Abstract
It is well known that the sequence of amino acids in proteins code for its tertiary structure. It is also known that there exists a relationship between sequence and the quaternary structure of proteins. The question addressed here is whether the nature of quaternary association can be predicted from the sequence, similar to the three-dimensional structure prediction from the sequence. The class of proteins called legume lectins is an interesting model system to investigate this problem, because they have very high sequence and tertiary structure homology, with diverse forms of quaternary association. Hence, we have used legume lectins as a probe in this paper to (1) gain novel insights about the relationship between sequence and quaternary structure; (2) identify the sequence motifs that are characteristic of a given type of quaternary association; and (3) predict the quaternary association from the sequence motif.
Keywords: computational protein structure analysis, quaternary association, oligomerization, legume lectins, graph-spectral method, interface amino acid clusters, conserved amino acid clusters
Lectins are carbohydrate-binding proteins that have high affinity and specificity for glycoconjugates, and have found applications in biological and biomedical research (Liener et al. 1986). Tertiary and quaternary structures of a significant number of lectins have been determined by X-ray crystallography (Loris et al. 1998; Vijayan and Chandra 1999; Svensson et al. 2002). Legume lectins are mainly β-sheet proteins, and hence, their quaternary interfaces are also formed between β-strands. All legume lectin monomers have highly similar sequences and share the same tertiary structure, with minor variations in loop lengths or lengths of strands. The monomer structure is characterized by the “jelly roll” motif present in many other proteins that is often associated with carbohydrate-binding activity (Loris et al. 1998; Vijayan and Chandra 1999). The “jelly roll” is characterized by the presence of three sets of antiparallel 3-sheets. There is a six-stranded flat “back” sheet, a curved seven-stranded “front” sheet, and a short sheet at the “top” of the molecule. The sheets are connected by several loops of varying lengths (Loris et al. 1998; Vijayan and Chandra 1999).
Most legume lectins are known to exist mainly as homodimers or homotetramers, with the tetramers being dimers of dimers. The striking feature about the legume lectins is that although all the monomers have similar tertiary structures, their modes of quaternary associations are very different. This study is aimed at understanding the factors responsible for the differences in the nature of the quaternary associations in legume lectins. The different kinds of quaternary structures seen in legume lectins include Canonical, ECorL-type, GS4-type, DBL-type, ConA-type, PNA-type, GS1-type, DB58-type, and Arcelin-5-type. All these are dimers or tetramers, except Arcelin-5, which exists as a monomer. It should be noted here that Arcelin-5 and Arce-lin-1 (both from Phaseolus vulgaris), which have been considered in this data set because of their similarity to legume lectins in sequence and tertiary structure, are not lectins because they do not bind sugars due to lack of crucial metal-binding residues. The dimers include Canonical, ECorL, GS4, and DB58 types and the tetramers include DBL, ConA, PNA, and GS1 types. The tetramers are generally dimers of dimers except in the case of PNA, which takes up an open quaternary structure (Banerjee et al. 1994). These nine quaternary structures consist of seven different types of dimeric interfaces, namely types II, X1, X2, X3 (handshake), X4 (back to back), and the unusual interfaces of PNA and GS1 (Manoj and Suguna 2001). The two unusual dimeric interfaces seen in PNA and GS1 are the only known cases of such interfaces, and hence, the name “unusual.” Most of these quaternary associations occur by the varied associations of the flat six-stranded “back” β-sheet. The Canonical mode of association, which is the most commonly observed mode of dimerization in legume lectins, occurs by the side-by-side arrangement of the back β-sheet to form a contiguous 12-stranded sheet. The other associations, namely ECorL, GS4, DB58, and ConA types, occur by the overlap of two back β-sheets on each other. The difference between these associations is the angle between the sheets during overlap. The unusual associations of PNA and GS1 mainly involve the top β-sheet and loop regions. The X1 interfaces of the DB58 dimer and DBL tetramer are also stabilized by a helix sandwiched between the two monomers.
Some preliminary analyses have already been carried out to correlate the sequence, tertiary structure, and quaternary structure in lectins. These studies include the correlation with the phylogenetic trees (Manoj and Suguna 2001), identification of conserved residues from multiple sequence alignment (Srinivas et al. 2001) or inspection of pair-wise interactions at the intersubunit interface(s) (Srinivas et al. 2001). Although the phylogenetic tree analysis is able to correlate the quaternary structure in many cases, the exact residues or sequence motifs responsible for the quaternary association cannot be deduced from such studies. These analyses have been unable to recognize the sequence motifs required for quaternary association, because the overall similarity in both sequence and tertiary structure is very high across the different quaternary associations to be able to distinguish those residues that are important for a particular type of quaternary association. Hence, the choice of the method of analysis should be such that it takes a global view of the interaction and not at the pair-wise level. The clustering algorithm based on graph spectral method (Kannan and Vishveshwara 1999), on the other hand, identifies the clusters of interacting amino acid residues in the protein structure. This method has been effectively used to analyze and understand the factors stabilizing a set of homodimeric protein interfaces (Brinda et al. 2002). In the present work, we have used this method to identify clusters of interacting residues at the interface of the legume lectin dimers and tetramers. Furthermore, this method is used in combination with multiple sequence alignment for identifying the sequential and structural determinants of quaternary association in legume lectins. This study is aimed at identifying the residues that characterize and stabilize the oligomeric interfaces of the legume lectins and also determine the nature of quaternary associations that the legume lectin monomers would take up. The present analysis provides valuable insights into the factors that affect the quaternary association in legume lectins, as can be seen clearly in the following sections. Moreover, the method has worked well in predicting the nature of quaternary association in lectins whose structures are unknown.
