Abstract
It is well established that the rate of formation of fibrils by amyloidogenic proteins is enhanced by the addition of preformed fibrils, a phenomenon known as seeding. We show that the efficiency of seeding fibril formation from solutions of hen lysozyme by a series of other proteins depends strongly on the similarity of their sequences. This observation is consistent with the importance of long-range interactions in stabilizing the core structure of amyloid fibrils and may be associated with the existence of a species barrier observed in the transmissible spongiform encephalopathies. In addition, it is consistent with the observation of a single dominant type of protein in the deposits associated with each form of amyloid disease.
Keywords: amyloid, lysozyme, seeding, cross-seeding, species barrier
The formation of amyloid fibrils from otherwise soluble peptides and proteins is associated with a variety of human disorders including Alzheimer’s disease, Type II diabetes, and the transmissible spongiform encephalopathies (TSEs) such as Creutzfeld-Jakob disease (Selkoe 2003). Similar fibrils can also be formed in vitro from a wide range of proteins and peptides, most of which are not associated with any known disease (Dobson 2003). The kinetics of formation of such structures can generally be increased very substantially by the addition of an aliquot of a solution containing preformed fibrils (Jarrett and Lansbury 1993). This process is similar to the well-established seeding phenomenon in crystallization and has been associated with the transmission of the TSEs and with the rapid development of disorders such as Alzheimer’s disease once clinical symptoms are detected (Jarrett and Lansbury 1993; Perutz and Windle 2001; Korth et al. 2003).
In this study, we have examined the specificity of the seeding of amyloid fibrils formed from hen lysozyme under low pH solution conditions described previously (Krebs et al. 2000). Five other proteins were used in the seeding experiments and were chosen to be increasingly different from hen lysozyme in sequence identity and structure. These were the I55T mutational variant (99.2% sequence identity to hen lysozyme), an analog of one of the mutations associated with lysozyme amyloidosis in humans (Pepys et al. 1993); turkey lysozyme (95% identity); human lysozyme (60% identity); human α-lactalbumin (36% identity); and bovine insulin (no identity). Of these proteins, human lysozyme and bovine insulin have been shown to form amyloid fibrils under conditions similar to those appropriate for hen lysozyme (Bouchard et al. 2000; Morozova-Roche et al. 2000).
Results and Discussion
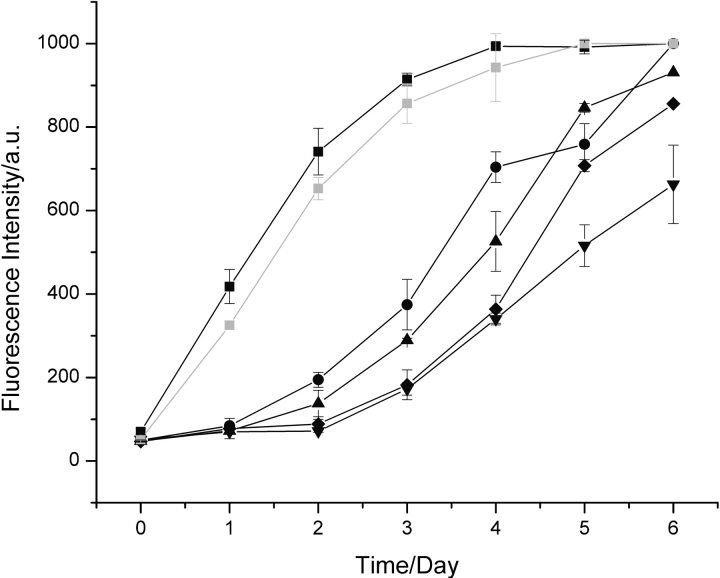
To define the seeding conditions, aliquots of solutions of preformed hen lysozyme fibrils were added to fresh solutions of the protein, and the kinetics of fibril formation were followed by means of thioflavin-T, a dye whose fluorescence increases significantly upon binding to amyloid fibrils (Fig. 1 ▶; Naiki et al. 1989). In the absence of added fibrils, hen lysozyme exhibits a lag phase of ~2 d under the chosen conditions, after which the fluorescence intensity starts to rise, indicative of the growth of amyloid fibrils. After ~6 d, the intensity remains approximately constant. Addition of aliquots containing preformed fibrils was found to result in a shortening of the lag phase indicative of seeding. When added fibrils reached 5% (v/v), the increase in thioflavin-T fluorescence showed no lag phase and the maximum fluorescence intensity was reached after only 3 to 4 d. Experiments also showed that nonfibrillar, seemingly amorphous aggregates do not significantly perturb the process of fibril formation.
