification is evaluated by determining the extent to which proteins of similar structure and function are grouped together through comparison with the SCOP and Pfam databases, respectively. In general, proteins with similar disulfide patterns have similar structure and function, even in cases of low sequence similarity, and we illustrate this with specific examples. Using a measure of disulfide topology complexity, we find that there is a predominance of less complex topologies. We also explored the importance of loss or addition of disulfides to protein structure and function by linking classification wheels through disulfide subpattern comparisons. This classification, when coupled with our disulfide database, will serve as a useful resource for searching and comparing disulfide patterns, and understanding their role in protein structure, folding, and stability. Proteins in the disulfide clusters that do not contain structural information are prime candidates for structural genomics initiatives, because they may correspond to novel structures.
Keywords: disulfide, classification, functional genomics, structural genomics, topology
In contrast to the tremendous volume of genome sequence information that has been generated over the last decade, the amount of information on the posttranslational modifications of proteins remains relatively modest. Formed by the cross-linking of cysteine residues, disulfide bridges comprise one type of posttranslational modification. Disulfide bridges are highly conserved structural features that play an important role in the stabilization, folding, and structure of proteins (Thornton 1981; Creighton 1988). In SwissProt (Boeckmann et al. 2003), 8.6% of sequences have disulfide annotations (van Vlijmen et al. 2004). Therefore, disulfide bonds constitute a commonly occurring posttranslational modification of proteins.
Experimental determination of disulfide bonds involves partially fragmenting the nonreduced form of a protein, isolating the fragments, and characterizing both the reduced and unreduced cystinyl forms (Ryle et al. 1955; Sanger 1959). The recent incorporation of mass spectrometry into disulfide determination techniques has increased both the accuracy and efficiency of the determination process (Gorman et al. 2002). The structural analysis of a protein by X-ray crystallography and NMR provides an alternative method for determining the disulfide bonds of a protein. Annotation on the disulfide bonds in proteins is accessible through the SwissProt sequence database. These annotations are based on experimental techniques, similar to those discussed earlier, or on inference from proteins with significant homology. In addition, the Protein Data Bank (PDB; Berman et al. 2000) contains disulfide information on proteins with three-dimensional structures. Finally, our recently published disulfide database, composed of 94,499 Swiss-Prot-extracted and inferred disulfide patterns, offers the most comprehensive repository of disulfide information (van Vlijmen et al. 2004).
Thornton (1981) performed one of the first broad analyses of disulfide bridges in proteins. Conducted on 128 proteins with either three-dimensional structures or sequences with known disulfide connectivity, the analysis identified the distribution of disulfides across different topologies, structural folds, and cystine conformations. The topological properties of disulfide bonding patterns were later explored in more depth by Benham and Jafri (1993) who grouped 208 distinct proteins, with and without structural information, by disulfide topology. The observed nonuniform distribution of disulfide topologies was attributed to disulfide bridge formation being a directed process. In a different study, Harrison and Sternberg (1996) created a clustering of small disulfide-rich β-sheet-containing folds on the basis of their cystine geometries. Mas et al. (1998, 2001) developed an automated way of aligning related disulfide-rich proteins by three-dimensional superposition of their disulfide bridges. The results reinforce the strong structural conservation of disulfides across related family members, even in cases of low sequence similarity.
In our previous study (van Vlijmen et al. 2004), we introduced a novel database of disulfide patterns based on a simple disulfide description, called the disulfide signature, which incorporates both the cysteine spacing and the disulfide topology of a protein. This description was computed for all proteins in the SwissProt database and then used to infer additional disulfide signatures for related protein domains in Pfam (Sonnhammer et al. 1997), thereby enabling a sevenfold increase in the amount of disulfide annotation. In this paper, we present a comprehensive classification of this expanded disulfide space and explore numerous cases of structurally and functionally homologous proteins grouping together in the absence of significant sequence similarity. We believe that our disulfide database and classification will serve as a valuable tool for searching, comparing, and understanding the role of disulfides in protein structure, folding, stability, and function.
Results
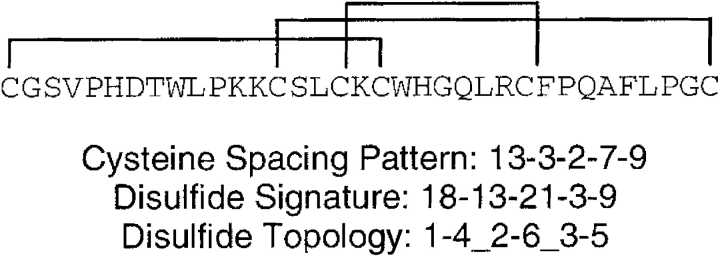
We represent disulfide topologies by using a string of paired numbers, where each number corresponds to the sequential order of cysteines in a domain. For example, the Cripto CFC domain depicted in Figure 1 ▶ has three disulfides with the topology 1–4_2–6_3–5, because the first cysteine is connected to the fourth cysteine, the second cysteine is connected to the sixth cysteine, and the third cysteine is connected to the fifth (Foley et al. 2003). Additionally, the protein has the cysteine spacing pattern 13–3–2–7–9, where the integers signify the number of residues occurring between sequential cysteines. In proteins where both the cysteine spacing and topology information is known, we use the disulfide signature representation. Incorporating both disulfide topology and cysteine spacing into a single string, the disulfide signature comprises 2N - 1 integers for a pattern with N disulfides. Odd-numbered positions in the string, which correspond to 18, 21, and 9 in the Cripto CFC domain, indicate the number of residues between pairs of cysteines forming disulfides. The even-numbered positions in the string, which correspond to 13 and 3 in the Cripto CFC domain, indicate the number of residues separating the first-occurring cysteines of consecutive disulfides. We use the term disulfide patterns to collectively refer to both disulfide signatures and cysteine spacing patterns.
Figure 1.
Disulfide patterns of the Cripto CFC domain. The disulfide signature incorporates both cysteine spacing and cysteine connectivity. The disulfide topology reflects the cysteine connectivity of the protein.
Disulfide database
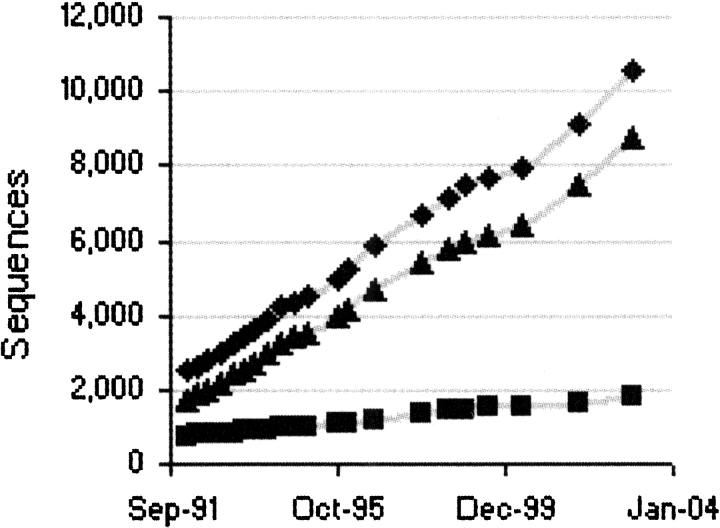
Over the past 10 yr, the number of SwissProt sequences with disulfide annotations has been growing in a linear manner (Fig. 2 ▶), with SwissProt Release 40.41 (revision, March 2003) containing 10,568 sequences with disulfide annotations. When these annotations were partitioned using Pfam Release 8.0 (revision, February 2003) domain boundaries, 16,736 independent disulfide patterns were generated. Of these partitioned disulfide patterns, 14,505 (87%) patterns were annotated in SwissProt as being at least partially inferred and the remaining 2231 (13%) were assumed to be experimentally determined. Using the inferring algorithms described in Materials and Methods, we were able to extrapolate an additional 77,763 disulfide patterns, increasing the size of the disulfide database sevenfold to a total of 94,499 patterns. A subset of 2934 patterns generated in the inferring process corresponded to SwissProt sequences that were previously either partially or completely lacking in their disulfide annotation. The remaining 74,829 patterns, ~95% of the inferred disulfide patterns, corresponded to TrEmbl sequences that were largely unannotated. Together, the SwissProtextracted and inferred disulfide patterns were distributed among 345 Pfam-A domain families and 288 Pfam-B domain families.
Figure 2.
Growth of disulfide annotations in SwissProt. The number of experimentally determined annotations, homology inferred annotations, and total disulfide annotations is shown in squares, triangles, and diamonds, respectively.
