Abstract
A model for the human δ opioid receptor has been generated via sequence alignment, structure building using the crystal structure of bovine rhodopsin as a template, and refinement by molecular dynamics simulation. The model building suggested that, in addition to the previously postulated interaction between D128 and Y308, an internal salt bridge also exists between residues D128 and R192, both of which are conserved in all the opioid receptors. The model and salt bridge were then shown to be stable during a 20-nsec simulation in a lipid bilayer. It is therefore proposed that both of these interactions play a role in stabilizing the inactive state of the receptor. The model is also used in an effort to rationalize many of the mutational studies performed on δ opioid receptors, and to suggest a plausible explanation for the differences between known δ opioid agonists and antagonists.
Keywords: molecular dynamics, bovine rhodopsin, GPCR, agonist-antagonist binding, activation mechanism
The opioid receptors are integral membrane proteins of the central nervous system implicated in mediating the analgesic effects of opium derived alkaloids, endogenous ligands such as the enkephalins, and their precursors (Akil et al. 1984; Kieffer et al. 1992; Chaturvedi et al. 2000; Smith and Lee 2003). In addition to analgesia, the opioids generate a multitude of effects, including euphoria, sedation, respiratory depression, muscle rigidity, and a potential for physical dependence (Chaturvedi et al. 2000). Pharmacological studies have shown the existence of at least three different classes of opioid receptors (μ, δ, andκ), which differ in their anatomical distributions and pharmacological profiles (Simonds 1988; Kieffer et al. 1992). A large number of synthetic peptides have been developed to bind to these receptors, and it has been shown that selective recognition at the receptor is facilitated by two aromatic rings and a positively charged N terminus (Feinberg et al. 1976; Smith and Griffin 1978; Chao et al. 1996; Shenderovich et al. 2000), whereas nonspecific recognition requires a protonated amine, two hydrophobic groups, and a centroid of aromatic ring (Filizola et al. 2001). To gain further insight into the differences among the various opioids receptors, and to understand the ligand-receptor interactions in detail, a knowledge of the receptor structure at the atomic level is desirable. Unfortunately, because of their size and the inherent obstacles in crystallizing complex membrane proteins, no experimental three-dimensional structure of an opioid receptor is currently available.
The cloning studies of δ (Evans et al. 1992; Kieffer et al. 1992), followed by μ (Chen et al. 1993) and κ (Meng et al. 1993), have demonstrated that these receptors belong to a G protein–coupled receptor (GPCR) super family (Wess 1998; Smith and Lee 2003) characterized by seven hydrophobic transmembrane (TM) helices (TM1–7) connected by alternating intracellular (ICL1–3) and extracellular loops (ECL1–3; Chabre 1985; Baldwin 1993). The N terminus is located on the extracellular side of the membrane, whereas the C terminus is on the intracellular side (Baldwin 1993). The GPCR acts as a link between the extracellular ligand and the intracellular G protein (Gilman 1987; Ji et al. 1998; Gether 2000; Fabian 2001; Christopoulos and Kenakin 2002; Gether et al. 2002; Wong 2002; Bissantz 2003; Karnik et al. 2003). Binding of an agonist to the inactive receptor leads to a structural change in the receptor primarily involving movement of helices III, VI, and VII (Gether 2000; Bissantz 2003; Decaillot et al. 2003; Karnik et al. 2003). The active state of the receptor can then couple to a G protein via interactions with the intracellular loops (and the C-terminal in some receptors), which then initiates the subsequent intracellular signaling cascade (Strader et al. 1994; Wess 1998; Gether 2000; Horn et al. 2000; Chan et al. 2003; Breitwieser 2004). The opioid receptors are members of family A GPCRs, which include rhodopsin-like receptors (Gether 2000). Several highly conserved features are observed in Family A receptors. These include the disulfide bond linking TM3 and ECL2, the DRY motif in TM3, and an NPxxY motif in TM7 (Baldwin et al. 1997; Gether 2000; Karnik et al. 2003). It is therefore believed that these residues have a significant role in the structure and function of the receptors.
Even though the molecular basis of binding and the activation mechanism for opioid receptors are still unknown, site-directed mutagenesis studies have provided insights into the various residues and possible interactions important for binding and activation (Gioannini et al. 1989; Kong et al. 1993; Fukuda et al. 1995; Befort et al. 1996a, b, 1999; Meng et al. 1996; Valiquette et al. 1996; Pepin et al. 1997; Chaturvedi et al. 2000; Xu et al. 2000; Hosohata et al. 2001; Decaillot et al. 2003). Here we will focus on the δ opioid receptor (DOR) as there is growing evidence that stimulation of the DOR mediates analgesia but without the addictive properties of morphine associated with μ receptors (Rapaka and Porreca 1991). Hence, the DOR is a prime target for drug design. The sequence numbers refer to the human DOR (hDOR).
Numerous studies have been performed to identify the residues involved in the binding of ligands to the DOR (Gioannini et al. 1989; Kong et al. 1993; Fukuda et al. 1995; Metzer and Ferguson 1995; Befort et al. 1996a, b; Meng et al. 1996; Valiquette et al. 1996; Pepin et al. 1997; Mosberg 1999; Chaturvedi et al. 2000; Xu et al. 2000). Even though the opioids share a large repertoire of ligands, a positively charged N atom is required for agonist binding (Feinberg et al. 1976; Smith and Griffin 1978; Chao et al. 1996; Shenderovich et al. 2000). This has led to the idea that a negatively charged amino acid side chain may be involved in a direct interaction with the cationic ligand via a salt bridge. Examination of the acidic residues in opioid receptors implicated an aspartic acid in TM3 (D128) as being the most likely anionic counterpart (Befort et al. 1996b). Interestingly, mutation studies have shown that the D128A mutation does not affect ligand binding, but does result in agonist independent activation, which cannot be further enhanced on agonist binding. In contrast, the corresponding D128N mutation results in reduced binding and an active receptor that is further activated by δ agonists. It is therefore believed that cationic opioids bind in the region of D128 via formation of an ionic interaction and that, although the Asp side chain is not essential for agonist binding in all opioid receptors, it is important for stabilization of the receptor in the inactive conformation (Befort et al. 1996a, b, 1999; Chaturvedi et al. 2000; McFadyen et al. 2002; Decaillot et al. 2003). A number of other residues have also been implicated in opioid binding (Gioannini et al. 1989; Kong et al. 1993; Fukuda et al. 1995; Metzer and Ferguson 1995; Befort et al. 1996a, b; Meng et al. 1996; Valiquette et al. 1996; Pepin et al. 1997; Mosberg 1999; Chaturvedi et al. 2000; Xu et al. 2000). However, the precise position of the binding pocket is still not clear.
The receptor undergoes a transition from inactive to active state on binding of an agonist. It is therefore assumed that the inactive state is stabilized by a series of interactions that have to be broken during the activation process. Hence, the determination of amino acid substitutions that lead to mutant receptors with significant agonist independent constitutive activity (CAM) is of particular interest. Site-directed mutagenesis studies have revealed a number of CAMs for the DOR (Befort et al. 1999; Decaillot et al. 2003). One of the proposed interactions considered to be broken during activation of DORs involves residues D128 and Y308 (Befort et al. 1999; McFadyen et al. 2002; Decaillot et al. 2003), based on the fact that disruption of a corresponding interaction between TM3 and TM7 is the primary trigger for the conformational change in opsins (Robinson et al. 1992; Bissantz 2003). Molecular modeling studies of DORs have also suggested a possible hydrogen bond between D128 and Y308, leading to stabilization of the inactive form of the receptor (Befort et al. 1999; Mosberg 1999; Decaillot et al. 2003). Mutations of D128 and Y308 have been investigated and shown to be important for signal transduction as well as ligand recognition at the DOR (Befort et al. 1996a, b, 1999; Chaturvedi et al. 2000; Decaillot et al. 2003). The D128A/N and Y308F/H mutants result in constitutive activation of the receptor (Befort et al. 1999; Decaillot et al. 2003). However, although the D128A mutant cannot be further activated by ligand, the Y308F mutant is further activated on agonist binding. The difference in behavior between the D128A and the Y308F mutants suggests that the proposed hydrogen bond is not the only interaction stabilizing the inactive state of the receptor, and that the Asp and/or Tyr must therefore be involved in additional interactions (Befort et al. 1999). The identification of CAMs in other TM helices also indicates that several additional residues are of importance for maintaining the receptor in the inactive state (Decaillot et al. 2003). In addition, a conserved disulfide bridge (C121 to C198) has been shown to be essential for ligand binding and maintaining the structural integrity of the receptor (Gioannini et al. 1989).
In the absence of a crystal structure of the DOR, several tertiary structure models have been proposed. The models are based on the rhodopsin projection maps or the bovine rhodopsin crystal structure, as well as comparison with the various mutation studies (Alkorta and Loew 1996; Strahs and Weinstein 1997; Pogozheva et al. 1998; Brandt et al. 1999; Filizola et al. 1999a,b; Chaturvedi et al. 2000; McFadyen et al. 2002; Bissantz et al. 2003; Decaillot et al. 2003). These models have been used in an effort to explain the mutational data concerning ligand binding and receptor activation. However, in doing so one must keep in mind that it is generally difficult to explain all the mutations with any particular model. This is due to the fact that mutational studies pertaining to the structure-function of the receptor are often based on a loss of function strategy. Unfortunately, a loss of function could also be due to changes in the trafficking efficiency of the receptor to the cell surface or receptor misfolding (Gether et al. 1997; Pepin et al. 1997).