Results and Discussion
Because the preliminary analysis of sequence and structure have not yielded any insights into the factors responsible for quaternary association in legume lectins, we have used a clustering algorithm to identify the residues that form a network of interactions across the dimeric interfaces of legume lectins, and thus are involved in the formation and stabilization of these interfaces. These are then correlated with the sequentially conserved residues (completely or partially conserved or conservatively mutated), obtained from multiple sequence alignments, to obtain a motif of sequentially and structurally conserved residues at the interfaces of these legume lectins. Such motifs have been obtained for most of the different kinds of quaternary associations seen in legume lectins. In the present investigation, a set of 39 legume lectin dimers obtained from the crystal structures of 28 legume lectins (shown in Table 1) have been analyzed using the graph-spectra–based clustering algorithm (Kannan and Vishveshwara 1999) and ClustalW–based multiple sequence alignments (Higgins et al. 1994) to identify the determinants of quaternary association in these legume lectins. We have tried to characterize (based on sequence and structure) the nine types of quaternary structures seen in legume lectins comprising the seven different dimeric interface types discussed earlier. The monomeric case of Arcelin-5 is also discussed.
Table 1.
Dataset, types of quaternary associatons in legume lectins, and their Sugar specificities
| Oligomerizations in legume lectins | |||||
| S. No. | TQSa | TDIb | State | PDB codes | Sugar specificity |
| 1. | Canonical | II (Canonical) | Dimer | 11oc,11gc,1les,1h9pAB,1g7yAB,1fx5,1fnyAB, 1fatAB,1dglAB,1bqp,1bjqAB,1azdAC,1avbf, 2cnaAC,1n47AB,1sbfAB,1qnwAB,1n3o, 1qmoAEBF,1dbnAC | Glc/Man/Fucose |
| 2. | DBLc | II+X1 | Tetramer | 1g7yACg,1fnyAC,1fatAC,1dbnAB,1sbfAC, 1qnwAC,1n47AD,1bjqAC | GlcNAc/Gal/GalNac/Complex/Sialyl lactose |
| 3. | ConAc | II+X2 | Tetramer | 1h9pAC,1dglAC,1azdAB,2cnaAB,1qmoAECG | Man/Glc |
| 4. | ECorL | X3 (Handshake) | Dimer | 1gz9,1f9k,1axy,1wbf | Gal/GalNAc |
| 5. | GS4 | X4 (Back to back) | Dimer | 1gsl,1hqlABh | Complex |
| 6. | GS1 | X4+Unusuald | Tetramer | 1hqlACd,h | G-α linked oligosaccharide |
| 7. | DB58 | X1 | Dimer | 1g7yACg | GalNAc |
| 8. | PNA | II+X4+Unusuale | Tetramer | 2pele | Gal |
| 9. | Arcelin-5 | — | Monomer | 1ioaf | —f |
a TQS: type of quaternary structure.
b TDI: type(s) of Dimeric interface(s).
c In the case of DBL and ConA type tetramers, the PDB codes of X1 and X2 dimers respectively are given, and those of the canonical dimers are included in type II category.
d,e The unusual types of dimeric interfaces seen in GS1 and PNA are analyzed separately.
f Arcelin1 (1avb) and Arcelin-5 (1ioa) do not bind to any sugar.
g DB58 (1g7y) exists as an X1 dimer as well as a II+X1 tetramer, and hence, is considered in both types.
h In the case of GS1 (1hql), the PDB code of the unusual interface is given, and that of the X4 interface has been included in the GS4-type category.
The clustering algorithm used in the present study utilizes the crystal structures of proteins to determine clusters of spatially interacting side chains in these protein structures using a graph spectral technique. The method also quantitatively evaluates the interactions amongst the cluster-forming residues using an interaction criterion, given in terms of percentage of the total interactions possible for a given residue type. The higher the value of the percentage interaction criterion, the stronger the interaction among the residues within a cluster (Kannan and Vishveshwara 1999). The graph-spectral algorithm also gives the eigen spectra for the cluster-forming residues, constituting the eigenvalues and their corresponding vector components. The vector components of the top eigenvalues give information regarding the centers of the clusters (Brinda et al. 2002; Vishveshwara et al. 2002). The higher the magnitude of the vector component, the greater is the contribution of the corresponding residue to the stability of the cluster. The details of the methodology are discussed in a later section.