Of the total 633 disulfide-containing Pfam families, 372 (59%) contained references to three-dimensional PDB structures. We noted earlier that 2231 (13%) disulfide patterns were assumed to have experimentally determined disulfides because of the lack of any explicit annotations suggesting otherwise. When we examine the Pfam domains of experimentally determined disulfide patterns for three-dimensional structural information, we find that 1609 (72%) experimentally determined disulfide patterns belong to Pfam domains with three-dimensional structural information. Therefore, we assume that at least the remaining 622 (28%) experimentally determined disulfide patterns are derived from alternate techniques such as partial digestion. However, we acknowledge that these values may be inaccurate because of inconsistencies in the SwissProt disulfide annotations, as discussed previously (van Vlijmen et al. 2004).
Classification wheels
We constructed depictions of the disulfide classification by using the described layout. The three-disulfide classification wheel is shown in Figure 3 ▶. All three tiers of the classification are immediately discernable with this representation. The number three placed in the center of the wheel signifies the first level of the disulfide classification—only disulfide patterns with three disulfides are classified in the wheel. The inner ring of ellipses, each representing a different topology observed within patterns of three disulfides, composes the second tier of the disulfide classification. As all 15 of the possible topologies for patterns with three disulfides are observed, the corresponding 15 ellipses are present in the inner ring.
Figure 3.
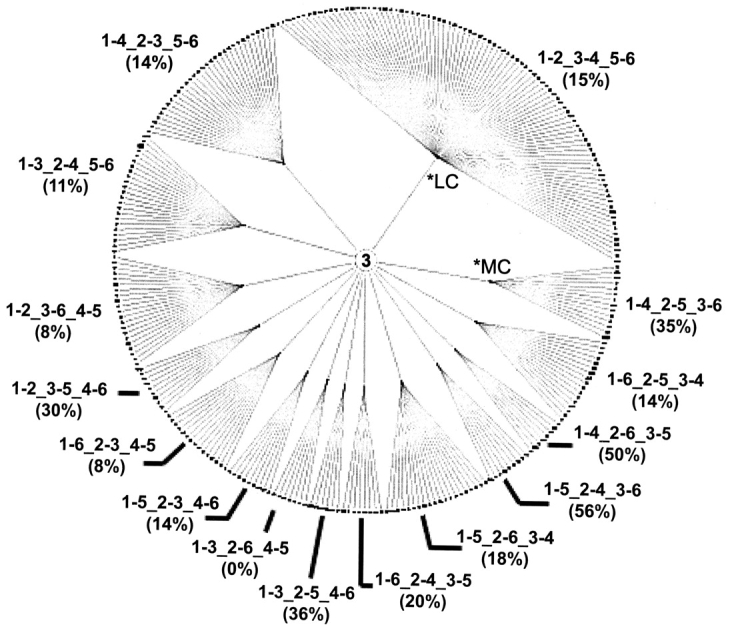
Three-disulfide classification wheel. All 15 disulfide topologies present in the inner ring of the wheel are arranged in ascending order of complexity beginning with the least complex topology (*LC) in the first quadrant of the circle and continuing in a counterclockwise direction to the most complex topology (*MC). The clusters of similar disulfide patterns are present on the outer ring of the classification wheel. In addition, the fraction (%) of clusters containing domains whose structure has been solved is shown for each topology.
Within a single topology, a large range of structures and functions are observed. For instance, the topology 1–2_3–4_5–6 contains families of proteins as diverse as eukaryotic aspartyl proteases and hemagglutinins. These families share no common structural or functional qualities, yet are classified together at the topology level because they share the same disulfide topology. As we will demonstrate, it is the third tier of the disulfide classification that enables protein domains with similar structures and functions to be classified together. Classifications based solely on disulfide topology perform poorly at uniting related protein domains. This additional third tier of the classification has not been previously reported in disulfide classification approaches.
The third tier of the disulfide classification is represented by the clusters forming the outer ring of the classification wheel. As described in Materials and Methods, each cluster was assigned a cluster identifier that consists of the length of the disulfide patterns in the cluster, the disulfide topology of the patterns in the cluster, and the cluster number within the classification wheel. For example, in a cluster with the identifier 3.1–3_2–4_5–6.121, the “3” indicates that each of the disulfide patterns contained in the cluster has three disulfides. The “1–3_2–4_5–6” indicates the disulfide topology of the patterns, and the last part, “121”, is the cluster’s assigned number within the three-disulfide classification wheel. All of the 287 clusters, dispersed across 15 topologies, in the three-disulfide classification wheel were assigned cluster identifiers. We determined that 60 of the 287 clusters (21%) contain disulfide patterns with exact sequence matches to three-dimensional structures (Fig. 3 ▶). Also, we note that the fraction of clusters with exact matches to structures ranges across the topologies. For example, the topology 1–5_2–4_3–6 has structural information for 56% of its clusters, whereas topology 1–3_2–6_4–5 has no clusters with structural information.
As observed in the three-disulfide classification wheel, the number of clusters per topology is not uniformly distributed across the different topologies. Because similar disulfide patterns are grouped together into a cluster, each cluster can be thought of as a distinct disulfide pattern. Therefore, the disulfide classification wheel representation enables one to easily identify a greater diversity of disulfide patterns within a particular topology by the increased number of clusters extending from the same topology. Moreover, the radial arrangement of the classification depiction allows one to recognize any trends in the diversity of disulfide patterns that may occur across the different topologies. Using the disulfide topology complexity measure defined in Materials and Methods, coupled with the counterclockwise ordering of topologies in increasing complexity, we observe that the first three least complex topologies exhibit the greatest diversity in disulfide patterns: 1–2_3–4_5–6 encompasses 31% of the clusters, 1–4_2–3_5–6 encompasses 11% of the clusters, and 1–3_2–4_5–6 encompasses 8% of the clusters. Alone, these three topologies make up half of the clusters in the three-disulfide classification wheel.
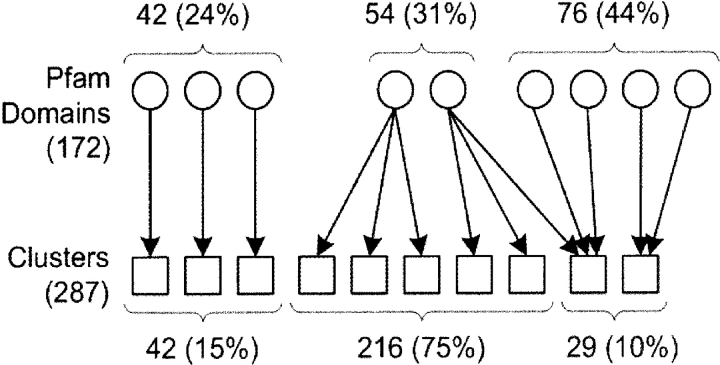
For 118 (42 plus 76) of the 172 Pfam domains (69%) represented in the three-disulfide classification wheel, all of the disulfide patterns belonging to a domain were found grouped together into a single cluster of the classification wheel (Fig. 4 ▶). Although multiple Pfam domains can be found in a single cluster, the grouping of related patterns into a single cluster indicates that the disulfide topologies and cysteine spacings are highly conserved within these domains. In the remaining 54 domains (31%), however, we found disulfide patterns split across multiple clusters and even multiple topologies (Fig. 4 ▶). This situation of related disulfide patterns having different topologies can occur when a novel disulfide incorporates itself into the fold of the protein, displaces another disulfide present in the fold, and changes the overall disulfide connectivity of the protein domain. From a cluster perspective, 258 (216 plus 42) of 287 clusters (90%) contain only a single Pfam domain. This suggests that most disulfide patterns are associated with a unique structure and function. Interestingly, the clusters with disulfide patterns from multiple Pfam domains arise because of significant similarities in the disulfide patterns. A detailed examination of the structural and functional implications of these clusters is presented later in this paper.
Figure 4.
Relationship between Pfam domains, represented with circles, and clusters, represented with squares, in the three-disulfide classification wheel. Three scenarios are illustrated: one-to-one, one-to-many, and many-to-one relationships of Pfam domains to clusters. The percentages at the top and bottom of the figure indicate the fraction of the 172 Pfam domains and 287 clusters, respectively, that participate in the different scenarios.