Initial GPCR models used a structure of the light sensitive bacteriorhodopsin as a template (Henderson et al. 1990; Grigorieff et al. 1996; McFadyen et al. 2002). However, although bacteriorhodopsin shares the seven-TM helix motif characteristic of GPCRs, it does not couple to G proteins, and later comparison with rhodopsin revealed marked differences in their three-dimensional structures (Henderson et al. 1990; Schertler et al. 1993; Unger et al. 1997; McFadyen et al. 2002). Subsequent efforts have used the rhodopsin projection maps (Alkorta and Loew 1996; Strahs and Weinstein 1997; Pogozheva et al. 1998; Filizola et al. 1999a,b) and, more recently, the crystal structure of rhodopsin to construct DOR models to explain the mutational data (Mc-Fadyen et al. 2002; Bissantz et al. 2003; Decaillot et al. 2003). Most of the recent models based on the rhodopsin crystal structure contain a hydrogen bond between D128 and Y308.
Molecular dynamics simulations of various DOR models with or without a ligand have also appeared in the literature. Strahs and Weinstein (1997) used molecular modeling to construct models for the helical regions of the three opioid receptors based on the low-resolution projection map of bovine rhodopsin. The models were stable for 2 nsec of molecular dynamics simulation, and some correlation between the motions of different helices was observed. Filizola et al. (1999b) investigated the details of the binding and activation mechanism of DORs using molecular dynamics simulations of the three opioid receptor models in the presence and absence of agonists. It was observed that the salt bridge located in the DRY motif, which is considered to play a central role in G-protein activation (Acharya and Karnik 1996; Lu et al. 1997; Scheer et al. 1997, 2000; Ballesteros et al. 1998; Rasmussen et al. 1999; Alewijnse et al. 2000), was maintained in the absence of bound ligand but was broken in models including a bound agonist (Filizola et al. 1999b). More recently, a simulation of the κ opioid receptor in a phospholipid bilayer was described in which different aspects of ligand binding were examined (Iadanza et al. 2002).
Even though the wealth of information gained over the past decade has improved our knowledge of the DOR, as well as other GPCRs, the binding and activation processes are still not fully understood. In particular, it is clear that the receptor is held in the inactive state by more than just the postulated D128–Y308 interaction. Hence, we have investigated the DOR for other possible interactions. A model of the DOR was generated by using homology modeling. The stability of the model was then established by molecular dynamics simulations for 20 nsec in a lipid bilayer. A key feature of the model is a proposed salt bridge between D128 of TM3 and R192 of ECL2, both of which are conserved between opioid receptors and across different species. This interaction is implicated as another possible stabilizing interaction, in addition to D128–Y308, holding the receptor in the inactive state. The final model is then used in an attempt to explain many of the known amino acid mutation studies for the DOR.
Results
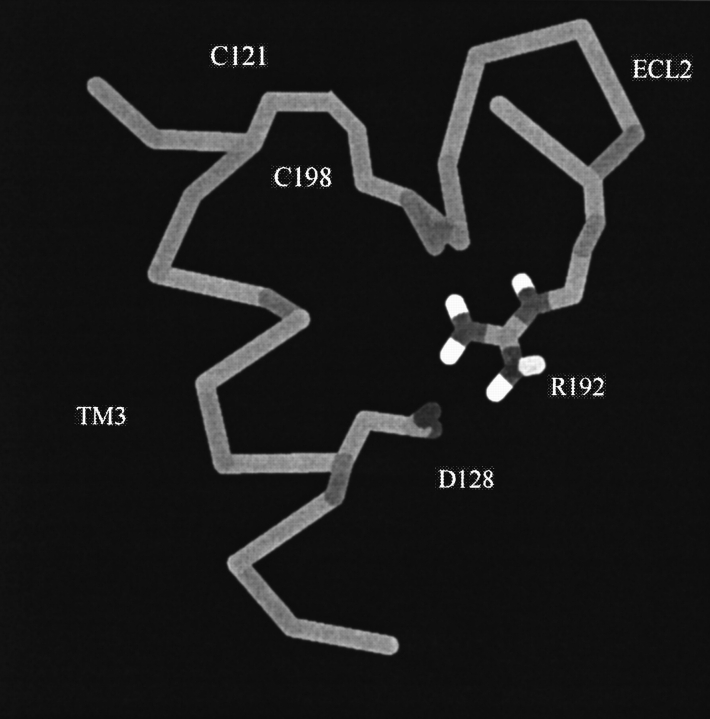
The hDOR was aligned with the sequence of bovine rhodopsin by using the GAP alignment as shown in Figure 1 ▶. Model helix fragments were built by using the hDOR sequence and then least squares fitted to the rhodopsin crystal structure (Protein Data Bank [PDB] code 1L9H). Details of the alignment and model building are presented in Materials and Methods. A careful visualization of the area around R192 in ECL2 indicated that the side chain Cβ atom was pointing into the protein and toward D128. ECL2 is almost completely buried in the protein and not exposed to the solvent. With this initial orientation, the charged Arg side chain could not reach the protein surface and be solvated. It was also noticeable that there were no other δ opioid conserved charged residues or hydrogen bonding groups in the vicinity of the Asp side chain. In addition, this region is quite nonpolar, suggesting it would not contain a significant number of water molecules. Therefore, as the distance between the main chain of the Asp and the Arg was reasonable to accommodate what is a very favorable interaction when both groups are desolvated, a salt bridge was introduced between D128 and R192. The initial salt bridge arrangement is displayed in Figure 2 ▶.
Figure 1.
Final sequence alignment for TM1–7 and ECL2 of bovine rhodopsin (bRho) and the hDOR used to build the receptor model. Vertical lines correspond to an alignment of identical residues, and the dots indicate degrees of similarity between aligned residues. Sequence similarities for the above fragments are 38% (TM1), 34% (TM2), 49% (TM3), 39% (TM4), 60% (TM5), 55% (TM6), 41% (TM7), and 33% (ECL2). The most conserved residue between all GPCRs in each helix is underlined (Ballesteros and Weinstein 1995). The highlighted residues (C121, D128, R192, C198, and Y308) are conserved among all the known opioid receptors and are of particular interest to this study.
Figure 2.
The D128-to-R192 salt bridge after model building and energy minimization. The salt bridge connects residues in TM3 and ECL2, which are also connected via a disulfide bridge (C121 to C198).
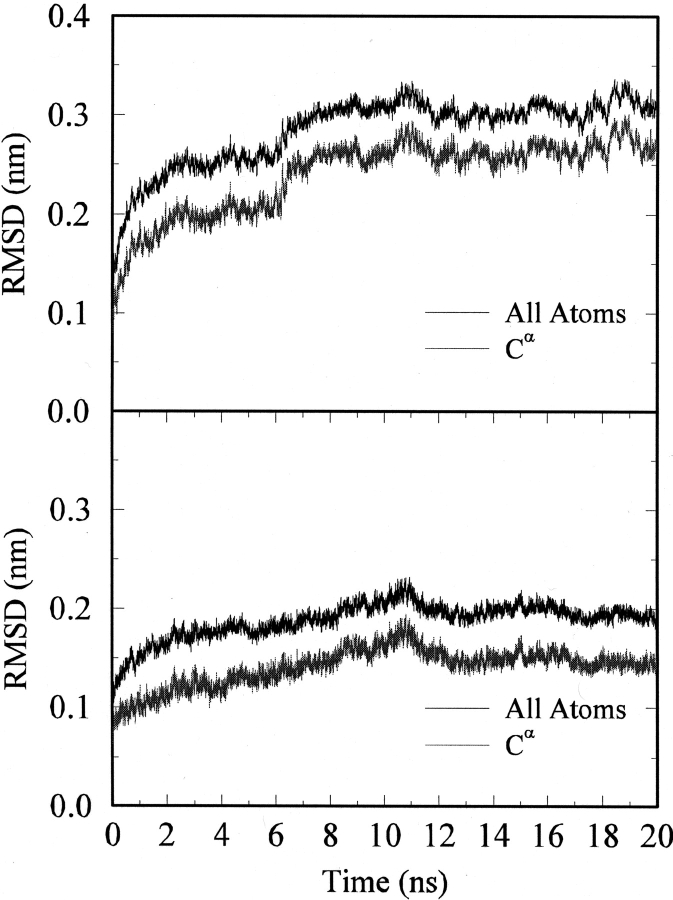
After model building and refinement, the receptor was simulated in an explicit lipid environment for 20 nsec in an effort to refine the structure and establish the stability of the model in general and the salt bridge in particular. The initial configuration of the system is shown in Figure 3 ▶. The root mean square deviation (RMSD) from the initial model-built structure is displayed in Figure 4 ▶ as a function of simulation time. The overall deviation increased with time for the first 10 nsec and then remained relatively constant, indicating that the structure was not changing in a systematic manner. The largest structural change occurred between 5 and 10 nsec and involved the rearrangement of ICL3. This was not too surprising as the corresponding loop in the rhodopsin crystal structure is poorly defined. The time history for the RMSD of the Cα atoms corresponding to the seven helices and ECL2 reached a maximum deviation of 0.15 nm after 10 nsec. This is reasonable for a model-built structure and suggested that the system had converged to a stable structure, or at least a stable local minimum, which was close to the starting structure. More importantly, the rearrangement of ICL3 indicated that unstable regions of the model would have been expected to show large fluctuations over this time period (Voordijk et al. 2000; Lei and Smith 2003).
Figure 3.