Figure 1.
Aggregation of hen lysozyme. Data are shown for the aggregation of 1 mM hen lysozyme at pH 2.0 and 65°C monitored by thioflavin-T fluorescence in the absence of added fibrils (down triangles) and following addition of 5% (v/v) (black squares), 2.5% (v/v) (circles), 1% (v/v) (triangles), and 0.5% (v/v) fibrils preformed from hen lysozyme at pH 2.0 (diamonds). Seeding with a 5% (v/v) aliquot of a solution of amyloid fibrils containing purified full-length hen lysozyme gave essentially identical kinetics (gray squares) to aliquots formed simply by incubation at pH 2.0 and 65°C in which a significant degree of fragmentation had occurred (see text).
The other proteins studied here also form well-defined fibrils at pH 2.0. Fibrils from human lysozyme and bovine insulin have been described previously (Bouchard et al. 2000; Morozova-Roche et al. 2000). Although bovine α-lactalbumin is known to form amyloid fibrils at low pH in both oxidized and partially reduced states (Goers et al. 2002), amyloid fibrils from the human form of the protein have not been described. Thus, along with those of I55T and turkey lysozyme, the aggregates of human α-lactalbumin formed under the low pH conditions used here were characterized by electron microscopy, Congo red binding, and CD spectroscopy (Fig. 2 ▶). All the fibrils formed were confirmed as exhibiting the typical morphology, secondary structure, and dye-binding properties of amyloid fibrils. The temperature and concentrations of solutions of the five proteins were adjusted such that the appearance of fibrils was on a timescale similar to that of hen lysozyme. Experiments revealed that addition of 5% (v/v) aliquots of solutions containing fibrils to monomeric solutions of the same proteins resulted in enhancements in the rate of formation of amyloid fibrils similar to those observed for hen lysozyme. These experiments confirmed that aliquots of the various protein solutions containing preformed amyloid fibrils have the ability to seed the formation of such fibrils in freshly dissolved solutions.
Figure 2.

Characterization of amyloid fibrils from the various proteins formed at pH 2.0. (Left) EM images of fibrils from (A) hen lysozyme, (B) I55T lysozyme, (C) turkey lysozyme, and (D) human α-lactalbumin, incubated at pH 2.0 using conditions described in Materials and Methods. The scale bar represents 200 nm. Characterization of the fibril solutions by Congo red (CR) absorption (center) and far-UV CD (right) are also shown for the same proteins: (squares) CR in buffer; (triangles) CR in the presence of fibrils; and CD spectra of freshly dissolved protein (squares) and protein fibrils (triangles).