Comparison of classification wheels
In Figure 5, A–C ▶, we present the classification wheels for disulfide patterns of two, four, and five disulfides, respectively. Although a smaller number of patterns are present in the two-, four-, and five-disulfide classification wheels, many important comparisons can be made with the three-disulfide classification wheel. The most striking feature observed across the wheels is the disulfide pattern diversity exhibited in the less complex topologies. The first few least complex topologies contain the greatest number of disulfide pattern clusters in the wheels. Also, we note that the fraction of clusters containing members with known three-dimensional structures for the two-disulfide and four- through nine-disulfide classification wheels ranges from 8% to 33%, comparable to that of the three-disulfide classification wheel.
Figure 5.
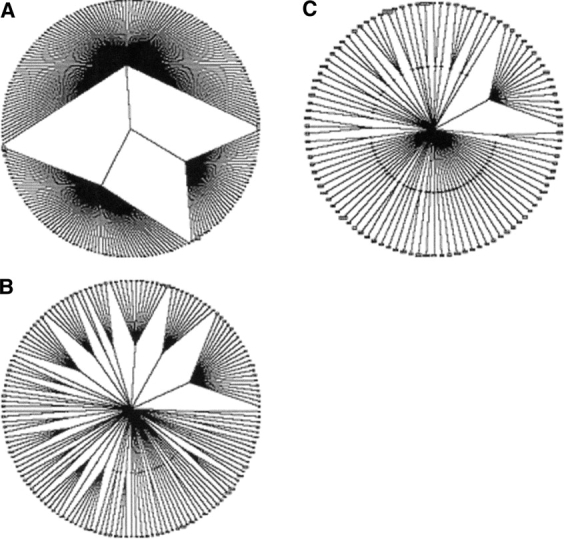
(A) Two-disulfide classification wheel. The inner rings of the wheels contain the observed topologies within the disulfide pattern length, and the outer rings contain the clusters formed in the clustering process. The organization of the wheel is the same as described for Figure 3 ▶. (B) Four-disulfide classification wheel. (C) Five-disulfide classification wheel.
As noted earlier in this section, all 15 of the possible three-disulfide topologies were observed in the database. For domains with four disulfides, 59 of the 105 (56%) possible disulfide topologies were represented by 3667 disulfide patterns in the database (Table 1). For domains with five disulfides, only 66 of the 945 (7%) possible topologies found over 1662 disulfide patterns were observed. For topologies with greater than five disulfides, <1% of the total theoretical topologies were observed. It should be noted, however, that the number of theoretical disulfide topologies increases exponentially with the number of disulfides. Benham and Jafri (1993) made some of these observations while exploring the topological properties of disulfide bonding patterns. However, a significant number of topologies were present in the database that were not previously noted by Benham and others (Thornton 1981; Benham and Jafri 1993). These new topologies were only found in topologies of more than three disulfides (Table 1), as all of the possible topologies for domains with one, two, or three disulfides were already observed. Interestingly, a few of the topologies recorded by Benham and Jafri were not found in our database. We suspect that these missing topologies are attributed to the disulfide annotations of multidomain proteins, because the Benham and Jafri analysis considered entire protein sequences rather than independent structural domains.
Table 1.
Selected clustering cutoffs and comparison of disulfide topologies observed in the disulfide database with those previously reported by Benham and Jafri (1993)
| # of disulfides | # of patterns | Clustering cutoff | # of clusters | Theoretical topologies | Topologies observed | Previously reported | New | Missing |
| 2 | 13,188 | 8 | 292 | 3 | 3 | 3 | 0 | 0 |
| 3 | 17,940 | 10 | 287 | 15 | 15 | 15 | 0 | 0 |
| 4 | 3667 | 15 | 154 | 105 | 59 | 15 | 44 | 1 |
| 5 | 1662 | 25 | 102 | 945 | 66 | 9 | 57 | 6 |
| 6 | 837 | 45 | 58 | 10,395 | 47 | 4 | 43 | 5 |
| 7 | 1038 | 50 | 36 | 135,135 | 32 | 3 | 29 | 2 |
| 8 | 629 | 50 | 29 | 2,027,025 | 25 | 1 | 24 | 2 |
| 9 | 1625 | 50 | 18 | 34,459,425 | 14 | 0 | 14 | 1 |
| 10 | 34 | 50 | 14 | 654,729,075 | 13 | 0 | 13 | 0 |
Many novel disulfide topologies have emerged in the disulfide database.
In the database, we identified multiple cases of proteins with nonplanar disulfide topologies, as defined by Benham and others (Kikuchi et al. 1986, 1988; Benham and Jafri 1993). Although only a scorpion neurotoxin was discovered previously, we found numerous proteins exhibiting nonplanar topologies from the RTI/MTI-2 protease inhibitor, γ thionin, transferrin, and long-chain scorpion toxin families. Moreover, a second, nonplanar disulfide topology 1–4_2–3_5–12_6–9_7–10_8–11_13–14 emerged in the database that had previously not been recorded. These findings demonstrate that our disulfide database, consisting of both the SwissProtextracted and inferred disulfide annotations, covers a significantly larger disulfide space.
Assessing the clustering cutoffs
As described in Materials and Methods, a detailed analysis of the cutoffs used in the clustering process was conducted to optimize the grouping of similar disulfide patterns. When more tolerant clustering cutoffs were applied, we observed significant variation in the disulfide patterns and we found multiple unrelated Pfam domains present in the same cluster. When we reduced the clustering cutoffs, less variation across the disulfide pattern coordinates was observed. Moreover, we found disulfide signatures separating such that only related sequences were found grouped together into the same cluster. On optimization of the clustering cutoffs for each wheel, similar, less varying clusters were created across all of the classification wheels. The clustering cutoff values selected for the different classification wheels are shown in Table 1.
We calculated the overlap between clusters by using the described techniques in order to assess how well the clusters separated. Each cluster was assigned a disulfide pattern range, defined by the minimum and maximum values observed for each position of the disulfide patterns encompassed within the clusters. For the two-disulfide classification wheel, we found that ~6% of the patterns in the wheel fit into the disulfide pattern ranges of more than one cluster in the wheel. This nontrivial overlap was not observed, however, in the other classification wheels. Although several of the disulfide pattern ranges overlapped slightly in the 3- through 10-disulfide classification wheels, we discovered only one example of a disulfide pattern fitting within the disulfide pattern ranges of two different clusters. No other overlaps were found in the 4- through 10-disulfide classification wheels. This indicates that our clusters are well separated for disulfide patterns with three or more disulfides. Moreover, this indicates that the classification of a given disulfide pattern with greater than two disulfides is unambiguous.
Structural comparison of similar disulfide patterns from different Pfam domains
In cases where multiple Pfam domains were grouped together into the same cluster, we consulted the Structural Classification of Proteins (SCOP), Revision 1.61 (Murzin et al. 1995) to assess the validity of the classification on the basis of structural arguments. For each of the clusters with multiple Pfam domains in the three-, four-, and five-disulfide classification wheels, we performed all of the possible pairwise structural comparisons between the PDBs of Pfam domains in a given cluster to identify the greatest level of structural similarity designated in SCOP (Table 2). We limited our measure of similarity to the first four levels of increasing similarity in SCOP: class, fold, superfamily, and family. When structural information was not available for a given Pfam domain, we ignored any pairwise comparisons involving that domain. We were thus able to perform 225 (27%) of the possible 838 pairwise comparisons. We found that Pfam domains that grouped together in the four- or five-disulfide classification wheels generally exhibited high structural similarity. Across the three-, four-, and five-disulfide classification wheels, more than half of the pairwise comparisons performed reflected structural similarities on at least the fold level. About 19% of the pairwise comparisons indicated structural similarities on the family or superfamily level, which strongly suggests common evolutionary origins (Murzin et al. 1995). The pairwise comparisons reflecting structural similarities on the fold level highlight the ability of the disulfide classification to group together structurally related proteins that would be otherwise difficult to relate without knowledge of their three-dimensional structures.
Table 2.