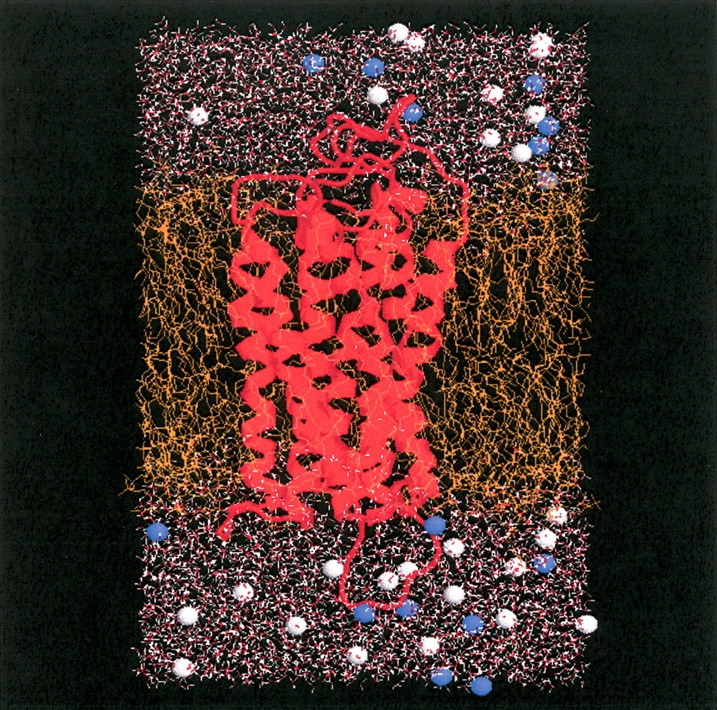
The starting configuration for the simulation of the hDOR in a lipid bilayer. The receptor is displayed as red ribbons, the POPC lipids are in orange, water is in red/white, and the blue/gray spheres indicate sodium or chloride ions.
Figure 4.
RMSD time histories obtained from the molecular dynamics simulation. (Top) The all atom RMSD (upper line) and Cα atom RMSD (lower line) of the receptor. (Bottom) The all atom RMSD (upper line) and Cα atom RMSD (lower line) for TM1–7 and ECL2 atoms.
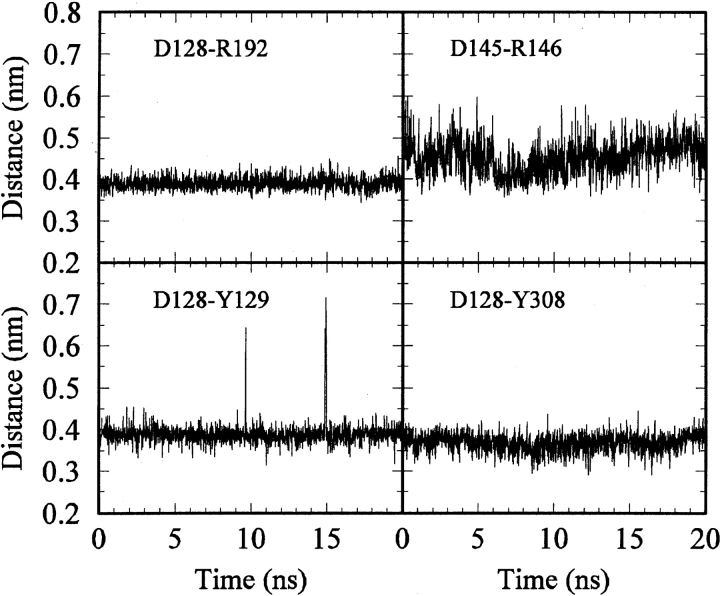
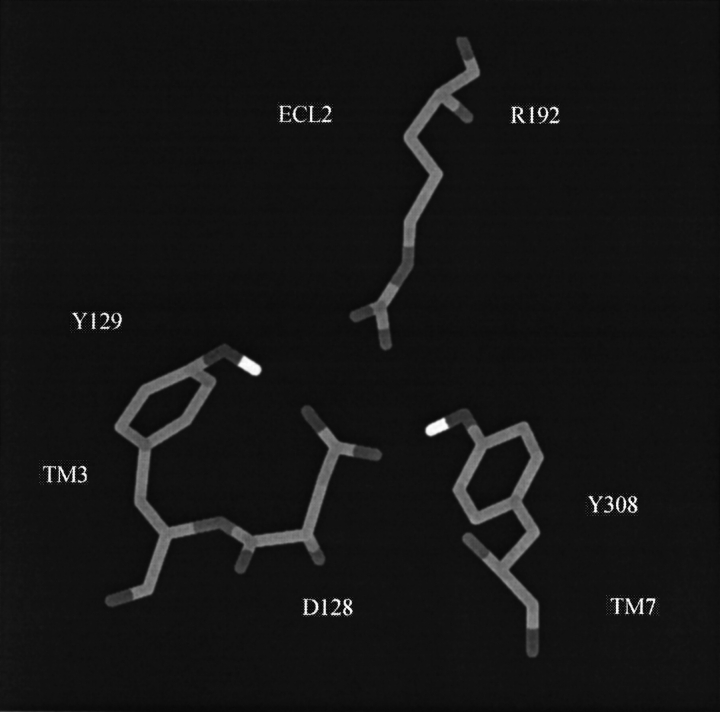
The salt bridge distance time histories are displayed in Figure 5 ▶. Both our postulated salt bridge and the known salt bridge (D145–R146) located in the DRY motif of family A receptors remained intact during the whole simulation.Average Cγ—Cζ distances of 0.40 and 0.45 nm were obtained for the D128–R192 and DRY salt bridges, respectively. From the distances and fluctuations observed in Figure 5 ▶, it appeared that the D128–R192 salt bridge was more stable than was the DRY salt bridge. This is expected based on their relative proximity to solvent. Other residues observed to interact strongly with D128 in the model were Y129 and Y308 (which was aligned with the Schiff base K296 in rhodopsin). The time histories corresponding to distances between the hydroxyl and carboxylate groups are also displayed in Figure 5 ▶. The interactions remained intact for the whole trajectory. Hence, the model building and simulation data indicated that D128 of TM3 interacted strongly with ECL2 (via R192) and TM7 (via Y308). Figure 6 ▶ displays a snapshot from the dynamics trajectory illustrating the above interactions.
Figure 5.
Time histories for selected distances obtained from the hDOR simulation. Salt bridge distances were defined by atoms Cγ and Cζ, whereas Asp-to-Tyr distances were defined by atoms Cγ and Oη.
Figure 6.
The final arrangement of the side chains in direct contact with the D128-to-R192 salt bridged side chains.
An analysis of the φ/ψ values for each residue was performed by using the PROCHECK program (Laskowski et al. 1993). The results for bovine rhodopsin crystal structure (chain A) and the final structure generated after the 20-nsec simulation are displayed in Figure 7 ▶. No residues were observed in the disallowed regions, and just seven amino acid residues were observed in the generously allowed regions of the Ramachandran plot. Analysis of structures generated after each nanosecond of simulation indicated that the same residue was never consistently observed in the generously allowed regions, and that the percentage of residues in favorable regions remained constant at 80%, with 97% observed in the favorable and additionally favorable regions. Although this is slightly lower than expected for good models of small globular water soluble proteins (usually ∼90%; Morris et al. 1992), it is very reasonable in comparison with the bovine rhodopsin crystal structure analysis (also 80% in favorable regions), and a previous model of the κ opioid receptor, which displayed 95% in favorable and additionally favorable regions (Iadanza et al. 2002). None of the residues in TM3 and ECL2 were observed in unfavorable regions of the Ramachandran plot.
Figure 7.
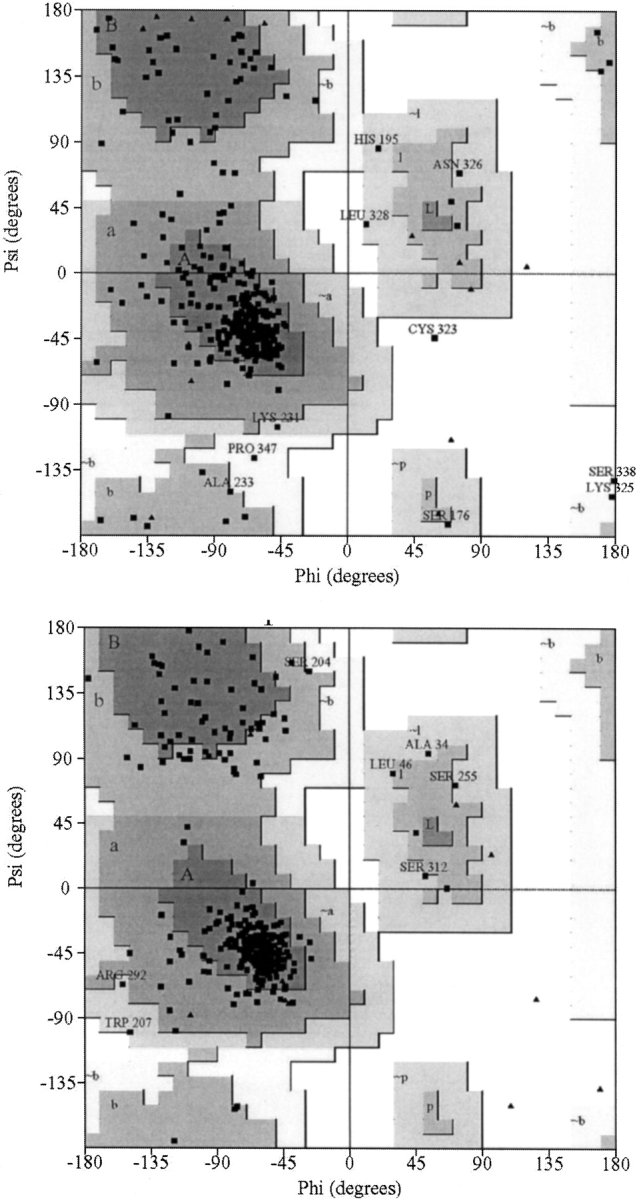
Ramachandran plots obtained by using the PROCHECK program. Results are displayed for chain A of bovine rhodopsin (top) and the final configuration obtained from the 20-nsec trajectory (bottom).