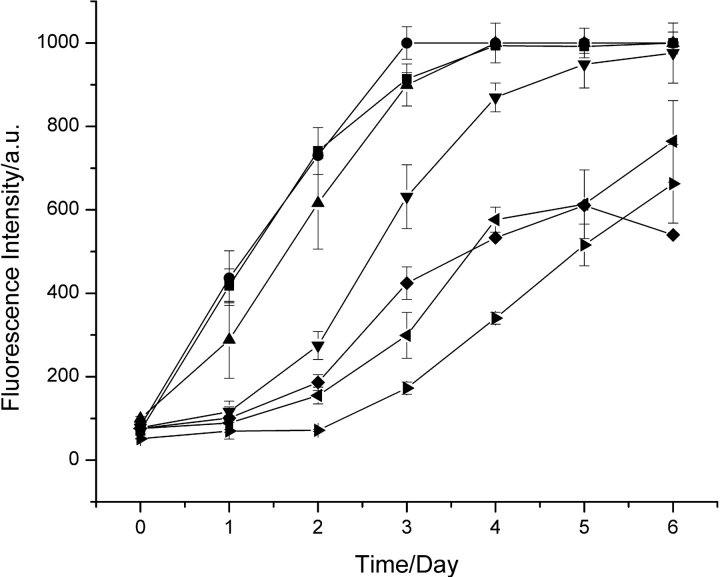
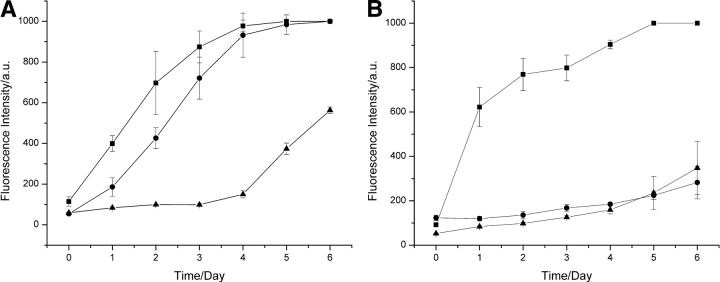
To investigate the specificity of the seeding phenomenon, 5% (v/v) aliquots of solutions containing amyloid fibrils formed from the different proteins were added to solutions of hen lysozyme, and the process of aggregation was again followed by fluorescence (Fig. 3 ▶). In the presence of a 5% (v/v) aliquot of a solution containing I55T lysozyme or turkey lysozyme fibrils, the aggregation behavior of hen lysozyme was essentially identical to that resulting from the addition of hen lysozyme fibrils. Addition of 5% (v/v) aliquots of solutions containing human lysozyme fibrils, however, resulted in the more rapid appearance of fibrils than in the absence of seeding, but a lag phase was still evident. Addition of aliquots of solutions containing amyloid fibrils formed from human α-lactalbumin or bovine insulin had no significant effect on the kinetics of fibril formation by hen lysozyme. Experiments were also carried out in which the aggregation was monitored following addition of 5% (v/v) aliquots of solutions containing hen lysozyme fibrils to solutions of the five other proteins (Fig. 4 ▶). The results were complementary to those described above, showing that a decrease in sequence identity between hen lysozyme and the protein in the fibrils that were added decreased the efficiency of seeding. Thus, in the case of turkey lysozyme, for example, there was a profound effect on the kinetics of fibril formation upon addition of hen lysozyme fibrils, a significantly smaller effect for human lysozyme, and no detectable effect for human α-lactalbumin or insulin.
Figure 3.
Seeding of hen lysozyme solutions with preformed fibrils from other protein. Data are shown for solutions of hen lysozyme incubated at pH 2.0 and 65°C (right triangles) and for similar solutions to which were added 5% (v/v) aliquots of solutions containing preformed fibrils from hen lysozyme (squares), I55T lysozyme (circles), turkey lysozyme (triangles), human lysozyme (down triangles), human α-lactalbumin (diamonds), and insulin (left triangles).
Figure 4.
Seeding of fibril formation by turkey lysozyme and human α-lactalbumin. Fibril formation in solutions of (A) turkey lysozyme and (B) human α-lactalbumin without seeding (triangles) and seeded with 5% (v/v) aliquots of preformed fibril solutions of the same protein (squares) or of hen lysozyme (circles).