Clusters containing multiple Pfam domains
| SCOP similarity | ||||||||
| Cluster # | Pfam domains present | Sequence identity (%) | Structural comparison pairs | None | cl | cf | sf | fa |
| 5-Disulfide classification wheel | ||||||||
| 25 | PB000034, PB004006, PB017918 | 14.5%–23.1% | 0 | — | — | — | — | — |
| 83 | PB004042, PB073771, PF00021, PF00087, PF01064 | 9.8%–54.5% | 3 | — | — | — | 67% | 33% |
| 4-Disulfide classification wheel | ||||||||
| 9 | PF00219, PF01033 | 14.8%–29.5% | 0 | — | — | — | — | — |
| 59 | PF00021, PF00053, PF00087, PF01064 | 10.3%–29.4%* | 6 | — | 50% | — | 33% | 17% |
| 100 | PB013405, PB036929, PF02819, PF05309 | 11.4%–63.9% | 3 | — | — | 67% | — | 33% |
| 101 | PF00537, PF05353 | 19.0%–23.8% | 1 | — | — | 100% | — | — |
| 149 | PB008170, PF00304, PF00537 | 6.5%–33.3% | 1 | — | — | — | 100% | — |
| 3-Disulfide classification wheel | ||||||||
| 3 | PB000034, PB008407, PB017282, PF00053, PF00086, PF01033 | 4.5%–32.4% | 3 | — | 67% | — | 33% | — |
| 10 | PB000034, PB007041 | 21.8%–23.3% | 1 | 100% | — | — | — | — |
| 90 | PB000320, PB058864, PB071582, PF00020, PF00246, PF00429, PF00713 | 7.9%–31.0% | 10 | 10% | 70% | 10% | — | 10% |
| 105 | PB074800, PF00020 | 55.2%–58.6% | 0 | — | — | — | — | — |
| 123 | PF00008, PF00053, PF00187, PF00219, PF00757, PF01826 | 6.2%–68.1%* | 45 | — | 36% | 33% | 16% | 16% |
| 146 | PB024067, PB055043, PF00057 | 12.1%–43.9%* | 1 | — | — | 100% | — | — |
| 148 | PF05337, PF02947 | 17.6%–21.9% | 0 | — | — | — | — | — |
| 152 | PF00087, PF00184 | 25.0%–26.6% | 1 | 100% | — | — | — | — |
| 188 | PB01046, PB011477, PB014575, PB0160009, PB022013, PB023815, PB038421, PB038777, PB047402, PB053988, PB054370, PB074066, PB074072, PB074098, PF00187, PF00299, PF00304, PF00451, PF00537, PF01097, PF01821, PF02048, PF02822, PF02950, PF02977, PF03488, PF03784, PF05196, PF05374 | 2.6%–92.6% | 136 | 23% | 10% | 54% | 10% | 3% |
| 194 | PF00019, PF00341 | 9.7%–29.4% | 1 | — | — | — | 100% | — |
| 199 | PB018619, PF00074 | 12.6%–24.1% | 0 | — | — | — | — | — |
| 203 | PB012724, PB024890, PF00200, PF05375 | 9.5%–30.2% | 3 | 67% | 33% | — | — | — |
| 219 | PB014575, PB037861, PB045373, PF00050, PF00088, PF00323, PF00711, PF00819, PF01147, PF04736 | 4.8%–40.5%* | 6 | — | 83% | — | — | — |
| 229 | PB002338, PB047330 | 12.2%–12.2% | 0 | — | — | — | — | — |
| 263 | PF00323, PF01549, PF03913 | 6.8%–38.9% | 3 | — | 100% | — | — | — |
| 274 | PB027670, PF00321 | 15.9%–15.9% | 0 | — | — | — | — | — |
| 280 | PF00024, PF01421 | 14.1%–22.8% | 1 | 100% | — | — | — | — |
Only matching clusters from the three-, four-, and five-disulfide classification wheels are shown here. A structural analysis of the clusters using SCOP is also included. cl, cf, sf, and fa indicate the first four levels of structural homology: class, fold, superfamily, and family. The range of sequence identities for sequences across different Pfam domains is also provided. An asterisk in the “Sequence identity” column indicates that the range was calculated on a randomly selected set of 15 sequences from the cluster.
We carefully explored examples of multiple Pfam domains and “NULL” domains clustering together for homologous structures or functions. In several cases, similarities between related proteins could not be found through sequence comparison means because no significant sequence similarity was present. We find that the homologies in these cases have been determined only through analyses of their three-dimensional structures. Interestingly, these structural relationships could have been made solely through comparisons of their disulfide patterns. A complete listing of the clusters containing multiple domains is shown in Table 2. For each cluster, a range of percent sequence identities across the Pfam domains present in the cluster is included. These identities are calculated by first aligning the disulfide-containing sequence domains of different domain families, using the Needleman-Wunsch algorithm (Needleman and Wunsch 1970). For clusters with >500 disulfide patterns (indicated in Table 2 with an asterisk), 15 sequences were randomly selected from each domain to be used in the sequence identity range calculation. In the following paragraphs, we explore several disulfide pattern clusters in detail to highlight some of the interesting cases found in the database.
Single domain family example: Cluster 3.1–3_2–4_5–6.121
As noted previously, the majority of Pfam domains (69%) represented in the three-disulfide classification wheel appear in a single cluster per domain basis. One of these families, the papain family cysteine proteases (PF00112), appears in cluster 121 of the three-disulfide classification wheel. The parallel plot of the disulfide signatures for this family (Fig. 6A ▶) illustrates the high degree of similarity observed among the related patterns. Of the 350 disulfide patterns grouped together in the cluster, 79% are inferred disulfide patterns generated from the inferring algorithms described in Materials and Methods. The remaining disulfide patterns are extracted directly from SwissProt. The disulfide patterns with defined domain boundaries in Pfam are annotated with the “PF00112” class assignment, and the patterns without defined boundaries are annotated with “NULL” class assignment. The SwissProt functional annotations for the “NULL” disulfide patterns indicate that the proteins are indeed related to the other sequence domains of the PF00112 family. A superposition of five representative three-dimensional structures associated with the patterns in this cluster is shown in Figure 6B ▶. The low average RMSD (1.32 Å ± 0.30 Å Cα atoms) of the superposition reflects the strong structural conservation across the cluster’s disulfide patterns.
Figure 6.
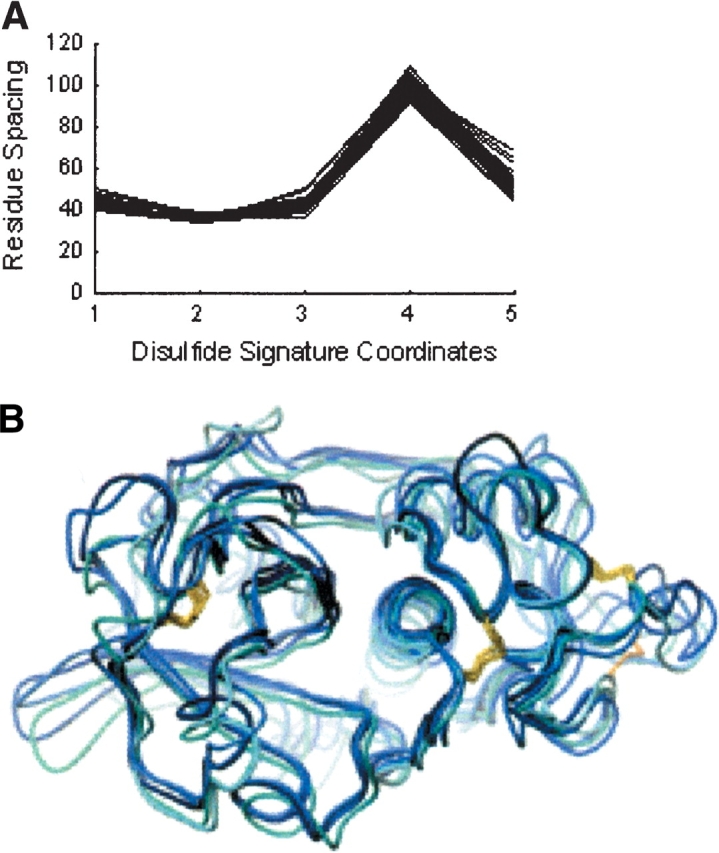
(A) Parallel-coordinate plot of the PF00112 family disulfide signatures contained in cluster 3.1–3_2–4_5–6.121. Axes parallel to the Y-axis are constructed for each coordinate of the disulfide signature and equally spaced across the X-axis. Disulfide signatures are represented by connecting the values of each sequential coordinate along its respective axis. (B) Superposition of representative three-dimensional PDB structures (1aec, 1bp4, 1f2c, 1k3b, 7pck) from the PF00112 family in cluster 3.1–3_2–4_5–6.121. The structures were superimposed by using the Ca atoms of the matching Cys residues. The different structures are colored in shades of blue. The side chains of the cysteines involved in disulfides are shown in yellow. 1k3b has a unique disulfide shown in orange on the bottom right and lacks the disulfide shown on the far left.