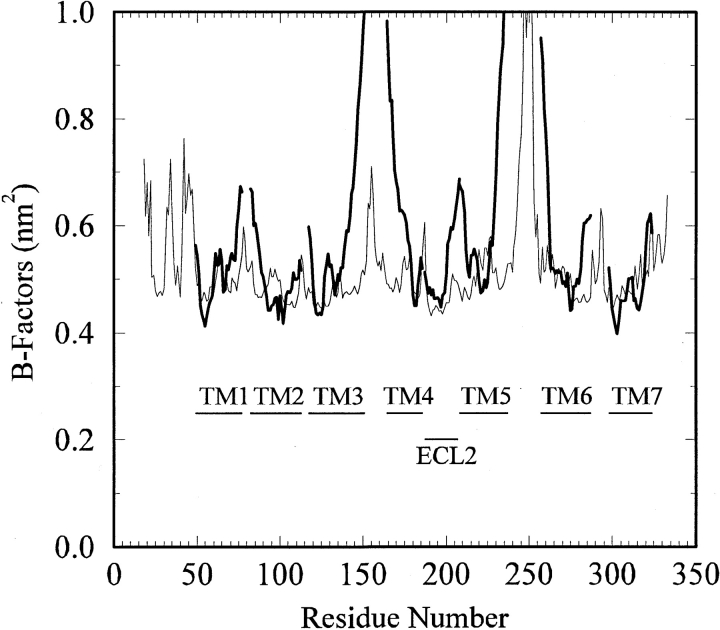
Another approach to assess the stability of the model is to quantify the atomic fluctuations in terms of B-factors (van Gunsteren and Mark 1998; Voordijk et al. 2000). The B-factors obtained from the simulation are presented in Figure 8 ▶ and compared with the corresponding values from the bovine rhodopsin crystal structure (Palczewski et al. 2000). The majority of the helical and ECL2 residues displayed low (0.1 nm2) Cα B-factors, indicating that these regions of the receptor were very stable during the simulation. Only residues located in loops or at the helix termini displayed significant flexibility. Interestingly, the two lowest B-factors corresponded to the Cα atoms of D128 and R192. Unfortunately, a direct comparison with rhodopsin is complicated by the presence of high B-factors obtained from the crystal refinement. The base line value of 0.4 nm2 suggests a significant degree of static disorder in the crystal. However, it was clear that ICL2 and ICL3 displayed significant motion during the simulation, in agreement with the rhodopsin B-factors. Furthermore, the B-factors determined here are in good agreement with the values observed for stable secondary structural elements in simulations of other membrane and globular proteins performed with the same force field (Stocker et al. 2000; Voordijk et al. 2000; Faraldo-Gomez et al. 2003; Lei and Smith 2003).
Figure 8.
B-factors for the Cα atoms obtained from the simulation (thin line) compared with the corresponding residues (TM1–7 and ECL2) for bovine rhodopsin (thick lines). The residue numbering system corresponds to that of the hDOR. The simulated data were determined after averaging over the final 10 nsec of the simulation and has been offset by 0.4 nm2 to counter the large static disorder contribution contained in the rhodopsin data.
Nine internal water molecules were included in the initial model. Five had diffused into the solvent by the end of the simulation. However, diffusion of the five waters into the bulk solvent did not appear to result in any significant structural changes in the model. The waters initially placed in the vicinity of the buried charged D95 residue remained in position for the whole simulation. In addition, the structural stability of the receptor model was also investigated by determining the fraction of helical residues, the orientation of each helix with the bilayer and with each other, and the solvent accessible surface area of the receptor, all as a function of simulation time (data not shown). None of these properties displayed any significant increases or decreases during the simulation. In summary, sequence alignment and modeling leads to a predicted model for the hDOR. Analysis of the model suggests the presence of a salt bridge between D128 and R192. Simulation of the model demonstrated that the overall structure, and the salt bridge in particular, were stable for >20 nsec.
Discussion
Structure refinement using molecular dynamics
It is known that simulation of a homology model does not necessarily improve the structure (Schonbrun et al. 2002).However, this is primarily based on the use of relatively short molecular dynamics runs. When refining a model-built structure, one would ideally prefer to perform multiple short simulations of the same system by using slightly different initial starting structures. These short simulations should still be long enough to allow for full relaxation of the initial structures to a common final structure, otherwise one will simply generate many different possible models with no way to distinguish which is correct. A recent study has shown that refinement of model-built protein structures in an explicit solvent environment does produce good agreement with experiment but often requires between 10 and 100 nsec of simulation time (Fan and Mark 2004). This is supported by the RMSD data presented in Figure 4 ▶, which indicates that the present model only converged to a stable structure after 10 nsec. Clearly, it is computationally prohibitive to perform multiple simulations of at least 10 nsec for a system of this size (25,602 atoms). Hence, we have chosen to perform a single, relatively long, simulation of the hDOR in an explicit solvent environment in an effort to validate the present model. We expect this approach to generate a very reasonable description of the seven helices and ECL2 (see below), with more uncertainty in the exact position of the remaining extracellular and intracellular loops.
Rationale for salt bridge formation
The major new feature of our proposed model for the hDOR is the presence of a salt bridge between D128 and R192. Buried salt bridges have been observed in protein structures, although the degree to which they contribute to protein stability is thought to be marginal (Waldburger et al. 1995). Our justification for a salt bridge in the hDOR is several-fold. First, the sequence alignment places both the Asp and Arg side chains (aligned with the buried A117 and E181 residues of bovine rhodopsin) in the interior of the protein. The alignment for both TM3 and ECL2 does not involve any insertions or deletions, and the same results are generated from both the GAP and BLAST algorithms. Therefore, the possibility of misalignment appears small. Second, in most situations in which a charged Asp or Arg side chain is buried in the interior of a protein, there is a significant interaction with either backbone carbonyl groups or other polar side chains (Borders et al. 1994; Giletto and Pace 1999; Winn et al. 2002). However, both the Asp and Arg groups are in close vicinity to the retinal binding site of rhodopsin. This is a nonpolar region, in both rhodopsin and our hDOR model, which is not expected to be significantly solvated. None of the previously proposed opioid receptor models have suggested the presence of solvent molecules in the vicinity of D128. Therefore, it seems improbable that both the Asp and Arg side chains could interact with either polar side-chain groups or backbone carbonyl groups (which are all involved in intrahelical hydrogen bonding). It seems more probable that the two charged side chains would interact strongly with each other in such an environment. Third, both the Asp and Arg residues are conserved between different opioid receptors and across different species. R192 is also the only positively charged residue that is close to D128 in the opioid receptor model and is conserved in all opioid receptors. Finally, the molecular dynamics simulation suggests that both the model, and the salt bridge in particular, were stable. In fact, the D128 to R192 salt bridge appeared to be more stable during the simulation than the known D145 to R146 salt bridge in the DRY motif.
Extracellular loop 2
The presence of a possible D128–R192 salt bridge clearly depends on a correct alignment of both residues. The alignment of rhodopsin and DOR TM3 is generally accepted (Pogozheva et al. 1998; Chaturvedi et al. 2000; Bissantz et al. 2003; Decaillot et al. 2003). At first sight, the alignment and structure of ECL2 is more problematic due to a high degree of expected flexibility. In fact, many previous models have not included ECL2, even though residues in this region are known to be important for binding (Li et al. 1996; Quock et al. 1999; Fabian 2001). However, on closer examination it is observed that ECL2 in rhodopsin is almost totally buried by other protein residues (Bissantz et al. 2003). This is also supported by the crystal B-factors in this region, which are more similar to the helical residues than other loop residues. The alignment of ECL2 between the receptor and rhodopsin was also very good, and no gaps were predicted. The present alignment of ECL2 is exactly the same as that proposed by a recent modeling study (Bissantz et al. 2003). These facts, and the low B-factors for ECL2 observed during the simulation, provide a reasonable degree of confidence in the position of R192.
Mechanism of DOR activation
The presence of a D128–R192 salt bridge, combined with the existing experimental mutation studies (Befort et al. 1999; Chaturvedi et al. 2000; Decaillot et al. 2003), leads to a plausible hypothesis for the first step in receptor activation. If it is considered that TM3 is mainly held in the inactive state by the presence of the known disulfide bridge (C121–C198), the assumed D128-to-Y308 interaction, and the postulated salt bridge, then binding of an agonist places the positively charged N-terminal amino group of the ligand in the vicinity of the negatively charged side chain of D128. It is possible that this is sufficient to break the D128–R192 and D128–Y308 interactions holding TM3 and TM7 in position. This would then allow TM3 and TM7 to move toward the cytoplasm in an effort to further accommodate the agonist (Decaillot et al. 2003; Karnik et al. 2003), with the disulfide acting as a molecular hinge. The net result is to expose the DRY motif and intracellular loops to the solvent so they can bind the G protein (Gether 2000; Chan et al. 2003; Karnik et al. 2003). Although this is clearly a simplified mechanism, we feel that it is a plausible one that captures the initial steps of the overall process from binding to activation in DORs. However, the exact motion of TM3 and TM7, and possibly additional helices, will involve changes in interactions with other helical side-chain residues. It is unclear from the current model exactly what these changes might be.