The high efficiency of cross-seeding between the I55T mutational variant and wild-type hen lysozyme is consistent with previous observations for single point mutations of human lysozyme (Morozova-Roche et al. 2000) and of α-synuclein (Wood et al. 1999) that proteins with very similar sequences can seed efficiently the formation of amyloid fibrils. The present results, however, demonstrate that as the sequence identity between the protein in the preformed amyloid fibrils and that in the solution to which they are added decreases, the efficiency of the seeding reaction decreases. Because hen lysozyme and human α-lactalbumin share substantially the same native state fold (McKenzie and White 1991) but addition of preformed fibrils of one to a solution of the other does not alter the kinetics of fibril formation, the efficiency of seeding does not appear to be associated with the degree of similarity of the native state structures. This conclusion is consistent with the idea that to form amyloid fibrils, proteins need to unfold, at least partially, to allow essentially unstructured segments of the polypeptide chain to form intermolecular interactions (Dobson 2001). The results are also consistent with observations that aggregate formation and cross-seeding in vitro, between unfolded proteins with moderate or low sequence identities are relatively inefficient. Examples include amyloid formation by α- and β-synucleins (78% sequence similarity) and α- and γ-synucleins (60% similarity; Biere et al. 2000) and inclusion body formation by P22 tailspike and P22 coat proteins (Speed et al. 1996).
The origin of the observed sequence specificity of the seeding process can be explained in general terms by consideration of the known structural characteristics of amyloid fibrils. For example, recent solid-state NMR studies reveal a high degree of crystal-like order within the amyloid core structure and well-defined conformational states for the majority of the side chains through the fibril (Jaroniec et al. 2004). It is clear that the core structure of the fibrils involves hydrogen bonding between extended β-strands to form β-sheets running in the direction of the fibrils (Sunde and Blake 1997). As the main chain structure is the same in all polypeptide chains, this core structure will be similar for all sequences. The side chains, however, influence both the propensity to form amyloid structures and the specific details of the structure, for example, the separation between the β-sheets (Fändrich and Dobson 2002) and the way protofilaments assemble to form fibrils (Chamberlain et al. 2000; Jiménez et al. 2001).
As with fully crystalline materials, therefore, the lowest energy structure of an amyloid fibril is likely to involve repetition of identical molecular conformations because, as is found in three-dimensional crystals, this maximizes long-range order. Seeding is therefore likely to be most efficient when identical molecules are in both the preformed fibrils and the solution to which they are added. Some relaxation of this constraint is, however, likely to occur because a degree of disorder can be tolerated even in the core structure of amyloid fibrils. For example, at least 1% of a foreign peptide can be randomly incorporated into insulin fibrils (MacPhee and Dobson 2000), and sequence variations are present in the β-sheet regions of spider silk (Thiel et al. 1997). Such observations are therefore consistent with the present findings that cross-seeding can occur, but decreases in efficiency as the difference between sequences increases. If the differences in sequence are located in regions of the polypeptide chain that are not involved in the core structure, however, the efficiency of seeding is likely to be much less affected. The conclusions drawn from these experiments resolve the apparent paradox between the generic nature of the fibril core structure and the relatively high sequence specificity of seeding observed in this and other studies (Jarrett and Lansbury 1992; Biere et al. 2000).
The observation of a lower efficiency of seeding by proteins of differing sequence is of particular interest in the context of the well-established species barrier in the TSEs; that is, that there is a lower efficiency of transmission between animals of different species compared with transmission between animals of a given species (Korth et al. 2003 and references therein). A similar species barrier has also been observed between different types of yeast prions (Chien et al. 2003 and references therein). Although it is clear that factors such as different strains and glycosylation patterns are also important (Chien et al. 2003; Korth et al. 2003), the present results suggest that the intrinsic effects of differences in sequence could be a highly significant contributor to the yeast and mammalian species barriers. More generally, the observed specificity is not sufficient to prevent closely similar proteins from seeding fibril formation; it is therefore possible that processes such as posttranslational or chemical modification can be important in initiating aggregation in vivo (Nilsson et al. 2002). In addition, fragments of proteins that originate from the region of the protein that is responsible for at least the initial aggregation process may be able to seed fibril formation by the intact protein (Krebs et al. 2000). An important consequence of the sequence specificity, however, is that it is consistent with the finding that proteins deposited in amyloid diseases are typically composed of a single primary component protein rather than resulting from the widespread recruitment of other amyloidogenic proteins present within the body.