Multiple related domain families example: Cluster 5.1–5_2–3_4–6_7–8_9–10.83
Six domain families cluster together in cluster 83 of the five-disulfide classification wheel. This situation of multiple Pfam domains grouping together into the same cluster occurs in <10% of the clusters of the 3- through 10-disulfide classification wheels. In this cluster, a few disulfide patterns are assigned to belong to “NULL” domain and therefore correspond to sequence segments not present in Pfam. Two Pfam-B domains, PB004042 and PB073771, appear in the cluster and are annotated in Pfam as related to the Pfam-A u-PAR/Ly-6 domain (PF00021), which also appears in the cluster. This situation of related sequences not coupled with their Pfam-A domain counterparts arises when sequences in the automatically generated Pfam-B alignments have not yet been manually reviewed and appended to their corresponding Pfam-A domains. The disulfide patterns from these Pfam-B domains consist mostly of sperm acrosomal proteins. Although no structural information exists for these proteins, the functional annotations indicate the presence of Ly-6 domains within the sequences. Moreover, the Swiss-Prot entries corresponding to these proteins do not contain any disulfide annotations: the disulfide patterns used in the clustering are derived from the disulfide inferring algorithms. The inclusion of these sequences in the cluster highlights the capacity of the inferred disulfide annotations to encompass a much greater disulfide space than that explicitly annotated in SwissProt.
A second Pfam-A domain, the snake toxin family (PF00087), and a third Pfam-A domain, Activin Receptor Types I and II extracellular domain (PF01064), are also grouped into the cluster. The snake toxin and u-PAR/Ly-6 domain families have a previously documented structural and functional relationship, yet lack any significant sequence similarity (Palfree 1996). The Activin Receptor family also lacks any significant sequence similarity with the other Pfam-A domain families in this cluster. PSI-BLAST searches with a reasonable cutoff (E-value <0.01) on the NR database were unsuccessful in reporting similarities between the three Pfam-A families when sequences from the Activin or snake toxin families were selected as the query sequences. However, PSI-BLAST searches using sequences from the u-PAR/Ly-6 domains were able to find related sequences from the Activin and snake toxin families.
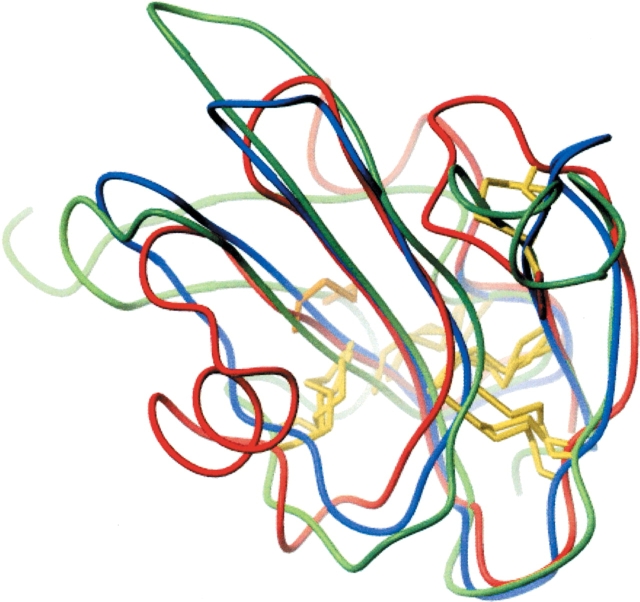
Both the Activin receptor domain family and the u-PAR/ Ly-6 domain family are extracellular domains of cell surface receptors. SCOP classifies the Activin Type II Receptors and u-PAR/Ly-6 domains together on the family level, implying that an evolutionary relationship exists between the two. Furthermore, superposition using Combinatorial Extension (Shindyalov and Bourne 1998) of representative structures from each domain family resulted in RMSD values ranging from 2.3 Å to 6.6 Å (Z-scores ranging from 3.1 to 3.3; Fig. 7 ▶). This cluster highlights the effectiveness of the disulfide classification in grouping together domain families with clear structural and functional homologies, despite the absence of significant sequence similarity.
Figure 7.
Superposition of representative PDB structures from the snake toxin (1cdq, red), u-PAR/Ly-6 domain (1f94, blue), and Activin Receptor Types I and II extracellular domains (1bte, green). Note the unique disul-fide of 1bte shown in orange. Compared with the two other structures, 1bte lacks the disulfide shown in the upper right part of the structure.
Multiple related domain families example: Cluster 3.1–4_2–5_3–6.194
In cluster 194 of the three-disulfide classification wheel, disulfide patterns from the TGF-β-like domain family and the platelet-derived growth factor family appear together. The corresponding sequences of these domains exhibit very low sequence similarity to one another (~11%), yet a structural and functional homology between these two protein families has been noted (Murray-Rust et al. 1993). This relationship was discovered only after three-dimensional structures from both protein families were determined. Combinatorial Extension applied to representative PDB structures from both families (1tfg and 1pdg, respectively) yields an RMSD score of 4.0 Å (Cα only) and a Z-score of 3.3. Both families are classified together at the SCOP superfamily level, which suggests a probable evolutionary relationship. Once again, the disulfide classification effectively groups together distantly related proteins using only disulfide spacing and cysteine connectivity information.
Multiple unrelated domain families example: Cluster 3.1–4_2–5_3–6.188
A large number of Pfam-A domains and automatically generated Pfam-B domains are found grouped together in cluster 188 of the three-disulfide classification wheel. A considerable diversity of protein functions is observed in the cluster. Only one other cluster, present in the four-disulfide classification wheel, exhibits the same vast diversity of protein functions as witnessed in this cluster. Some of the protein families represented in the cluster—the scorpion toxins, omegatoxins, muconotoxins, plant lectins, and defensins—have long been known to share common structural and functional relationships; however, the other domain families present in the cluster—the proteinase inhibitors, cyclotides, antistatins, and conotoxins—do not have any homologous relationships with one another. Sequence similarity between proteins of the related domains was typically low, ranging from 8% to 33%. PSI-BLAST searches with an E-value cutoff of .01 were unable to report relationships between the related protein families in almost all of the cases.
A prominent feature of the disulfide patterns in this cluster is the relatively short length of the protein domains (average 40 residues). The disulfide patterns in the cluster therefore reflect closely spaced cysteines with little freedom to vary across the different domain families. This cluster reveals that a small fraction of unrelated sequences are inevitably clustered together because of their short sequences and limited variability in cysteine spacing.
Analysis of loss and gain of disulfides within a Pfam domain family
While exploring related disulfide patterns, we found that disulfide patterns from the same Pfam domain family often varied in the number of disulfides. Therefore, we tabulated the relative loss or gain of disulfides across all of the sequences within a domain family for all Pfam domains appearing in the database. The most represented number of disulfides per sequence within a family was designated as the reference number of disulfides for that family. The change in the number of disulfides for patterns in a family was calculated relative to the reference number of disulfides for that family. Across all of the Pfam domains represented in the disulfide database, ~10% of the disulfide patterns per family lost or gained one disulfide when compared with the reference value. The frequency of patterns losing or gaining two disulfides was ~2%, and the frequency for shifts of three or more disulfides was <1%. Numerous examples of disulfide patterns both losing one disulfide and gaining another were also observed in the database. These exchanges of disulfides, a net change of zero disulfides for the domain, often accompanied changes in the overall disulfide topology of the domain as well. In these types of situations, it may be difficult to recognize similarities between patterns by using the disulfide pattern similarity measure (equation 1); however, by comparing the appropriate subsets of these disulfide patterns, relationships between patterns can often be revealed, as is shown in the following example.
Linking the three-, four-, and five-disulfide classification wheels
Using the techniques outlined in Materials and Methods, we formed links between clusters of different classification wheels. The links formed between the three-, four-, and five-disulfide classification wheels are illustrated in Figure 8A ▶. These connected graphs or extended clusters were generated to accommodate for differences in the number of disulfides across related disulfide patterns. The trypsin family domain (PF00089), for example, exhibits significant diversity in its disulfide patterns. The SwissProt sequence CFAD_HUMAN contains a trypsin domain with the disulfide signature 16–97–66–31–16–25–25. Similarly, the sequence CATG_HUMAN contains a trypsin domain with the disulfide signature 16–93–65–30–14. When comparing the disulfide pattern subsets of CFAD_HUMAN with the CATG_HUMAN domain, we find that the subset containing the first three disulfides has a high similarity (dmn = 4.69) to the CATG_HUMAN disulfide pattern. Because the similarity score between the two patterns is less than the clustering cutoff used in the three-disulfide classification wheel (10), the clusters containing both of these sequences are linked together by our linking algorithm. We also observed links between the CFAD_HUMAN disulfide pattern and patterns where the first, second, or third disulfide was removed.
Figure 8.