Nature, types, and states of quaternary association in lectins
As mentioned earlier, the seven types of dimeric interfaces constitute the nine types of quaternary structures seen in legume lectins. Table 1 gives the details of the different types of quaternary structures seen in legume lectins and the combinations of dimeric interfaces seen in these quaternary structures along with their sugar specificities. The nine different kinds of quaternary structures seen in legume lectins include Canonical, ECorL-type, GS4-type, DBL-type, ConA-type, PNA-type, GS1-type, DB58-type, and Arcelin-5-type (monomeric) and the different kinds of dimeric interfaces seen in these nine types of quaternary structures include types II (canonical), X1 (DB58-type), X2 (noncanonical interface of ConA), X3 (ECorL-type, handshake), X4 (GS4-type, back to back), and the unusual interfaces of PNA and GS1. The legume lectin or the “jelly-roll” fold can exist as monomers (as in the case of Arcelin-5) or dimers (like Canonical, ECorL, GS4, and DB58 types) or tetramers (such as DBL, ConA, PNA, and GS1). The tetramers are essentially dimers of dimers, and hence, we see a combination of dimeric interfaces in the tetramers. For example, the DBL tetramer is a combination of Canonical and DB58 types, whereas the ConA type is a combination of Canonical and a different type of dimeric interface that is seen only in the tetramers of this type. The PNA tetramer is made up of Canonical, GS4 and an unusual type of interface seen only in PNA. GS1 type is essentially a tetramer with GS4 type and an unusual type of interface, again seen only in this case. The ECorL dimer (X3) is the only dimer that is not seen in tetramers, and the unusual interfaces of PNA and GS1 and the noncanonical dimeric interface of ConA (X2) are seen only in tetramers and not in dimers. All the others, namely, Canonical (II), GS4 (X4), and DB58 (X1), occur as both dimers and tetramers. Moreover, the DB58 dimer is the only known example of an X1 dimer (X1 dimer in solution and II + X1 tetramer in crystal structure). All other legume lectins with an X1 interface, including DBL, which is highly homologous to DB58, are known to exist as II + X1 tetramers in solution as well as in crystal structure.
Figure 1A–E ▶ shows the major five types of dimeric interfaces (II, X1, X2, X3, X4) seen in legume lectins. It is clear from Figure 1 ▶ that the difference between these five interfaces is mainly owing to the orientation of the β-faces at the interfaces, although the overall tertiary structure of the monomer is very similar in all of them. However, these associations are essentially brought about by some specific residues in the tertiary structure of the monomers. This study focuses on the identification of the residues that impart such kind of specificity to the quaternary association of the monomers with similar tertiary structure.
Figure 1.
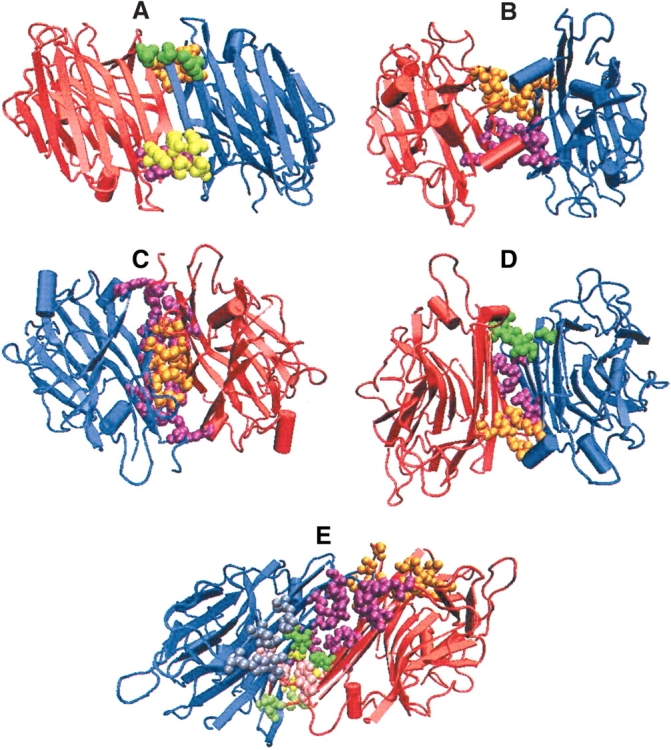
Examples of the five major types of interfaces in legume lectins. A–E represents the three-dimensional structure of the legume lectins belonging to these five types of dimeric interfaces. The monomer chains are represented in the form of cartoon diagrams in blue and in red color, respectively. Although the tertiary structures are similar, in all the five cases, their quaternary associations are different as seen in the figure. The interface cluster-forming residues are represented as van der Waals’ spheres. Each cluster is colored differently to differentiate them in the three-dimensional space. (A) Canonical—type II (1fatAB at 6% cutoff); (B) ECorL-type—X3 (1axy at 5% cutoff); (C) GS4-type—X4 (1gsl at 6% cutoff); (D) DB58-type—X1 (1qnwAC at 4% cutoff); (E) Noncanonical interface of ConA-type—X2 (1dglAC at 4% cutoff).