Materials and methods
Solvents and solutions
With the exception of I55T lysozyme, all proteins and chemicals were of analytical grade, purchased from Sigma and used without further purification. The I55T hen lysozyme mutant was expressed and purified in the fungus Aspergillus niger by Andrew Spencer (Archer et al. 1990; Spencer et al. 1999). Protein solutions were prepared by weighing out the appropriate amount of protein and dissolving it in the solvent of choice, followed by adjustment of the pH using HCl or NaOH. Protein concentrations were verified by absorption measurements at 280 nm.
Sequence alignments
Protein sequences were obtained from the SWISS-PROT database (via http://www.expasy.ch) and manually aligned.
Fibril formation
All protein solutions were incubated at pH 2.0 in 100-μL aliquots in thermocycler tubes (MoBiTec) at the following concentrations and temperatures: hen lysozyme, 1 mM and 65°C; I55T lysozyme, 1 mM and 40°C; turkey lysozyme, 1.4 mM and 65°C; human lysozyme, 0.7 mM and 57°C; human α-lactalbumin, 0.7 mM and 57°C; bovine insulin, 1 mM and 37°C. For seeding experiments, 0.5% to 5% (v/v) aliquots of solutions containing preformed amyloid fibrils were added to the solutions prior to incubation. All experiments were repeated a minimum of three times.
Bovine insulin is known to remain substantially unmodified under these conditions (Nilsson and Dobson 2003). Hen lysozyme fibrils prepared in this way were found to contain fragmented as well as full-length protein (Frare et al. 2004; M.R. Nilsson, M.R.H. Krebs, and C.M. Dobson, unpubl.). Fibrils could, however, be formed containing up to 90% full-length lysozyme, as shown by SDS-PAGE gel, by repetitive seeding of fresh solutions of hen lysozyme. Fibrils from such samples were found to be indistinguishable from the fibrils formed as described above in the seeding experiments.
Characterization of the fibrils: EM, Congo red, and CD
Samples were studied by EM by placing a 3.5-μL aliquot of the fibril solution on a formvar- and carbon-coated grid (Agar Scientific Ltd.). After 30 sec, two drops of 15 μL of a 2% (w/v) uranyl acetate solution were added. After 30 sec, the droplet was removed by tipping the grid a quarter turn and leaving it to air dry. The grids were checked in a Jeol JEM1010 microscope at magnifications of typically 25,000×.
For Congo red binding assays, a Congo red solution (1 mM in 150 mM NaCl, 5 mM NaH2PO4/Na2HPO4 at pH 7.4) was made fresh on the day of the experiment and filtered through a 0.2-μm filter prior to use. A 450–700-nm spectrum of the buffer (985 μL) plus fibril solution (10 μL) was subtracted from a spectrum of buffer (985 μL) plus fibril solution (10 μL) plus CR (5 μL). This spectrum was compared with a spectrum of Congo red (985 μL) plus CR (5 μL) that had been background-corrected by subtracting a spectrum containing only buffer. A shift of the maximum Congo red absorbance from 480 nm to 490–500 nm and the formation of a shoulder around 540 nm were taken as indicative of the presence of amyloid fibrils (Klunk et al. 1989).
Prior to evaluating the secondary structure content of the fibrils by CD, nonaggregated protein was removed from fibril solutions using Centricon filters (100 kDa cutoff; Amicon, Millipore). The solution was diluted 50-fold and transferred to a 1-mm path length cuvette, and four scans were recorded on a Jasco J720 CD apparatus (Jasco Ltd.), scanning at 50 nm/min with a response time of 1 sec.