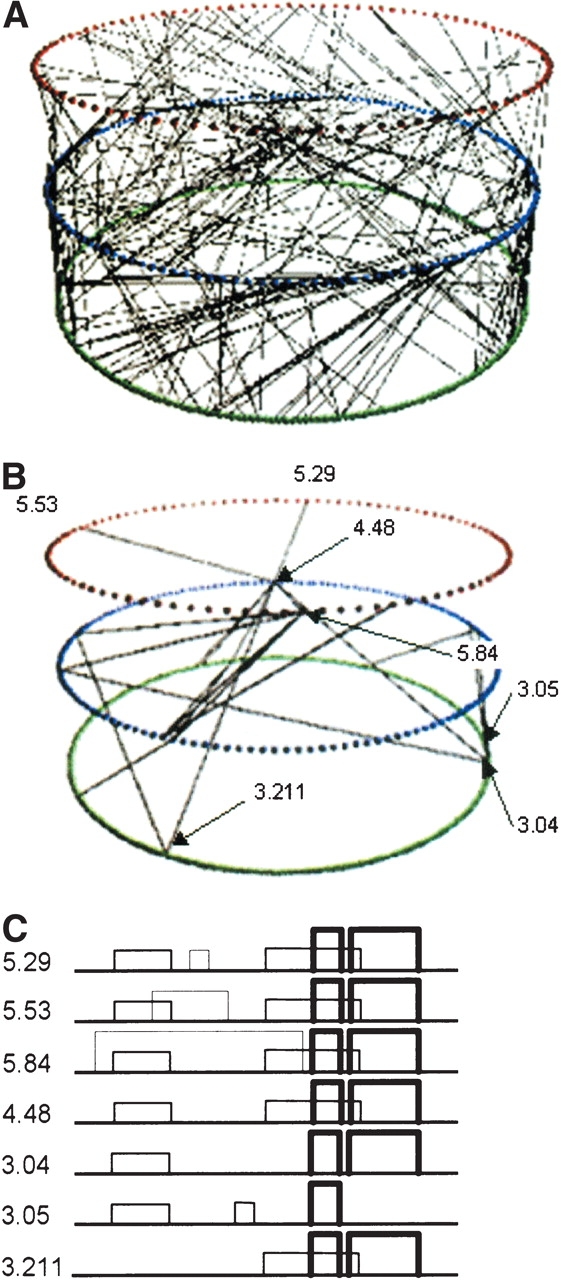
(A) Representation of linking between the three-, four-, and five-disulfide classification wheels. The three-disulfide classification wheel is shown in green, the four-disulfide classification wheel is shown in blue, and the five-disulfide classification wheel is shown in red. (B) The subset of links in panel A that form the subgraph encompassing the trypsin family. (C) Comparison of different disulfide patterns in the subgraph containing trypsin family members.
Examining the other trypsin family disulfide patterns, we find disulfide patterns from the trypsin family are distributed among eight clusters in the three-disulfide classification wheel, seven clusters in the four-disulfide classification wheel, and four clusters in the five-disulfide classification wheel. Within a classification wheel, clusters were also found to occur across different topologies. The subgraph searching algorithms described in Materials and Methods were applied to isolate the networks of connected clusters containing trypsin family members. Of the 38 separate networks of connected clusters present across the three-, four-, and five-disulfide classification wheels, the subgraph search tool found only one network that contained trypsin family members. Moreover, this single network did not encompass any other Pfam domains and successfully united 18 of the 19 trypsin family clusters (357 of 367 trypsin family disulfide patterns) across the different classification wheels. The cluster links associated with this subgraph are shown in Figure 8B ▶. In Figure 8C ▶, we illustrate representative disulfide patterns for a small subset of the clusters. The latter two disulfides, indicated with a thick line width, are highly conserved across these clusters. The disulfide pattern for cluster 3.1_2–3_4–5_6.5 (shown as “3.05”) is the only pattern lacking one of the latter two disulfides. This cluster is the only one that contains trypsin family members but was not linked together into the trypsin subgraph. The variation observed in this family illustrates the importance of exploring disulfide patterns with different numbers of disulfides when searching for related proteins.
Discussion
We have created a novel classification of disulfide bonding patterns that effectively groups together proteins with related structures and functions. Constructed with three levels of organization, the disulfide classification is the first broad approach to understanding disulfide space.
Our disulfide classification builds on previously reported approaches in several important regards. First, our classification is applied to disulfide patterns from the recently reported disulfide database (van Vlijmen et al. 2004), which increased the number of annotated disulfide patterns from 16,736 to 94,499, and thus covers a significantly greater disulfide space than previous approaches. This is evident from the 224 disulfide topologies present in the disulfide database that were not observed by Benham and others (Table 1; Thornton 1981; Benham and Jafri 1993). The second factor distinguishing our classification from previous approaches is the implementation of the disulfide signature as the third level of organization, which essentially incorporates the spacing between cysteines. Whereas previous approaches performed poorly at grouping related proteins, the inclusion of disulfide signatures in the classification results in a correct grouping of proteins with related structures and function. The majority of clusters are associated with proteins from a single Pfam family, which suggests a strong relationship between disulfide patterns and protein structure and function. In clusters that contain representatives from multiple Pfam families, we also observed a clear grouping of proteins with related structure and function, even in cases of low sequence similarity. Table 2 shows the range of pairwise sequence similarities found within clusters containing multiple Pfam domains. Here, we see multiple cases of the disulfide classification in which sequences are correctly clustered together despite very low sequence similarity (<20%). In addition, we found that more than half of the clusters containing multiple Pfam domains had structural similarities on at least the SCOP fold level (Table 2). While this manuscript was being completed, Chuang et al. (2003) published a paper in which the relationship between the disulfide pattern and protein structure was explored for 3134 protein structures from the PDB database. Although Chuang et al. (2003) use a different measure of disulfide similarity, their findings support the connection between disulfide pattern and structure described here. Because our analysis used the significantly larger database of 94,499 disulfide-containing domains, the vast majority of which are not represented in the PDB, we were able to establish the strong relationship between disulfide patterns and protein function. Also, we developed a means to find relationships between disulfide subpatterns of proteins, which was not reported by Chuang et al. We have highlighted the importance of this type of analysis with the trypsin family example, where we were able to show clusters of related disulfide patterns with different numbers of disulfides and disulfide topologies connected together through their subpatterns.
As the grouping of similar disulfide patterns depends on the clustering cutoffs used in the classification, we validated the selected clustering cutoffs in terms of separability among the clusters and structural overlap in clusters with multiple Pfam domains represented. We found ample separation between clusters and we are confident that the selected clustering cutoffs accurately partition the disulfide patterns.
As noted in our previous work (van Vlijmen et al. 2004), we made several assumptions regarding the SwissProt disulfide annotations. Because of the lack of any explicit definition for experimentally determined disulfides, we assumed disulfides without any similarity annotations such as “By Similarity,” “Potential,” or “Probable” to be experimentally determined. To clarify the source and reliability of the disulfide information, we propose a restructuring of the disulfide annotation standard in SwissProt. We believe that disulfide annotations should be accompanied by references to the source from which they are obtained. In the case of disulfides that are inferred through homology with another protein, the homologous protein should be referenced in the annotation. Moreover, the functional roles of disulfides should be reflected in the annotations when applicable—for example, the regulatory function of disulfides in thioredoxin (Yano et al. 2002).
As shown in the Results, it is common for proteins within the same family to drop or add one or more disulfides. It is therefore important to relate clusters with different numbers of disulfides and correspondingly different classification wheels. We implemented an algorithm to link clusters of patterns across different topologies and classification wheels. The example of the trypsin family inhibitors clearly illustrates the effectiveness of this algorithm in uniting 357 of 367 related disulfide patterns across the multiple classification wheels. The resulting network of connected clusters encompasses many disulfide patterns whose relationship is not obvious in pairwise comparisons. We are currently exploring the correspondence of connectivity in these disulfide networks to evolutionary distance.
In addition to broadly classifying disulfide space, the di-sulfide classification wheels can be used as an aid in curating disulfide-containing Pfam domains. The second example described in the Results, cluster 5.1–5_2–3_4–6_7–8_9–10.83, illustrates the ability of the classification to associate two automatically generated Pfam-B domains with a related Pfam-A domain. Similarly, disulfide-annotated protein domains that are present in SwissProt but have not yet been included into Pfam (thus assigned to the “NULL” domain in the disulfide database) are clustered together with their corresponding Pfam domains using the disulfide classification. As noted previously, this grouping of related disulfide-containing domains occurs even in cases of low sequence similarity.