Considering the various states of oligomerizations seen in legume lectins, we find that they exist as monomers, dimers, and tetramers, as mentioned above. The only monomeric case is Arcelin-5 (Hamelryck et al. 1996), while all others are either dimers or tetramers. These dimers and tetramers are generally homooligomers where all the chains in the proteins are identical in nature. Nevertheless, there are some proteins that are heterooligomeric in nature. For example, DB58 is a heterodimer whereas DBL is a heterotetramer (Buts et al. 2001). The difference between the two chains of the heterodimers of DB58 and DBL (which is a dimer of a heterodimer) is that one chain is truncated by 12 residues at its C terminus. This removal of 12 C-terminal residues is effected by a posttranslational modification after the synthesis of the complete chain (Buts et al. 2001). The presence of the additional 12 residues at the C-terminal of one of the chains seems to be of significance from the quaternary structure point of view, because these residues form a helix that gets sandwiched between the two monomers (one with the C-terminal helix and the other without) involved in X1 dimerization, and thus stabilizes the X1 dimeric interface. However, this C-terminal helix is not compulsory for the formation of X1 type interface, because many of the other legume lectins like Bark lectin (1fny), MAL (1dbn), and UEA-II (1qnw) form X1 interfaces without this C-terminal helix region. Hence, this region is not present in the sequence motif required for X1 type, according to our present analysis.
As mentioned earlier, legume lectins occur mainly as dimers or tetramers, which are generally dimers of dimers (Manoj and Suguna 2001). Considering the seven different types of interfaces, it is found that not all seven types occur as both dimers and tetramers in nature. Some specificities between the types and states of oligomerization are seen in the legume lectins. To understand this, we first studied the overall region in the sequence and structure that contributes to the different interface types. We find that the region in the sequence and structure, contributing to the type II interface is completely different from the region contributing to the X1, X2, X3, and X4 types of interfaces. Type II region is determined by some specific residues in the N-terminal region of 1–60 residues and the C-terminal region of 210–240 residues. But the X1, X2, X3, X4 interface types are contributed by the 70–80 region and the 160–200 region in the protein sequence. Further, the existence of these lectins as pure dimers or tetramers depends upon whether they have the residues required to form two different types of interfaces. Moreover, the determinants of types X1, X2, X3, and X4 occur at the same region in the sequence; they are mutually exclusive, and hence, cannot coexist. However, they can exist in combination with the type II interface because the type II is contributed from a different region in the sequence. The unusual interface of GS1 is contributed from the N-terminal region (10–40) and C-terminal region (220–230), whereas that of PNA includes some residues in the 70–80 region and 150–160 region apart from the contributions from the N and C terminal regions (20–30 and 220–230). Therefore, from pure sequence perspective, the type II and the unusual interfaces of PNA and GS1 can occur in combination with any of the other four interface types (X1, X2, X3, and X4) in tetramers.
Consensus patterns specific for each interface type
The identification of the consensus patterns of residues for the different interface types seen in legume lectins has been carried out by mapping the structurally conserved residues identified in the interface clusters on to the multiple sequence alignments as described in the Materials and Methods section. This could be done only for five out of the seven interface types because the two unusual interfaces of PNA and GS1 are the only known examples of each type, and hence, a multiple sequence alignment for these two interface types cannot be obtained. However, the interface cluster analyses of these two unusual interfaces have been carried out using the available crystal structures, and the details of these are presented in a later section. Therefore, the consensus sequence patterns could be obtained only for the five interface types (II, X1, X2, X3, and X4) that have more than one example each. Hence, these five dimeric interface types and their consensus patterns are discussed in detail in this section and the next.
The multiple sequence alignments of the five dimeric interface types are shown in Figure 2A–E ▶. Due to space constraints, only a few examples in each case and specific sequence regions involved are shown in the alignment. The residues identified in the consensus pattern are shown in bold and are underlined. Table 2 summarizes the major results of this analysis. It gives in detail the residues and patterns required for each of these five interface types as identified from Figure 2 ▶. It is evident that the presence of a particular residue type at a particular region can be responsible for deciding what kind of interface the lectin sequence forms. For example, in the 60–70 region, there is a sequence motif “SFX,” where X can be one of the following residues: Tyr, Glu, or Asp. The type of interface (X4/GS4, X3/EcorL, or X2/ConA) adopted depends on the nature of this residue X. Similar pattern specificities exist in the other regions of the lectin sequence also as can be understood from Figure 2 ▶ and Table 2. Although the consensus pattern for the type II interface involves only five residues as can be seen in Figure 2 ▶, we find that many residues in the N- and C-terminal regions of these proteins are extensively involved in the interface clusters of the type II interface. There are many negatively charged and aromatic residues in the N terminus that take part in the interface cluster formation in all the type II interfaces, but these are not always the same residues from the aligned sequence perspective, and hence, their absence in the consensus pattern.