Thioflavin-T assay
For the thioflavin-T assay, 2.5 mM solutions of thioflavin-T were made up in buffer (150 mM NaCl, 10 mM NaH2PO4/Na2HPO4 at pH 7.4) and filtered through a 0.2-μm filter. This stock solution was diluted 50-fold into buffer and used for testing the samples for the presence of amyloid fibrils by adding 995 μL of this solution to the 5-μL aliquots of sample. The solutions were left to stir for 1 min in the cuvette in the dark, after which the fluorescence was measured for 1 min. A baseline measurement of just the 50-fold diluted stock solution was also taken. The fluorescence was measured by excitation at 440 nm (5 nm slit width) and observing the emission at 482 nm (10 nm slit width) on a Perkin Elmer LS50b fluorimeter.
Acknowledgments
M.R.H.K. is supported by the EPSRC and acknowledges scholarships from 3M and St. Catherine’s College, Oxford. The research of L.A.M.-R. is supported by the Swedish Medical Research Council. The research of C.M.D. is supported in part by a Programme Grant from the Wellcome Trust. The research of C.V.R. is supported in part by the Royal Society. C.V.R. and C.M.D. acknowledge a grant from the BBSRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04707004.
References
- Archer, D.B., Jeanes, D.J., MacKenzie, D.A., Brightwell, G., Lambert, N., Lowe, G., Radford, S.E., and Dobson, C.M. 1990. Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Biotechnol. 8 741–745. [DOI] [PubMed] [Google Scholar]
- Biere, A.L., Wood, S.J., Wypych, J., Steavenson, S., Jian, Y., Anafi, D., Jacobsen, F.W., Jarosinski, M.A., Wu, G.-M., Louis, J.C., et al. 2000. Parkinson’s disease associated α-synuclein is more fibrillogenic than β- and γ-synuclein and cannot cross-seed its homologs. J. Biol. Chem. 275 34574–34579. [DOI] [PubMed] [Google Scholar]
- Bouchard, M., Zurdo, J., Nettleton, E.J., Dobson, C.M., and Robinson, C.V. 2000. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 9 1960–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, A.K., MacPhee, C.E., Zurdo, J., Morozova-Roche, L.A., Hill, H.A.O., Dobson, C.M., and Davis, J.J. 2000. Ultrastructural organization of amyloid fibrils by atomic force microscopy. Biophys. J. 79 3282–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, P., DePace, A.H., Collins, S.R., and Weissman, J.S. 2003. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature 424 948–951. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 2001. The structural basis of protein folding and its link with human disease. Phil. Trans. Roy. Soc. Lond. B 256 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Protein folding and misfolding. Nature 426 884–890. [DOI] [PubMed] [Google Scholar]
- Fändrich, M. and Dobson, C.M. 2002. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21 5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frare, E., Polverino de Laureto, P., Zurdo, J., Dobson, C.M., and Fontana, A. 2004. A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. (in press). [DOI] [PubMed]
- Goers, J., Permyakov, S.E., Permyakov, E.A., Uversky, V.N., and Fink, A.L. 2002. Conformational prerequisites for α-lactalbumin fibrillation. Biochemistry 41 12546–12551. [DOI] [PubMed] [Google Scholar]
- Jaroniec, C.P., MacPhee, C.E., Bajaj, V.S., McMahon, M.T., Dobson, C.M., and Griffin, R.G. 2004. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc. Natl. Acad. Sci. 101 0711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett, J.T. and Lansbury, P.T.J. 1992. Amyloid fibril formation requires a chemically discriminating nucleation event: Studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry 31 12345–12352. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 73 1055–1058. [DOI] [PubMed] [Google Scholar]