Comparison with mutational studies of DORs
There are several known constitutively active mutants of the DOR family that can be rationalized by the above model. The D128A mutation leads to a constitutively active mutant (Befort et al. 1999; Decaillot et al. 2003). From the proposed mechanism, it is apparent that a D128A mutation would result in elimination of the salt bridge, causing TM3 and TM7 to move toward the intracellular region, thereby resulting in increased activation. Interestingly, the D128N mutation produces basal activity intermediate between the wild type and the D128A mutant (Befort et al. 1999; Decaillot et al. 2003). In this case, it is plausible that the presence of Asn results in a weakened interaction with R192, which shifts the equilibrium toward the active form of the receptor. It has also been observed that mutation of the Tyr residues Y129 in TM3 and Y308 in TM7 (Y129F, Y129A, Y308F, and Y308H) also lead to CAMs (Befort et al. 1999; Decaillot et al. 2003). In our model, the side-chain hydroxyl groups of both Y129 and Y308 interact directly with the D128 side chain. These residues are probably involved in maintaining TM3 and TM7 in the inactive arrangement and possibly in weakening the salt bridge so that agonist binding is sufficient to induce the required conformational change. However, other studies have suggested that Y129 interacts with residues on TM4 (Strahs and Weinstein 1997), TM5 (Strahs and Weinstein 1997; Filizola et al. 1999b), or TM6 (Pogozheva et al. 1998; Befort et al. 1999). We cannot rule out these possibilities as they provide a more compelling argument for the CAM generated by the Y129F mutation. The Y308F mutation results in partial activation of the receptor, which is increased by agonist binding (Befort et al. 1999). In terms of our model, the loss of the D128–Y308 hydrogen bond interaction would shift the equilibrium toward the active state, but an agonist is required to achieve full activation by breaking the salt bridge. Interestingly, the Y308H mutation results in full activation (Decaillot et al. 2003). A possible explanation for this mutation would require a charged His (as it is buried and close to D128), which effectively mimics the N terminus of an agonist, leading to salt bridge rupture. Removal of the di-sulfide bond connecting TM3 and ECL2 is also known to be important for binding and structural stability (Gioannini et al. 1989; Shahrestanifar et al. 1996; Chaturvedi et al. 2000). As mentioned previously, not all CAMs can be explained by any model. The most significant CAM that cannot be easily explained is the C328R mutant (Decaillot et al. 2003). This is located at the cytoplasmic end of TM7 and is far removed from any of the suggested opioid binding sites.
Differences between DOR agonists and antagonists
The proposed model can also be used to help explain some of the difference between DOR agonists and antagonists. The generally accepted pharmacophores for agonist binding are a charged N-terminal amino group and two aromatic rings (Feinberg et al. 1976; Smith and Griffin 1978; Chao et al. 1996; Shenderovich et al. 2000). In the endogenous en-kephalins (YGGFX, X = L or M), these are thought to be provided by the Tyr and Phe residues. Most antagonists fall into two categories. They either have N-terminal substitution by alkyl or allyl groups (Feinberg et al. 1976; Portoghese et al. 1990; Salvadori et al. 1997; Schiller et al. 2003), or no charged N terminus (Schiller et al. 2000, 2003). We hypothesize that allyl or alkyl substitution results in a ligand that can still bind to the receptor but is incapable of providing the interaction necessary to break the proposed salt bridge. Clearly, the absence of any N-terminal charge would have a similar effect. Hence, our initial mechanism assumes that the aromatic groups contribute the most to the binding process (Befort et al. 1996b), whereas the presence of a correctly positioned charged primary amino group is required for activation via competition for D128 and salt bridge disruption. A further class of δ antagonists has been designed around the Tyr-Tic sequence (Schiller et al. 1992). Here it is observed that Tyr-L-Tic behave as δ selective antagonists but Tyr-D-Tic sequences are nonselective agonists (Schiller et al. 1992). It is plausible that the difference in chirality results in different placements of the charged N terminus, with respect to the proposed salt bridge, leading to the different receptor responses. Unfortunately, the model does not provide any immediate clues as to the binding and mechanism of inverse agonists.
Conclusions
A model for the DOR has been generated from sequence alignment, model building, and molecular dynamics simulation. It is proposed that a salt bridge structure exists between conserved residues in DORs, corresponding to residues D128 and R192 in the hDOR. The salt bridge is implicated in helping to maintain the receptor in the inactive conformation primarily by stabilizing the arrangement of TM3. The model was observed to be stable in a membrane environment for >20 nsec and suggests a reasonable explanation for many of the available single amino acid mutation data, as well as the difference between agonist and antagonist behavior of known ligands.
By using the approach outlined here, we have the most confidence in the seven helices and ECL2, in particular TM3, TM5, TM6, and ECL2, all of which displayed high similarity to rhodopsin. Furthermore, TM3 and ECL2 have no insertions or deletions and display low experimental and simulated B-factors. Hence, we consider the final structure to be a good model for DORs. Even so, the model and the proposed initial step in mechanism still have to be verified by experimental studies. The most obvious mutation site would be R192. Mutations of R192 have not appeared in the literature or the opioid receptor mutation database (Horn et al. 2003). The effects of Lys and Ala substitution could provide valuable information as to the role of R192. However, the R192A mutation might be problematic, with potential for structural changes in the receptor. In principle, one should be able to simulate a known activating mutant, and/or a bound agonist, to determine if a conformational change is observed that supports the proposed model. In practice, it is not currently possible to reach the timescales required (probably microseconds) for a system of this size.
Although the above model can be used to explain many of the known features of DORs, and to make some predictions, there are still many aspects of the receptor that are not explained. The binding site interactions are not obvious from the model. The mechanism of sodium activation is unknown, although it is thought to involve D95 (Kong et al. 1993). The exact nature of the active conformation and the movement of TM3 and the other helices still needs to be elucidated. In addition, relatively little is known concerning the interaction with G proteins (Arnis et al. 1994; Burstein et al. 1998; Gether 2000). However, we believe the proposed model represents a good starting point for many of these studies.
Materials and methods
Human DOR
The model includes residues 18–333 of the human DOR sequence as obtained from the National Center for Biotechnology Information database (account no. I38532). For simplicity, the model did not include glycosylation of residues 18 and 33, or palmitoylation of residue 333. Furthermore, although it is known that the opioid receptors exist as dimers (Cvejic and Devi 1997), we have restricted the study to a monomer as follows: (1) The structure of the dimer is unknown, (2) there is no compelling evidence for cooperative binding effects, and (3) it is computationally more efficient to study the monomer for such a large system. All previous simulations of opioid receptors have focused on receptor monomers (Strahs and Weinstein 1997; Filizola et al. 1999b; Iadanza et al. 2002). The hDOR sequence was aligned with bovine rhodopsin by using the GAP algorithm (Genetics Computer Group 1991). The initial alignment (25% identity, 51% similarity) was then modified to remove any gaps within the seven helical segments of rhodopsin in an effort to optimize the hydrogen bonding within the helices (Ballesteros and Weinstein 1995). This was achieved by shifting residues on the left or the right of the most conserved residue in each helix (Fig. 1 ▶), resulting in a small decrease in the quality of the alignment (22% identity, 44% similarity). Hence, all gaps (typically only one or two residues) were assumed to be in the loop regions connecting each helix. Particular attention was paid to the positioning of Pro residues in the helices, as they can lead to helix kinking and potential problems in the tertiary structure. A careful inspection of the helical Pro residues indicated that all except P103 were aligned with Pro residues in the bovine rhodopsin sequence. P103 is located in TM2 and was closely aligned with G89 and G90 in rhodopsin. The bovine rhodopsin crystal structure displays a helical kink in the region of residues 89 and 90, and consequently, the carbonyl group of G89 is not involved in intrahelical hydrogen bonding (Stenkamp et al. 2002). Therefore, it seems reasonable to assume that a similar kink in TM2 is induced by P103 in the hDOR.
Structure building
Model helix fragments were built by using the hDOR sequence and then least squares fitted to the rhodopsin crystal structure (PDB code 1L9H). Helix kinking was maintained by using helix segments corresponding to the sequences before and after the kink. The intracellular and extracellular loops were then placed close to the appropriate helix termini by manually fitting to the rhodopsin structure using the alignment as a guide. In regions where gaps appeared in the alignment, the two terminal loop residues were placed as close to the corresponding terminal helix residues as possible. The side chain of every residue was then examined individually and rotated to occupy the same region of space as the corresponding side chain in rhodopsin. For residues aligned with Gly in rhodopsin, the most commonly observed side-chain conformation was used (Nayeem and Scheraga 1994).
Initial structure refinement
The structure generated above was then refined by energy minimization and simulation. All residues were assigned their standard protonation states at pH 7, and histidines were kept neutral, resulting in a total charge of +11 on the receptor. None of the three histidines (H152, H278, and H301) were located in regions that would be expected to significantly alter their pKas. Nine resolved internal water molecules are observed in the bovine rhodopsin crystal structure (residues W964, W2000, W2004, W2015, W2017, W2020, W2024, W2027, W2028 for chain A) and were included in our model. In particular, two of the internal waters (W2015 and W2017) help to stabilize the buried D95 (D83 in bovine rhodopsin). Two additional internal waters were not included in our model. The first was removed due to the presence of the side chain of R192, which overlapped with W2014. The second was removed due to the alignment of L102 in hDOR with G90 in rhodopsin, which caused steric overlap of the Leu side chain with W2021. Energy minimization and molecular dynamics simulation were then performed by using the vacuum 43b1 force field (used to approximate the effect of solvent). During the minimization and simulation stages, a series of conformational constraints were applied to help retain the overall arrangement of the helices while refining the loops and side-chain conformations. The Cα atoms of all the residues displayed in Figure 1 ▶ (TM1–7 and ECL2) were positionally restrained using a force constant of 25000 kJ/mole/ nm2. In addition, all peptide T bonds were constrained to remain in the trans conformation by using a harmonic potential and a force constant of 1.0 kJ/mole/deg2. By using these constraints, 1000 steps of steepest descent energy minimization were performed followed by 1 nsec of simulation at 300 K. The final structure was then used as a starting structure for the simulation in a full lipid bilayer environment.