The classification of disulfide patterns is also a useful resource for structural genomics efforts. Disulfide-containing proteins are good targets for structural elucidation, given the fact that 72% of known disulfide-containing proteins have their own or a family member’s structure determined. The majority of disulfide pattern clusters are associated with a single Pfam domain family, in many cases having a unique structure. As indicated in the Results, the majority of the clusters in the three-, four-, and five-disulfide classification wheels do not have references to any structural information. These clusters correspond to two possibilities. In the first, no structures have been solved for any of the family members of the Pfams represented in the cluster. These protein domains are important targets for structural studies, but they can easily be identified by querying the Pfam database. The second case involves clusters with members that have at least one Pfam family member with a known structure, who is a member of a different cluster. This situation occurs when the disulfide topology is different or when the spacings between the cysteines are sufficiently different to place it in a different cluster. The proteins from each of these clusters are also prime targets for structural determination, because they contain disulfide topologies and/or spacings that are not yet represented in the PDB and may have novel structures or structural features. The identification of these targets requires the disulfide classification presented in this paper.
The disulfide classification presented here can be used to annotate protein domains for function and/or structure by using only disulfide patterns. This may prove useful in cases where sequence comparisons are ambiguous and X-ray or NMR studies are difficult but experimental disulfide determination is feasible. Association of the experimental pattern with one of the clusters will usually assign a unique function and structure, as most clusters correspond to a single Pfam family or to a group of related Pfam families. In summary, we have created a classification of disulfide patterns that effectively groups together distantly related proteins and is useful for understanding the role of disulfides in protein structure and function.
Materials and methods
Disulfide bond similarity
As described previously (van Vlijmen et al. 2004), we explore disulfide bond similarity through disulfide topology and cysteine spacing. We define a measure of disulfide pattern similarity, such that for two disulfide patterns m and n, the disulfide pattern similarity is equal to the pairwise Euclidean distance, dmn (equation 1).
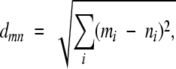 |
(1) |
where the index i sums over all integers in the disulfide pattern. Smaller distance values suggest higher degrees of similarity between patterns. As described previously (Fig. 1 ▶), disulfide patterns can correspond to disulfide signatures or to cysteine spacing patterns. Also, we arbitrarily define the “length” of a disulfide pattern as equal to the number of disulfides that the pattern represents. The disulfide pattern similarity measure (equation 1) can only be applied to disulfide patterns of the same length and correspondingly same number of disulfides. Similarity between patterns with different numbers of disulfides is evaluated using an approach based on comparing subpatterns of the disulfide patterns. This technique is presented later in this section.
Disulfide pattern database
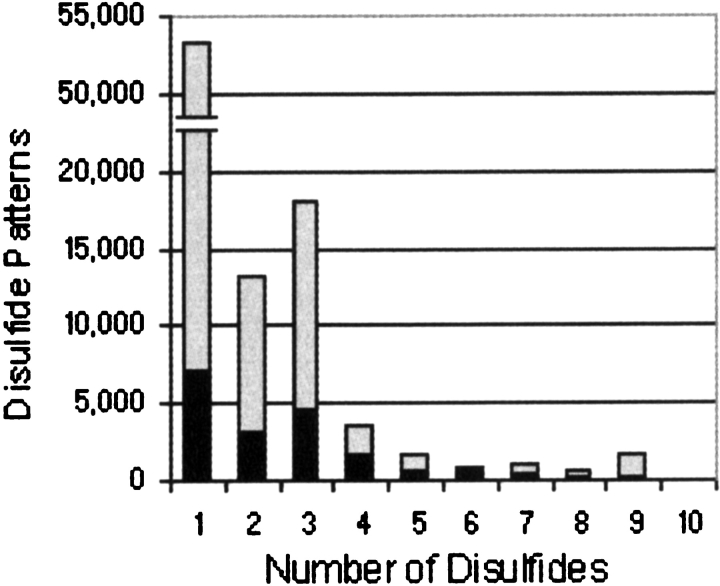
Using previously published methods (van Vlijmen et al. 2004), we constructed an initial database of disulfide patterns extracted from SwissProt annotations. Both experimentally determined and inferred disulfide annotations were incorporated into the database; however, interchain and ambiguous disulfide annotations were ignored. SwissPfam, a component of the Pfam database, was used to identify segments of disulfide-containing sequences that corresponded to Pfam-A or Pfam-B domains. Both Pfam-A and Pfam-B multiple alignments contain SwissProt and TrEmbl sequences; however, Pfam-A alignments are hand curated and Pfam-B alignments are automatically generated. As SwissProt sequences often contain multiple Pfam domains, the SwissProt-extracted disulfide patterns were subdivided according to the Pfam domain segments from which they originate. Only disulfides where both cysteines of the disulfides occur completely inside or outside Pfam domains were retained; all other disulfides were regarded as interdomain and discarded. The disulfides in a sequence occurring outside Pfam domains were grouped together across each individual sequence, assigned as belonging to the “NULL” domain, and appended to the database as independent disulfide patterns. A total of 2514 “NULL” domain disulfide patterns were generated in this way. Next, the Pfam multiple alignments with disulfide-containing sequence domains were used to infer additional disulfide patterns. This was achieved by first identifying the pairs of columns in each alignment that contained cysteines that participated in forming disulfides. All of the sequences in an alignment were then scanned for cysteines occurring in both columns of each column pair. Pairs of cysteines that satisfied these conditions were assumed to form disulfides and also inserted into the database. These inferred disulfide patterns were distinguished by appending “X” to the end of the Pfam family from which they were derived. Inferred patterns that exhibited any ambiguities such as two or more disulfides sharing a common cysteine were ignored. Including both Swiss-Protextracted and inferred annotations, 40,750 (43%) disulfide patterns in the database contain two or more disulfides (Fig. 9 ▶).
Figure 9.
Distribution of the number of disulfides per domain in the disulfide database. SwissProt-extracted disulfide annotations are shown in black, and inferred disulfide annotations are shown in gray.
Disulfide pattern classification
The classification of disulfide patterns has a structure consisting of three tiers. The first tier of the classification involves separating disulfide patterns by the number of disulfides. We restricted the classification to disulfide patterns with more than a single disulfide. The second tier of the classification entails partitioning disulfide patterns by their disulfide topologies. Only disulfide topologies observed in the database are considered at this level; theoretical topologies not observed thus far are ignored. The final tier of the classification involves grouping disulfide patterns on the basis of their similarity to one another, as defined by the pairwise distance dmn (equation 1). This is accomplished by applying the single-linkage, hierarchical clustering algorithm available with MatLab (Version 6.5, Release 13; Mathworks, Inc.) to the disulfide signatures of proteins sharing the same disulfide topology.
The clustering cutoffs used in generating the clusters were individually selected for each disulfide pattern length. Hierarchical-tree dendrograms of the disulfide pattern similarities were generated to aid in the selection of an appropriate cluster cutoff. In addition, parallel-coordinate plots of individual clusters’ disulfide signatures, where each position of a disulfide signature was regarded as a coordinate, were generated to visualize variation across the patterns. Higher, more tolerant cutoffs resulted in greater variation within a cluster, whereas smaller, more constraining cutoffs resulted in less variation. This process of applying a clustering cutoff, examining the resulting clusters, and revising the cutoff value was iteratively applied until an optimal cutoff value was attained. We define the optimal cutoff as the point where the grouping of related disulfide patterns (those sharing the same Pfam domain) is maximized and the grouping of unrelated disulfide patterns is minimized. Sufficient resolution existed between related and unrelated disulfide patterns to enable such an approach. Once determined, the cutoff was uniformly applied to all topologies with the same number of disulfides. The overlap between the formed clusters was calculated to evaluate how well the selected clustering cutoff separated the clusters. Each cluster was designated a band or range of values, called the disulfide pattern range, for each position in the disulfide pattern string. The range was defined by the minimum and maximum values at the same position across other disulfide patterns in a cluster. Next, disulfide patterns from other clusters, sharing the same topology, were tested for inclusion within the disulfide pattern range of a given cluster. This process was repeated for all clusters of the same number of disulfides.