Figure 2.
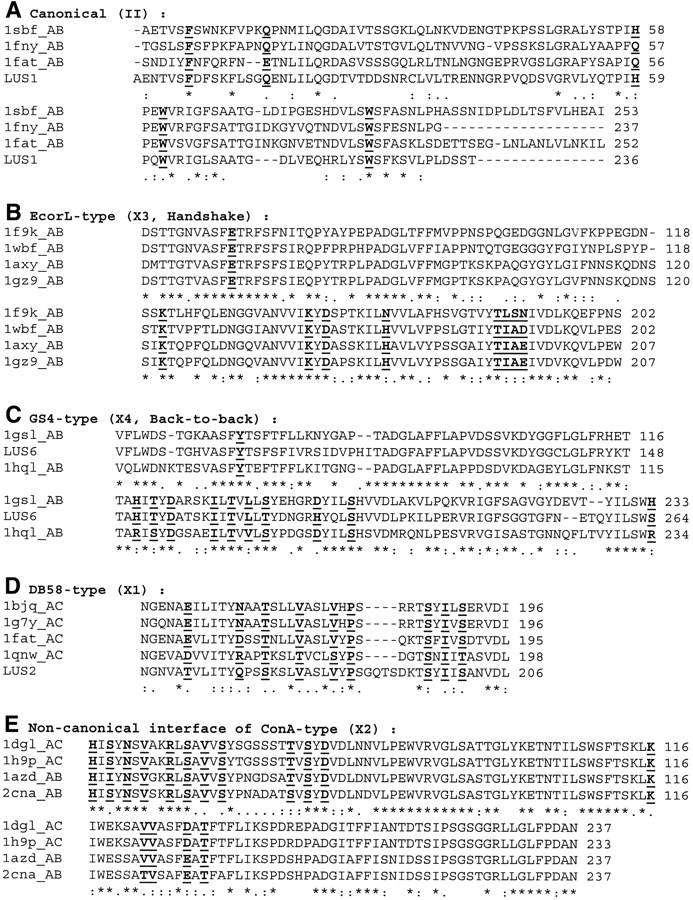
Multiple sequence alignments of different legume lectin interface types. (A) Canonical (II); (B) ECorL-type (X3); (C) GS4-type (X4); (D) DB58-type (X1); (E) Noncanonical interface of ConA-type (X2). For want of space, only some examples in each case and the important regions involved are shown in the figure, that is, N terminus and C terminus, region 50–80, and region 150–200. The alignment also includes an example of a lectin with unknown structure (LUS), wherever available. The residue number of each protein in the aligned region is given at the end of each line. The complete conservation, partial conservation, and conservative mutations that occur in the sequence alignment are shown as “*”, “.”, and “:”, respectively. The residues, which are either completely or partially conserved or conservatively mutated in sequence and are also present in the interface clusters of the legume lectins belonging to a particular category, are shown in bold and underlined.
Table 2.
Consensus patterns in the five interface types of legume lectins obtained from multiple sequence alignments and interface cluster-forming residues
|
_: Nonconserved residue in the flanking sequence.
/: Residues that have undergone mutations occurred at a position, giving rise to partial conservation or conservative mutations.
Residues forming the consensus motif are underlined and shown in bold and italics.
Figure 1A–E ▶, shows the interface cluster-forming residues in the major five dimerization types (II, X1, X2, X3, and X4) in the context of their overall structures. The positions and relative orientations of some of the interface cluster-forming residues in these five dimerization types are shown in Figure 3A–E ▶. Even though each interface may have many interface clusters, only one cluster in each case has been shown in the figure. The coloring scheme differentiates the structurally conserved ones from the others. The golden and pink residues are the structurally conserved, interface cluster-forming residues, from the two separate chains of the dimer respectively. Also shown in the figure are the positions of the vector components of the cluster forming residues, in the top eigenvalues. We can see that the vector components of the golden and pink colored residues are always high in magnitude, indicating that these conserved residues at the interface clusters, contribute significantly to the stability of the interface. The vector component magnitude correlates well with the structural conservation in all these cases. Further analysis shows that these cluster residues with high vector component magnitude in the top eigenvalues are conserved in sequence as well. Hence, the consensus of the sequentially and structurally conserved interface residues involve residues with high vector component magnitude in the top eigenvalues, indicating that these residues contribute largely to the interface formation and stabilization. Hence, it is significant that such residues form the determinants of quaternary association in legume lectins.
Figure 3.