Receptor in a lipid bilayer
The lipid bilayer consisted of solvated 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) molecules. The initial equilibrated bilayer contained 8 × 8 × 2 POPC molecules. The hDOR was placed in the center of the lipid bilayer in such a way that the extracellular and intracellular loops were at the lipid water interface, and the long axes of the helices (Z-axis) were perpendicular to the bilayer axis. Any lipid molecules that were in steric contact with the receptor were then removed (a total of 26 lipid molecules). The system was then solvated with water, and random water molecules were replaced by sodium or chloride ions to achieve electroneutrality and a physiological ionic strength of 0.15 M. The resultant system consisted of 316 protein residues, 102 POPC molecules, 5716 water molecules, 15 sodium ions, and 26 chloride ions, for a total of 25,602 atoms in a rectangular box of approximate dimensions 6.227 × 6.102 × 9.207 nm. The system was then energy minimized and simulated at 300 K for 1 nsec by using the 45a3 force field and the positional and peptide T constraints described above. Finally, all constraints were removed and the system simulated for a further 20 nsec.
Molecular dynamics protocol
All molecular dynamics simulations were performed by using the GROMOS96 program (Scott et al. 1999) and the SPC water model (Berendsen et al. 1981). The time step was 2 fsec, and SHAKE (Ryckaert et al. 1977) was used to constrain all bond lengths with a tolerance of 10−4 nm. A twin range cutoff of 0.8 nm/1.4 nm was used, and the nonbonded pair list was updated every 10 steps. Long-range electrostatics were treated by using the Poisson-Boltzmann reaction field approach (Tironi et al. 1995), with a reaction field permittivity for SPC water of 54 (Smith and van Gunsteren 1994). This treatment of electrostatic interactions has been shown to be free from major cutoff artifacts and to provide comparable results to the Ewald technique (Tironi et al. 1995). The simulations were performed under conditions of constant temperature (300 K) and constant pressure (1 atm) using the weak coupling approach (Berendsen et al. 1984). Configurations were saved every 1 psec for analysis.
Acknowledgments
This work was supported by the NSF and the ACS Petroleum Research Fund. We thank Indira Chandrasekhar and Wilfred F. van Gunsteren for providing coordinates of an equilibrated POPC bilayer, and Henry I. Mosberg for providing us with the coordinates of his model of the DOR.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04720304.
References
- Acharya, S. and Karnik, S.S. 1996. Modulation of GDP release from transducin by the conserved Glu134–Arg135 sequence in rhodopsin. J. Biol. Chem. 271 25406–25411. [DOI] [PubMed] [Google Scholar]
- Akil, H., Watson, S.J., Young, E., Lewis, M.E., Khachaturian, H., and Walker, J.M. 1984. Endogenous opioids: Biology and function. Annu. Rev. Neurosci. 7 223–255. [DOI] [PubMed] [Google Scholar]
- Alewijnse, A.E., Timmerman, H., Jacobs, E.H., Smit, M.J., Roovers, E., Cotecchia, S., and Leurs, R. 2000. The effect of mutations in the DRY motif on the constitutive activity and structural instability of the histamine H2 receptor. Mol. Pharmacol. 57 890–898. [PubMed] [Google Scholar]
- Alkorta, I. and Loew, G.H. 1996. A 3D model of the δ opioid receptor and ligand-receptor complexes. Protein Eng. 9 573–583. [DOI] [PubMed] [Google Scholar]
- Arnis, S., Fahmy, K., Hofmann, K.P., and Sakmar, T.P. 1994. A conserved carboxylic acid group mediates light-dependent proton uptake and signaling by rhodopsin. J. Biol. Chem. 269 23879–23881. [PubMed] [Google Scholar]
- Baldwin, J.M. 1993. The probable arrangement of the helices in G protein–coupled receptors. EMBO J. 12 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, J.M., Schertler, G.F.X., and Unger, V.M. 1997. An α-carbon template for the transmembrane helices in the rhodopsin family of G-protein–coupled receptors. J. Mol. Biol. 272 144–164. [DOI] [PubMed] [Google Scholar]
- Ballesteros, J.A. and Weinstein, H. 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein–coupled receptors. Meth. Neurosci. 25 365–428. [Google Scholar]
- Ballesteros, J.A., Kitanovic, S., Guarnieri, F., Davies, P., Fromme, B.J., Konvicka, K., Chi, L., Millar, R.P., Davidson, J.S., Weinstein, H., et al. 1998. Functional microdomains in G-protein coupled receptors. J. Biol. Chem. 273 10445–10453. [DOI] [PubMed] [Google Scholar]
- Befort, K., Tabbara, D.K., Maigret, B., and Kieffer, B.L. 1996a. Role of aromatic transmembrane residues of the δ opioid receptor in ligand recognition.J. Biol. Chem. 271 10161–10168. [DOI] [PubMed] [Google Scholar]
- Befort, K., Tabbara, L., Bausch, S., Chavkin, C., Evans, C., and Kieffer, B.L.1996b. The conserved aspartate residue in the third putative transmembrane domain of the δ opioid receptor is not the anionic counterpart for cationic opiate binding but is a constituent of the receptor binding site. Mol. Pharmacol. 49 216–223. [PubMed] [Google Scholar]
- Befort, K., Zilliox, C., Filliol, D., Yue, S.Y., and Kieffer, B.L. 1999. Constitutive activation of the δ opioid receptor by mutations in transmembrane domains III and VII. J. Biol. Chem. 274 18574–18581. [DOI] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., and Hermans, J. 1981. Interaction models for water in relation to protein hydration. In Intermolecular forces (ed. B. Pullman), pp. 331–342. D. Reidel Publishing, Dordrecht, The Netherlands.
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem.Phys. 81 3684–3690. [Google Scholar]
- Bissantz, C. 2003. Conformational changes of G-protein coupled receptors during their activation by agonist binding. J. Recept. Signal Trans. 23 123–153. [DOI] [PubMed] [Google Scholar]
- Bissantz, C., Bernard, P., Hilbert, M., and Rognan, D. 2003. Protein-based virtual screening of chemical databases, II: Are homology models of G-protein coupled receptors suitable targets? Proteins 50 5–25. [DOI] [PubMed] [Google Scholar]
- Borders, C.L., Broadwater, J.A., Bekeny, P.A., Salmon, J.E., Lee, A.S., Eldridge, A.M., and Pett, V.B. 1994. A structural role for arginine in proteins: Multiple hydrogen bonds to backbone carbonyl oxygens. Protein Sci. 3 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, W., Golbraikh, A., Tager, M., and Lendeckel, U. 1999. A molecular mechanism for the cleavage of a disulfide bond as the primary function of agonist binding to G-protein coupled receptors based on theoretical calculations supported by experiments. Eur. J. Biochem. 261 89–97. [DOI] [PubMed] [Google Scholar]
- Breitwieser, G.E. 2004. G protein–coupled receptor oligomerisation. Circ. Res. 94 17–27. [DOI] [PubMed] [Google Scholar]
- Burstein, E.S., Spalding, T.A., and Brann, M.R. 1998. The second intracellular loop of the m5 muscarinic receptor is the switch which enables G-protein coupling. J. Biol. Chem. 273 24322–24327. [DOI] [PubMed] [Google Scholar]
- Chabre, M. 1985. Trigger and amplification mechanism in visual phototransduction. Ann. Rev. Biophys. Biophys. Chem. 14 331–360. [DOI] [PubMed] [Google Scholar]
- Chan, A.S.L., Law, P.Y., Loh, H.H., Ho, P.N.N., Wu, W., Chan, J.S.C., and Wong, Y.H. 2003. The first and third intracellular loops together with carboxy terminal tail of the δ opioid receptor contribute toward functional interaction with G α 16. J. Neurochem. 87 697–708. [DOI] [PubMed] [Google Scholar]
- Chao, T.M., Perez, J.J., and Loew, G.H. 1996. Characterization of the bioactive form of linear antagonists at the ω-opioid receptor. Biopolymers 38 759–768. [DOI] [PubMed] [Google Scholar]
- Chaturvedi, K., Christoffers, K.H., Singh, K., and Howells, R.D. 2000. Structure and regulation of opioid receptors. Biopolymers 55 334–346. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Mestek, A., Liu, J., Hurley, J.A., and Yu, L. 1993. Molecular cloning and functional expression of a m-opioid receptor from rat brain. Mol. Pharmacol. 44 8–12. [PubMed] [Google Scholar]
- Christopoulos, A. and Kenakin, T. 2002. G protein–coupled receptor allosterism and complexing. Pharmacol. Rev. 54 323–374. [DOI] [PubMed] [Google Scholar]
- Cvejic, S. and Devi, L.A. 1997. Dimerization of the δ opioid receptor: Implication for a role in receptor internalization. J. Biol. Chem. 272 26959–26964. [DOI] [PubMed] [Google Scholar]
- Decaillot, F.M., Befort, K., Filliol, D., Yue, S., Walker, P., and Kieffer, B.L. 2003. Opioid receptor random mutagenesis reveals a mechanism for G protein–coupled receptor activation. Nat. Struct. Biol. 10 629–636. [DOI] [PubMed] [Google Scholar]
- Evans, C.J., Keith Jr., D.E., Morrison, H., Magendzo, K., and Edwards, R.H. 1992. Cloning of a δopioid receptor by functional expression. Science 258 1952–1955. [DOI] [PubMed] [Google Scholar]
- Fabian, G. 2001. Heterotrimeric G-proteins and their role in opioid receptor function. Acta Biologica Szegediensis 45 13–21. [Google Scholar]
- Fan, H. and Mark, A.E. 2004. Refinement of homology-based protein structures by molecular dynamics simulation techniques. Protein Sci. 13 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraldo-Gomez, J.D., Smith, G.R., and Sansom, M.S.P. 2003. Molecular dynamics simulations of the bacterial outer membrane protein fluA: A comparative study of the ferrichrome-free and bound states. Biophys. J. 85 1406–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., Creese, I., and Synder, S.H. 1976. The opiate receptor: A model explaining structure-activity relationships of agonists and antagonists. Proc.Natl. Acad. Sci. 73 4215–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filizola, M., Carteni-Farina, M., and Perez, J.B. 1999a. Molecular modeling study of the differential ligand-receptor interaction at the μ, α and κ opioid receptors. J. Comput. Aided Mol. Design 13 397–407. [DOI] [PubMed] [Google Scholar]
- Filizola, M., Laakkonen, L., and Loew, G.H. 1999b. Three-dimensional modeling, ligand binding and activation studies of the cloned mouse δ, μ and κ opioid receptors. Protein Eng. 12 927–942. [DOI] [PubMed] [Google Scholar]
- Filizola, M., Villar, H.O., and Loew, G.H. 2001. Molecular determinants of nonspecific recognition of δ, μ and κ opioid receptors. Bioorg. Med. Chem. 9 69–76. [DOI] [PubMed] [Google Scholar]
- Fukuda, K., Kato, S., and Mori, K. 1995. Location of regions of the opioid receptor involved in selective agonist binding. J. Biol. Chem. 270 6702–6709. [DOI] [PubMed] [Google Scholar]
- Genetics Computer Group. 1991. Program manual for the GCG package, version 7. Genetics Computer Group, Madison, WI.