- Jiménez, J.L., Tennent, G., Pepys, M.B., and Saibil, H.R. 2001. Structural diversity of ex-vivo amyloid fibrils studied by cryo-electron microscopy. J. Mol. Biol. 311 241–247. [DOI] [PubMed] [Google Scholar]
- Klunk, W.E., Pettegrew, J.W., and Abraham, D.J. 1989. Two simple methods for quantifying dye–substrate binding. J. Histochem. Cytochem. 37 1293–1297. [DOI] [PubMed] [Google Scholar]
- Korth, C., Kaneko, K., Groth, D., Heye, N., Telling, G., Matrianni, J., Parchi, P., Gambetti, P., Will, R., Ironside, J., et al. 2003. Abbreviated incubation time for human prions in mice expressing a chimeric mouse–human prion protein transgene. Proc. Natl. Acad. Sci. 100 4784–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, M.R.H., Wilkins, D.K., Chung, E.W., Pitkeathly, M.C., Chamberlain, A.K., Zurdo, J., Robinson, C.V., and Dobson, C.M. 2000. Formation and seeding of amyloid fibrils from wild type hen lysozyme and a peptide fragment from the β-domain. J. Mol. Biol. 300 541–549. [DOI] [PubMed] [Google Scholar]
- MacPhee, C.E. and Dobson, C.M. 2000. Formation of mixed fibrils demonstrates the generic nature and potential utility of amyloid nanostructures. J. Am. Chem. Soc. 122 12707–12713. [Google Scholar]
- McKenzie, H.A. and White, F.H. 1991. Lysozyme and α-lactalbumin: Structure, function and interrelationships. Adv. Prot. Chem. 41 175–315. [DOI] [PubMed] [Google Scholar]
- Morozova-Roche, L.A., Zurdo, J., Spencer, A., Noppe, W., Receveur, V., Archer, D.B., Joniau, M., and Dobson, C.M. 2000. Amyloid fibril formation and seeding by wild-type human lysozyme and its disease-related mutational variants. J. Struct. Biol. 130 339–351. [DOI] [PubMed] [Google Scholar]
- Naiki, H., Higuchi, K., Hosokawa, M., and Takeda, T. 1989. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, Thioflavine T. Anal. Biochem. 177 244–249. [DOI] [PubMed] [Google Scholar]
- Nilsson, M.R., Driscoll, M., and Raleigh, D.P. 2002. Low levels of asparagine deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: Implications for the study of amyloid formation. Protein Sci. 11 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys, M.B., Hawkins, P.N., Booth, D.R., Vigushin, D.M., Tennent, G.A., Soutar, A.K., Totty, N., Nguyen, O., Blake, C.C., Terry, C.J., et al. 1993. Human lysozyme gene mutations cause hereditary systemic amyloidosis. Nature 362 553–557. [DOI] [PubMed] [Google Scholar]
- Perutz, M.F. and Windle, A.H. 2001. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature 412 143–144. [DOI] [PubMed] [Google Scholar]
- Selkoe, D.J. 2003. Folding proteins in fatal ways. Nature 426 900–904. [DOI] [PubMed] [Google Scholar]
- Speed, M.A., Wang, D.I.C., and King, J. 1996. Specific aggregation of partially folded peptide chains: The molecular basis of inclusion body composition. Nat. Biotechnol. 14 1283–1287. [DOI] [PubMed] [Google Scholar]
- Spencer, A., Morozova-Roche, L.A., Noppe, W., MacKenzie, D.A., Jeenes, D.J., Joniau, M., Dobson, C.M., and Archer, D.B. 1999. Expression, purification and characterization of the recombinant calcium-binding equine lysozyme secreted by the filamentous fungus Aspergillus niger: Comparison with the production of hen and human lysozyme. Protein Expr. Purif. 16 171–180. [DOI] [PubMed] [Google Scholar]
- Sunde, M. and Blake, C.C.F. 1997. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Prot. Chem. 50 123–159. [DOI] [PubMed] [Google Scholar]
- Thiel, B.L., Guess, K.B., and Viney, C. 1997. Non-periodic lattice crystals in the hierarchical microstructure of spider (major ampullate) silk. Biopolym. 41 703–719. [DOI] [PubMed] [Google Scholar]
- Wood, S.J., Wypych, J., Steavenson, S., Louis, J.-C., Citron, M., and Biere, A.L. 1999. α-Synuclein fibrillogenesis is nucleation dependent. J. Biol. Chem. 274 19509–19512. [DOI] [PubMed] [Google Scholar]