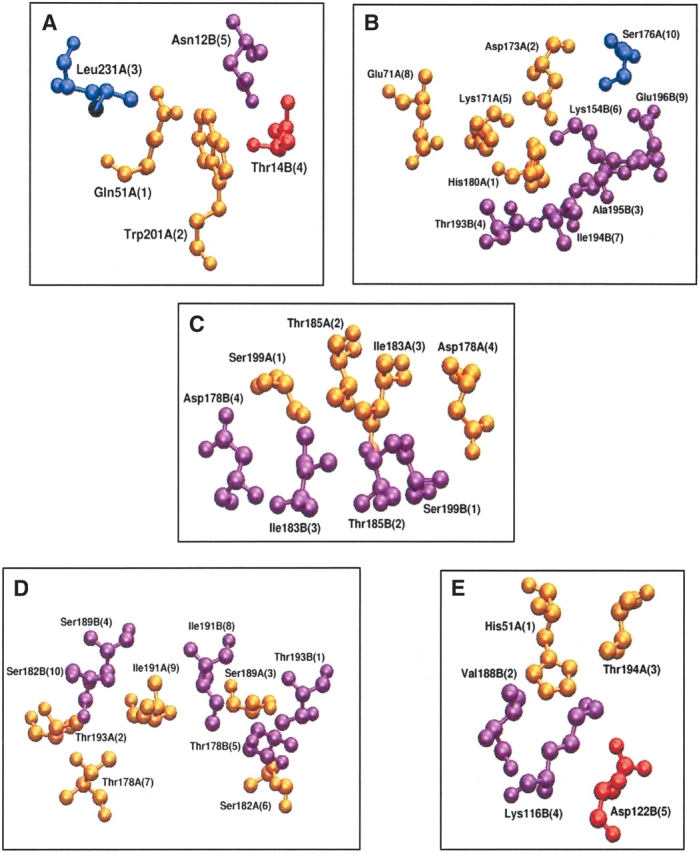
Interface cluster-forming residues in the five major types of legume lectin interfaces. (A) Canonical—type II (1fatAB at 6% cutoff); (B) ECorL-type—X3 (1axy at 5% cutoff); (C) GS4-type—X4 (1gsl at 6% cutoff); (D) DB58-type—X1 (1qnwAC at 4% cutoff); (E) Noncanonical interface of ConA-type—X2 (1dglAC at 4% cutoff). The representations are in a ball-and-stick model in the entire figure. Due to constraint of space, only one interface cluster is shown in each example, although there may actually be more than one interface cluster. The coloring scheme is as follows: blue, residues from chain A that are not conserved in the interface clusters of other proteins of the same interface type; red, residues from chain B/C/D that are not conserved in the interface clusters of other proteins of the same interface type; gold, residues from chain A that are conserved (in sequence and structure by total or partial conservation or conservative mutation) in the interface clusters of other proteins of the same interface type; magenta, residues from chain B/C/D that are conserved (in sequence and structure by total or partial conservation or conservative mutation) in the interface clusters of other proteins of the same interface type. The residue number, name and chain are indicated in the figure. The position of the vector components of the top eigenvalues corresponding to these interface cluster-forming residues is also indicated within parentheses. We see that the residues colored in gold and in magenta have higher vector component magnitude than the ones colored in blue and in red, indicating that the residues that are important for the stability of the cluster and the interface (indicated by high vector component magnitude) and the ones that are conserved in the interface clusters (shown in gold and magenta) correlate very well.
Differences between the consensus patterns of each interface type
A quick look at the consensus patterns of the five major dimeric interface types (Fig. 2A–E ▶) shows that there are specific signature motifs in sequence for each dimerization type, as elucidated in Table 2. Residues forming the consensus pattern are shown in bold and are underlined. We can see that there are exclusive sequence patterns that signify each of the five dimerization types. The difference between the patterns responsible for each dimeric interface type can be understood from Figure 2 ▶ and Table 2. For example, the presence of “T V S Y D” pattern in the 190 region of the sequence indicates an X2 interface (noncanonical interface of ConA), whereas “S Y I V S” in the same region indicates an X1 interface (DB58). Similarly, the presence of “V I K Y D” pattern in the 170 region indicates an X3 interface (ECorL), whereas “H I T Y D” in the same region indicates an X4 interface (GS4). Such specific signature sequence motifs have been seen for all the five interface types for which consensus patterns could be obtained. These motifs determine and differentiate each interface type from the other.
We have compared and contrasted the sequence patterns thus obtained, so as to understand the oligomerization specificities of the interface types. The residues responsible for type II interface are present in most of the tetramer forming sequences (because one type of the dimeric interfaces in the tetrameric interfaces can be a type II interface, as discussed earlier) as well as in the exclusive dimers forming type II interfaces, as expected. These residues and patterns are absent in the GS4 or X4 dimer (1gsl) and the GS1 or X4 + unusual tetramer (1hql), thus ruling out the possibility of a type II interface in these cases. These X4 forming sequences have only the pattern required for the X4 type, and hence form either exclusive X4 dimers or tetramers with an X4 and an unusual interface type (because type II is ruled out). The reason for GS4 remaining a dimer while GS1 tetramerizes using this unusual interface is discussed in a later section. Some of the sequences that form X3 dimers (1axy and 1gz9) do have all the residues and patterns required for the type II interface as well, along with those required for the X3 type. Yet, these form exclusive X3 interfaces only, and no type II interface (which can be formed on tetramerization) is seen in these cases. The other X3 forming sequences (1wbf and 1f9k) have exclusive X3 patterns and type II patterns are absent in these cases and so they form exclusive X3 dimers. Hence, X3 type seems to remain a dimer only, irrespective of whether it has patterns required for type II or not.