- Gether, U. 2000. Uncovering molecular mechanisms involved in activation of G protein–coupled receptors. Endocrine Rev. 21 90–113. [DOI] [PubMed] [Google Scholar]
- Gether, U., Ballesteros, J.A., Seifert, R., Sanders-Bush, E., Weinstein, H., and Kobilka, B.K. 1997. Structural instability of a constitutively active G-protein coupled receptor. J. Biol. Chem. 272 2587–2590. [DOI] [PubMed] [Google Scholar]
- Gether, U., Asmar, F., Meinild, A.K., and Rasmussen, S.G.F. 2002. Structural basis for activation of G-protein coupled receptors. Pharmacol. Toxicol. 91 304–312. [DOI] [PubMed] [Google Scholar]
- Giletto, A. and Pace, N. 1999. Buried, charged, non-ion-paired aspartic acid 76 contributes favorably to the conformational stability of ribonuclease T1.Biochemistry 38 13379–13384. [DOI] [PubMed] [Google Scholar]
- Gilman, A.G. 1987. G proteins: Transducers or receptor-generated signals. Ann.Rev. Biochem. 56 615–649. [DOI] [PubMed] [Google Scholar]
- Gioannini, T.L., Liu, Y.F., Park, Y.H., Hiller, J.M., and Simon, E.J. 1989. Evidence for the presence of disulfide bridges in opioid receptors essential for ligand binding: Possible role in receptor activation. J. Mol. Recog. 2 44–48. [DOI] [PubMed] [Google Scholar]
- Grigorieff, N., Ceska, T.A., Downing, K.H., Baldwin, J.M., and Henderson, R. 1996. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259 393–421. [DOI] [PubMed] [Google Scholar]
- Henderson, R., Baldwin, J.M., and Ceska, T.A. 1990. An atomic model for the structure of bacteriorhodopsin. J. Mol. Biol. 213 899–929. [DOI] [PubMed] [Google Scholar]
- Horn, F., van der Wenden, E.M., Oliveira, L., IJzerman, A.P., and Vriend, G. 2000. Receptor coupling to G proteins: Is there a signal behind the sequence? Proteins 41 448–459. [DOI] [PubMed] [Google Scholar]
- Horn, F., Bettler, E., Oliveira, L., Campagne, F., Cohen, F.E., and Vriend, G. 2003. GPCRDB information system for G protein–coupled receptors.Nucleic Acids Res. 31 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosohata, Y., Varga, E., Stropova, D., Li, X., Knapp, R.J., Hruby, V.J., Rice, K.C., Nagase, H., Roeske, W., and Yamamura, H.I. 2001. Mutation W284L of the human δopioid receptor reveals agonist specific receptor conformations for G protein activation. Life Sci. 68 2233–2242. [DOI] [PubMed] [Google Scholar]
- Iadanza, M., Holtje, M., Ronsisvalle, G., and Holtje, H.D. 2002. κ-opioid receptor model in a phospholipid bilayer: Molecular dynamics simulations.J. Med. Chem. 45 4843–4846. [DOI] [PubMed] [Google Scholar]
- Ji, T.H., Grossmann, M., and Ji, I. 1998. G protein–coupled receptors, I: Diversity of receptor-ligand interactions. J. Biol. Chem. 273 17299–17302. [DOI] [PubMed] [Google Scholar]
- Karnik, S.S., Gogonea, C., Patil, S., Saad, Y., and Takezako, T. 2003. Activation of G protein–coupled receptors: A common molecular mechanism.Trends Endocrinol. Metab. 19 431–437. [DOI] [PubMed] [Google Scholar]
- Kieffer, B.L., Befort, K., Gaveriaux-Ruff, C., and Hirth, C.G. 1992. The δ-opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc. Natl. Acad. Sci. 89 12048–12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H., Raynor, K., Yasuda, K., Moe, S.T., Portoghese, P.S., Bell, G.I., and Reisine, T. 1993. A single residue, aspartic acid 95, in the δopioid receptor specifies selective high affinity agonist binding. J. Biol. Chem. 268 23055–23058. [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Mass, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the steriochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Lei, H. and Smith, P.E. 2003. The effects of internal water molecules on the structure and dynamics of chymotrypsin inhibitor 2. J. Phys. Chem. B 107 1395–1402. [Google Scholar]
- Li, X., Varga, E., Stropova, D., Zalewska, T., Malatynska, E., Knapp, R.J., Roeske, W., and Yamamura, H.I. 1996. δOpioid receptor: The third extra-cellular loop determines naltrindole selectivity. Eur. J. Pharmacol. 300 R1–R2. [DOI] [PubMed] [Google Scholar]
- Lu, Z., Curtis, C.A., Jones, P.G., Pavia, J., and Hulme, E. 1997. The role of aspartate-arginine-tyrosine triad in the m1 muscarinic receptor: Mutations of aspartate 122 and tyrosine 124 decrease receptor expression but not abolish signalling. Mol. Pharmacol. 51 234–241. [DOI] [PubMed] [Google Scholar]
- McFadyen, I., Metzer, T., Subramanian, G., Poda, G., Jorvig, E., and Ferguson, D.M. 2002. Molecular modeling of opioid receptor-ligand complexes. Progress Med. Chem. 40 107–135. [DOI] [PubMed] [Google Scholar]
- Meng, F., Xie, G.X., Thompson, R.C., Mansour, A., Goldstein, A., Watson, S.J., and Akil, H. 1993. Cloning and pharmacological characterization of a rat κ opioid receptor. Proc. Natl. Acad. Sci. 90 9954–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F., Ueda, Y., Hoversten, M.T., Thompson, R.C., Taylor, L., Watson, S.J., and Akil H. 1996. Mapping the receptor domains critical for the ligand binding selectivity of δopioid receptor. Eur. J. Pharmacol. 311 285–292. [DOI] [PubMed] [Google Scholar]
- Metzer, T. and Ferguson, D.M. 1995. On the role of extracellular loops of opioid receptor in conferring ligand selectivity. FEBS Lett. 375 1–4. [DOI] [PubMed] [Google Scholar]
- Morris, A.L., MacArthur, M.W., Hutchinson, E.G., and Thornton, J.M. 1992. Steriochemical quality of protein structure coordinates. Proteins 12 345–364. [DOI] [PubMed] [Google Scholar]
- Mosberg, H.I. 1999. Complementarity of δopioid ligand pharmacophore and receptor models. Biopolymers 51 426–439. [DOI] [PubMed] [Google Scholar]
- Nayeem, A. and Scheraga, H,A. 1994. A statistical analysis of side-chain conformations in proteins: Comparison with ECEPP predictions. J. Protein Chem. 13 283–296. [DOI] [PubMed] [Google Scholar]
- Palczewski, K., Kumasaka, T., Hori, T., Behnke, C.A., Motoshima, H., Fox, B.A., Trong, I.L., Teller, D.C., Okada, T., Stenkamp, R.E., et al. 2000. Crystal structure of rhodopsin: A G protein–coupled receptor. Science 289 739–745. [DOI] [PubMed] [Google Scholar]
- Pepin, M., Yue, S.Y., Roberts, E., Wahlestedt, C., and Walker, P. 1997. Novel “restoration of function” mutagenesis strategy to identify amino acids of the δopioid receptor involved in ligand binding. J. Biol. Chem. 272 9260–9267. [DOI] [PubMed] [Google Scholar]
- Pogozheva, I.D., Lomize, A.L., and Mosberg, H.I. 1998. Opioid receptor three-dimensional structures from distance geometry calculations with hydrogen bonding constraints. Biophysical J. 75 612–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoghese, P.S., Sultana, M., and Takemori, A.E. 1990. Design of peptidomimetic δopioid receptor antagonists using the message-address concept.J. Med. Chem. 33 1714–1720. [DOI] [PubMed] [Google Scholar]
- Quock, R.M., Burkey, T.H., Varga, E., Hosohata, Y., Hosohata, K., Cowell, S.M., Slate, C.A., Ehlert, F.J., Roeske, W.R., and Yamamura, H.I. 1999. The δopioid receptor: Molecular pharmacology, signal transduction, and the determination of drug efficacy. Pharmacol. Rev. 51 503–532. [PubMed] [Google Scholar]
- Rapaka, R.S. and Porreca, F. 1991. Development of δopioid peptides as non-addicting analgesics. Pharmacol. Res. 8 1–8. [DOI] [PubMed] [Google Scholar]