Visualizing the disulfide classification
The graphing toolkit GraphViz (AT&T Research Labs) was used to create visual depictions of the disulfide pattern classification. The representations, referred to as the “classification wheels,” were arranged in a wheel shape composed of two concentric rings of nodes connected by lines extending radially outward. The two rings correspond to the latter two tiers of the classification. Separate wheels were constructed for each disulfide pattern length from 2 through 10 disulfides. The wheels were labeled in the center with a number indicating the length of the disulfide patterns present in the classification wheel. Elliptical nodes were constructed for each of the observed topologies within the specific disulfide pattern length and placed in the inner concentric ring. Topologies on the classification wheel were ordered by complexity, such that less complex topologies were present in the first quadrant of the wheel and progressively more complex topologies would appear in a counterclockwise fashion. Our definition of disulfide topology complexity differs from Benham and others (Kikuchi et al. 1986, 1988; Benham and Jafri 1993), as it is primarily dependent on two factors: the total number of intersections and overlaps occurring between cysteine pairs. An intersection occurs when a cysteine of one disulfide pair (x1, x2) lies in-between the cysteines of another disulfide pair (a1,a2),
| (2) |
Similarly, an overlap of disulfide pairs occur when one disulfide pair (x1,x2) is completely encompassed within another disulfide pair (a1,a2),
| (3) |
Every topology observed in a given classification wheel was assigned a complexity score, defined as the sum of the number of intersections and overlaps, and ranked against other topologies sharing the same number of disulfides. Topologies with the same complexity score were delineated first by symmetry, as defined by Benham and Jafri (1993), and finally alphanumerically. Nonsymmetrical topologies were considered more complex than symmetrical topologies. This approach does not definitively separate one topology’s complexity from another; however, it effectively separates less complex topologies from more complex ones such that general trends between the two may be observed.
The clusters of similar disulfide patterns generated from the clustering process were represented with rectangles placed in the outer concentric ring of the classification wheel. Each cluster was given an annotation that included the cluster identifier and details about the contents of the cluster. The cluster identifier is made up of three components: the length of the disulfide patterns represented, the disulfide topology under which the cluster belongs, and the cluster number. The values for these three descriptors are separated by periods and concatenated together to form the cluster identifier string. For example, in the cluster identifier 3.1–3_2–4_5–6.121, the “3” indicates that each of the disulfide patterns contained in the cluster has three disulfides, and the “1–3_2–4_5–6” reveals the topology of the patterns present in the cluster. The last part of the cluster identifier, “121”, is the cluster’s assigned number within the three-disulfide classification wheel. Other information present in the annotation includes the distribution of Pfam domains represented in the cluster as well as the consensus disulfide patterns computed for the cluster. The consensus disulfide patterns, defined by the average for each position of the disulfide pattern strings contained within a cluster, were calculated for both disulfide signatures and cysteine spacing patterns. Lastly, references to available structural information, PDB or Homology-derived Secondary Structures of Proteins; (Sander and Schneider 1991), were also included in the annotation. These references were obtained from either SwissProt or Pfam structural annotations.
Linking classification trees
Links between clusters of different disulfide pattern lengths were constructed forming connected graphs, which were regarded as extended clusters. These links were only generated between clusters of pattern length N - 1 or N - 2 and a cluster of pattern length N. The links thus correspond to the elimination of one or two disulfides, respectively. The links between the clusters were determined by first generating all N - 1 and N - 2 length subpatterns for every N length disulfide pattern. The subpatterns were then compared with the classified patterns of corresponding length in the N - 1 or N - 2 classification wheels. The disulfide topology constraint was maintained in these comparisons such that only patterns of equivalent topologies to the subset patterns were compared. If the similarity score calculated between a subpattern and a classified pattern was below the cutoff used in the hierarchical clustering of the respective N - 1 or N - 2 classification wheels, a link was drawn between the cluster from which the subpattern originated and the cluster containing the classified pattern. This technique was recursively applied to all disulfide patterns of length 3 through 10. In the case of the disulfide patterns with three disulfides, only the N - 1 subpatterns were generated, as the disulfide classification is only applied to patterns with two or more disulfides. The discrete networks of connected clusters formed in the linking process were then determined and information about the encompassed disulfide patterns (i.e., Pfam distribution, structural information) was generated.
Acknowledgments
We thank L. Narasimhan and K. Guruprasad for initial discussions on disulfide bond analysis. We thank Sarah Papp for calculating the clusters that contain structural information. Also, we thank A. Lukashin and R. Fuchs for discussions on clustering algorithms.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04613004.
References
- Benham, C.J. and Jafri, M.S. 1993. Disulfide bonding patterns and protein topologies. Protein Sci. 2 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O’Donovan, C., Phan, I., et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.C., Chen, C.Y., Yang, J.M., Lyu, P.C., and Hwang, J.K. 2003. Relationship between protein structures and disulfide-bonding patterns. Proteins 53 1–5. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. 1988. Disulphide bonds and protein stability. Bioessays 8 57–63. [DOI] [PubMed] [Google Scholar]
- Foley, S.F., van Vlijmen, H.W.T., Boynton, R.E., Adkins, H.B., Cheung, A.E., Singh, J., Sanicola, M., Young, C., and Wen, D. 2003. The CRIPTO/FRL-1/CRYPTIC (CFC) domain of human Cripto. Functional and structural insights through disulfide structure analysis. Eur. J. Biochem. 270 3610–3618. [DOI] [PubMed] [Google Scholar]
- Gorman, J.J., Wallis, T.P., and Pitt, J.J. 2002. Protein disulfide bond determination by mass spectrometry. Mass Spectrom. Rev. 21 183–216. [DOI] [PubMed] [Google Scholar]
- Harrison, P.M. and Sternberg, M.J. 1996. The disulphide β-cross: From cystine geometry and clustering to classification of small disulphide-rich protein folds. J. Mol. Biol. 264 603–623. [DOI] [PubMed] [Google Scholar]
- Kikuchi, T., Nemethy, G., and Scheraga, H.A. 1986. Spatial geometric arrangements of disulfide-crosslinked loops in proteins. J. Comput. Chem. 7 67–88. [Google Scholar]
- ———. 1988. Spatial geometric arrangements of disulfide-crosslinked loops in nonplanar proteins. J. Comput. Chem. 10 287–294. [Google Scholar]
- Mas, J.M., Aloy, P., Marti-Renom, M.A., Oliva, B., Blanco-Aparicio, C., Molina, M.A., de Llorens, R., Querol, E., and Aviles, F.X. 1998. Protein similarities beyond disulphide bridge topology. J. Mol. Biol. 284 541–548. [DOI] [PubMed] [Google Scholar]
- Mas, J.M., Aloy, P., Marti-Renom, M.A., Oliva, B., de Llorens, R., Aviles, F.X., and Querol, E. 2001. Classification of protein disulphide-bridge topologies. J. Comput. Aided Mol. Des. 15 477–487. [DOI] [PubMed] [Google Scholar]
- Murray-Rust, J., McDonald, N.Q., Blundell, T.L., Hosang, M., Oefner, C., Winkler, F., and Bradshaw, R.A. 1993. Topological similarities in TGF-β 2, PDGF-BB and NGF define a superfamily of polypeptide growth factors. Structure 1 153–159. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247 536–540. [DOI] [PubMed] [Google Scholar]
- Needleman, S.B. and Wunsch, C.D. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48 443–453. [DOI] [PubMed] [Google Scholar]
- Palfree, R.G. 1996. Ly-6-domain proteins—New insights and new members: A C-terminal Ly-6 domain in sperm acrosomal protein SP-10. Tissue Antigens 48 71–79. [DOI] [PubMed] [Google Scholar]
- Ryle, A.P., Sanger, F., Smith, L.F., and Kitai, R. 1955. The disulphide bonds of insulin. Biochem. J. 60 541–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, C. and Schneider, R. 1991. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9 56–68. [DOI] [PubMed] [Google Scholar]
- Sanger, F. 1959. Chemistry of insulin: Determination of the structure of insulin opens the way to greater understanding of life processes. Science 129 1340–1344. [DOI] [PubMed] [Google Scholar]
- Shindyalov, I.N. and Bourne, P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11 739–747. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L., Eddy, S.R., and Durbin, R. 1997. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 28 405–420. [DOI] [PubMed] [Google Scholar]
- Thornton, J.M. 1981. Disulphide bridges in globular proteins. J. Mol. Biol. 151 261–287. [DOI] [PubMed] [Google Scholar]
- van Vlijmen, H., Gupta, A., and Singh, J. 2004. A novel database of disulfide patterns and its application to the discovery of distantly related homologs. J. Mol. Biol. 335 1083–1092. [DOI] [PubMed] [Google Scholar]
- Yano, H., Kuroda, S., and Buchanan, B.B. 2002. Disulfide proteome in the analysis of protein function and structure. Proteomics 2 1090–1096. [DOI] [PubMed] [Google Scholar]