The tetramers involving X1 and X2 interface types (DBL and ConA types) always have the type II pattern also conserved in them along with the patterns required for either X1 or X2, indicating that these sequences are capable of tetramerizations. At one face they use the X1 or X2 type of interface to interact with the other monomer, while they use the other face of the monomer to form a type II interface with another monomer, thus forming a dimer of a dimer, that is, a tetramer with two types of interfaces. Hence, the X1 and X2 tetramers always have a type II interface along with either a X1 or X2 type. This fact is confirmed by the presence of two consensus patterns (type II and X1 or X2) in their sequences. However, the DB58 dimer is an exception because it is known to exist as an X1 dimer in solution and as type II + X1 tetramer in crystal structure (Buts et al. 2001). We find that it has the consensus patterns required for both II and X1 interface types. The peanut lectin (PNA, 2pel) is known to exist as a tetramer with two X4 interfaces, one type II interface and one unusual interface, leading to an open quaternary structure (Banerjee et al. 1994). As expected, PNA has most of the residues required for the X4 and type II interfaces.
Arcelin-5 has been crystallized as a monomer, but Arce-lin-5 oligomers are known to exist in solution under some specific conditions (Hamelryck et al. 1996). We also know that Arcelin-1, which has 62% sequence identity to Arcelin-5, exists as a canonical dimer. However, a single residue insertion in the 10–15 residue region of Arcelin-5 has been shown to inhibit the formation of the Arcelin-1 type canonical dimer (Hamelryck et al. 1996). We have checked for the presence of any of the five consensus patterns in the Arce-lin-5 monomer. The absence of all the five consensus patterns could justify the existence of Arcelin-5 as a monomer. Our analysis, however, shows that Arcelin-5 has the complete sequence pattern required for type II interface (5/5) and most of the residues (8/9) required for X3 type as well. Hence, it might be capable of forming II or X3 interface types under suiz conditions. However, the positions of the X3 residues in Arcelin-5 are not completely conserved, and the charged residues in the consensus pattern are conservatively mutated to other polar residues or replaced by nonpolar residues, thus leading to a net loss of charged interactions when compared to the normal X3 interfaces. Moreover, some of the bigger residues have been mutated to smaller ones like K to N, I to V and T to S, which may also lead to loss of some of interactions required for interface stability. Thus, X3 interface is likely to be highly destabilized in this case. However, the residues required for the type II interface are completely conserved in the sequence of Arcelin-5. Although our consensus pattern for type II interface involves only five residues from the sequence, we know that the N- and C-terminal regions of the sequence are extensively involved in the type II interface. So, any subtle change in these regions such as the single residue insertions mentioned by Hamelryck and colleagues could destabilize the formation of the type II interface. Hence, it is likely that both the type II and X3 interfaces will be considerably destabilized in the case of Arcelin-5. However, the possibility of the existence of type II dimers (like in Arcelin-1) cannot be completely ruled out because Arcelin-5 oligomers are known to exist under suitable conditions and the Arce-lin-5 sequence has all the residues present in the consensus pattern of the type II interface.
The type II canonical interface seems to be a basic kind of interface present in most of the tetramers and dimers, and the sequence pattern responsible for type II interface seems to be present in some non-type II dimers and monomers as well. Hence, the presence of the consensus sequence pattern of type II in legume lectin sequences seem to be the necessary condition for type II formation but not the sufficient condition for the same. Therefore, type II is a universal interface in legume lectins, with its sequence pattern being present in most of them, whether or not they actually form type II interfaces. However, the presence of other sequence patterns in the lectin sequence determines the nature, type, and state of oligomerization in these lectins. This analysis has thus answered most of our questions regarding why some sequences form dimers while the others form tetramers, and why some interface types have specific states of oligomerizations. The following two sections dealing with the unusual interfaces seen in some legume lectins and the legume lectins with multiple oligomeric states, respectively, will bring further clarity to the preferences in nature, types, and states of quaternary associations of these legume lectins, thus enhancing our understanding of the factors responsible for the same.