- Rasmussen, S.G.F., Jensen, A.D., Liapakis, G., Ghanouni, P., Javitch, J.A., and Gether, U. 1999. Mutation of a highly conserved aspartic acid in the β 2 adrenergic receptor: Constitutive activation, structural instability, and conformational rearrangement of transmembrane segment 6. Mol. Pharmacol. 56 175–184. [DOI] [PubMed] [Google Scholar]
- Robinson, P.R., Cohen, G.B., Zhukovsky, E.A., and Oprian, D.D. 1992. Constitutively active mutants of rhodopsin. Neuron 9 719–725. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.P., Ciccotti, G., and Berendsen, H.J.C. 1977. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-Alkanes. J. Comput. Phys. 23 327–341. [Google Scholar]
- Salvadori, S., Balboni, G., Guerrini, R., Tomatis, R., Bianchi, C., Bryant, S.D., Cooper, P.S., and Lazarus, L.H. 1997. Evolution of the Dmt-Tic pharmacophore: N-terminal methylated derivatives with extraordinary δopioid antagonist activity. J. Med. Chem. 40 3100–3108. [DOI] [PubMed] [Google Scholar]
- Scheer, A., Fanelli, F., Costa, T., and De Benedetti, P.G. 1997. The activation process of the α1B-adrenergic receptor: Potential role of protonation and hydrophobicity of a highly conserved aspartate. Proc. Natl. Acad. Sci. 94 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, A., Costa, T., Fanelli, F., De Benedetti, P.G., Mhaouty-Kodja, S., Abuin, L., Nenniger-Tosata, M., and Cotecchia, S. 2000. Mutational analysis of the highly conserved arginine within the Glu/Asp-Arg-Tyr motif of the α1b adrenergic receptor: Effects on receptor isomerisation and activation. Mol. Pharmacol. 57 219–231. [PubMed] [Google Scholar]
- Schertler, G.F.X., Villa, C., and Henderson, R. 1993. Projection structure of rhodopsin. Nature 362 770–772. [DOI] [PubMed] [Google Scholar]
- Schiller, P.W., Nguyen, T.M.D., Weltrowska, G., Wilkes, B.C., Marsden, B.J, Lemieux, C., and Chung, N.N. 1992. Differential stereochemical requirements of δvs. μ opioid receptors for ligand binding and signal transduction: Development of a class of potent and highly δ-selective peptide antagonists.Proc. Natl. Acad. Sci. 89 11871–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller, P.W., Berezowska, I., Nguyen, T.M.D., Schmidt, R., Lemieux, C., Chung, N.N., Falcone-Hindley, M.L., Yao, W., Liu, J., Iwama, S., et al. 2000. Novel ligands lacking a positive charge for the δand μ opioid receptors. J. Med. Chem. 43 551–559. [DOI] [PubMed] [Google Scholar]
- Schiller, P.W., Weltrowska, G., Nguyen, T.M.D., Lemieux, C., Chung, N.N., and Lu, Y. 2003. Conversion of δ, κ and μ receptor selective opioid peptide agonists into δ, κ and μ selective antagonists. Life Sci. 73 691–698. [DOI] [PubMed] [Google Scholar]
- Schonbrun, J., Wedemeyer, W.J., and Baker, D. 2002. Protein structure prediction in 2002. Curr. Opin. Struct. Biol. 12 348–354. [DOI] [PubMed] [Google Scholar]
- Scott, W., Hunenberger, P.H., Tironi, I.G., Mark, A.E., Billeter, S.R., Fennen, J., Torda, A.E., Huber, T., Kruger, P., and van Gunsteren, W.F. 1999. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 103 3596–3607. [Google Scholar]
- Shahrestanifar, M., Wang, W.W., and Howells, R.D. 1996. Studies on inhibition of μ and δopioid receptor binding by dithiothreitol and N-ethylmaleimide.J. Biol. Chem. 271 5505–5512. [DOI] [PubMed] [Google Scholar]
- Shenderovich, M.D., Liao, S., Qian, X., and Hruby, V.J. 2000. A three-dimensional model of the δ opioid pharmacophore: Comparative molecular modeling of peptide and nonpeptide ligands. Biopolymers 53 565–580. [DOI] [PubMed] [Google Scholar]
- Simonds, W.F. 1988. The molecular basis of opioid receptor function. Endocrine Rev. 9 200–212. [DOI] [PubMed] [Google Scholar]
- Smith, D.G. and Griffin, J.F. 1978. Conformation of leu enkephalin from X-ray diffraction: Features important for recognition at opiate receptor. Science 199 1214–1216. [DOI] [PubMed] [Google Scholar]
- Smith, A.P. and Lee, N.M. 2003. Opioid receptor interactions: Local and nonlocal symmetric and asymmetric, physical and functional. Life Sci. 73 1873–1893. [DOI] [PubMed] [Google Scholar]
- Smith, P.E. and van Gunsteren, W.F. 1994. Consistent dielectric properties of the simple point charge and extended simple point charge water models at 277 and 300K. J. Chem. Phys. 100 3169–3174. [Google Scholar]
- Stenkamp, R.E., Teller, D.C., and Palczewski, K. 2002. Crystal structure of rhodopsin: A G-protein–coupled receptor. ChemBioChem 3 963–967. [DOI] [PubMed] [Google Scholar]
- Stocker, U., Spiegel, K., and van Gunsteren, W.F. 2000. On the similarity of properties in solution or in the crystalline state: A molecular dynamics study of hen lysosome. J. Biomol. NMR 18 1–12. [DOI] [PubMed] [Google Scholar]
- Strader, C.D., Fong, T.M., Tota, M.R., Underwood, D., and Dixon, R.A.F. 1994. Structure and function of G protein–coupled receptors. Ann. Rev. Biochem. 63 101–132. [DOI] [PubMed] [Google Scholar]
- Strahs, D. and Weinstein, H. 1997. Comparative modeling and molecular dynamics studies of theδ , κ and μ opioid receptors. Protein Eng. 10 1019–1038. [DOI] [PubMed] [Google Scholar]
- Tironi, I.G., Sperb, R., Smith, P.E., and van Gunsteren, W.F. 1995. A generalized reaction field method for molecular dynamics simulations. J. Chem.Phys. 102 5451–5459. [Google Scholar]
- Unger, V.M., Hargrave, P.A., Baldwin, J.M., and Schertler, G.F.X. 1997. Arrangement of rhodopsin transmembrane α helices. Nature 389 203–206. [DOI] [PubMed] [Google Scholar]
- Valiquette, M., Vu, H.K., Yue, S.Y., Wahlestedt, C., and Walker, P. 1996. Involvement of Trp-284, Val-296, Val-297 of the human δ opioid receptor in binding of δ selective ligands. J. Biol. Chem. 271 18789–18796. [DOI] [PubMed] [Google Scholar]
- van Gunsteren, W.F. and Mark, A.E. 1998. Validation of molecular dynamics simulations. J. Chem. Phys. 108 6109–6116. [Google Scholar]
- Voordijk, S., Hansson, T., Hilvert, D., and van Gunsteren, W.F. 2000. Molecular dynamics simulations highlight mobile regions in proteins: A novel suggestion for converting a murine VH domain into a more tractable species. J. Mol. Biol. 300 963–973. [DOI] [PubMed] [Google Scholar]
- Waldburger, C.D., Schildbach, J.F., and Sauer, R.T. 1995. Are buried salt bridges important for protein stability and conformational specificity? Nat.Struct. Biol. 2 122–128. [DOI] [PubMed] [Google Scholar]
- Wess, J. 1998. Molecular basis of receptor/G-protein-coupling selectivity.Pharmacol. Ther. 80: 231–264. [DOI] [PubMed] [Google Scholar]
- Winn, P.J., Ludemann, S.K., Gauges, R., Lounnas, V., and Wade, R.C. 2002. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. 99 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S.K. 2002. G Protein selectivity is regulated by multiple intracellular regions of GPCRs. Neurosignals 12 1–12. [DOI] [PubMed] [Google Scholar]
- Xu, W., Chen, C., Huang, P., Li, J., de Riel, J.K., Javitch, J.A., and Liu-Chen, L. 2000. The conserved cysteine 7.38 residue is differentially accessible in the binding-site crevices of the μ, δ, and κ opioid receptors. Biochemistry 39 13904–13915. [DOI] [PubMed] [Google Scholar]