Characterization of the unusual interfaces of PNA and GS1
The quaternary structures of PNA (II + X4 + unusual) and GS1 (X4 + unusual) show the presence of an unusual interface in each, which has not been seen in any other legume lectin (Banerjee et al. 1994; Tempel et al. 2002). These interfaces have not been characterized like the other dimeric interfaces, and the consensus pattern for these interfaces have not been obtained because we do not know of more than one such example, and hence, a multiple sequence alignment cannot be obtained for these cases. Nevertheless, because the crystal structures of these single unusual interfaces are available, we have determined the interface clusters in the interfaces using the graph spectral method, and have compared these cluster forming residues with other related legume lectins. The results are presented below.
The monomer of Griffonia simplicifolia lectin1 or GS1 (1hql), has the residues required for the X4 interface in its sequence. Hence, three of the other interfaces (X3, X2, or X1) are ruled out because the determinants of all these four types occur in the same region of the sequence, so the presence of one excludes the presence of the other. The only other possibility for tetramerization is the type II interface. But on careful analysis, we find that this protein does not have the sequence motif required for the type II interface. Hence, the only way it can tetramerize is by forming an unusual type of interface. The cluster obtained at this interface at 6% cutoff includes the residues W10 (other chain), T26, G28 (other chain), Q31, T35, F75, Y226, and L228. We cannot conclusively say that these are the required residues for this unusual interface, but because these are forming interacting clusters at the interface, they seem to play a role in stabilizing this interface. GS4 is highly homologous to GS1, but remains as an X4 dimer and does not tetramerize using this unusual interface. Hence, we compared the sequences of GS1 and GS4 to check for the presence of the residues involved in the interface clusters of the unusual interface of GS1 in GS4. Six out of the eight residues involved in the unusual interface of GS1 are conserved in GS4 also. The only mutations are T26I and F75L, where there is a loss of the polar side chain when threonine is mutated to isoleucine, and also a loss of the bulky aromatic side chain when phenylalanine is mutated to leucine. A careful look at the interface cluster of the unusual GS1 interface shows that the F75 is involved in an aromatic π–π stacking interaction with the W10 of the other monomer, thus stabilizing the interface and T26 is involved in hydrogen bonding with Q31 of the same monomer, thus stabilizing this interface cluster. These interactions are lost when F75 is mutated to leucine and T26 is mutated to isoleucine. These losses are likely to destabilize the unusual interface in GS4, thereby preventing it from forming the GS1-like tetramer. Hence, GS4 exists only as an X4 dimer.
The peanut lectin PNA (2pel), has been found to be a special case in all the previous analyses of legume lectins (Banerjee et al. 1994). It is a homotetramer with two X4 dimers tetramerizing to form a type II interface and an unusual interface, leading to an open quaternary structure. The interface cluster analysis of the type II and X4 interfaces of PNA and the multiple sequence alignments of this lectin with other type II and X4 dimers show that PNA has most of the residues required for the type II (5/5) and X4 (8/11) interface types. Hence, it is capable of forming both type II and X4 interfaces. The interface cluster analysis of the unusual interface of PNA at 6% cutoff yields two interface clusters, namely, (1) L27, Q33, and S28 (other monomer), and (2) V160, R221, N31, E72 (other monomer), K74 (other monomer), and G158 (other monomer). Here again, this is not the conclusive pattern for the interface, but are the cluster-forming residues involved in intersubunit interactions. A comparison of the sequences of other type II and X4 dimers with the PNA sequence shows that most of the interface cluster residues involved in the unusual PNA interface are absent in these sequences, and therefore, these type II and X4 dimers are unable to tetramerize using this unusual interface.
Examples of legume lectins with multiple oligomeric states
Some lectins are known to have unusual quaternary association properties, where they are known to exist in more than one oligomeric state under different conditions. For example, one of the lectins from Dolichos biflorus (DB58) considered in the present data set, 1g7y, is known to be an X1 dimer in solution, whereas it is a type II + X1 tetramer in the crystal structure (Buts et al. 2001). This lectin is highly homologous to DBL (another lectin from the same source), which exists as a type II + X1 tetramer in solution as well as crystal. It has been proposed that both DB58 and DBL are capable of existing as dimers and tetramers in solution, and that there is a dynamic equilibrium between the two oligomeric states in both these cases, where there is a strong preference for tetramers in case of DBL and dimers in case of DB58 (Buts et al. 2001). This preference of dimeric state in DB58 in solution has been attributed to a single amino acid substitution in the sequence (P14S) causing the destabilization of the type II interface in DB58. Hence, the type II interface of DB58 does not exist in solution although it is seen under certain conditions as in the crystal structure. Moreover, the existence of this type II interface in the crystal structure means that it is not sterically prohibited. In the present analysis, we have looked at the strengths of the interface clusters in both these interfaces in the crystal structure to examine why this protein exists as a dimer in solution. We find that the interface clusters at the X1 interface are formed at a very high interaction cutoff (12%), whereas the type II interface clusters are formed at a lower cutoff of 6%. Moreover, at 6% cutoff, the X1 interface is much stronger, in terms of both the number of interface clusters and the number of residues in the interface clusters, than the type II interface. Therefore, the X1 interface is more stabilized than the type II interface, and hence, the existence of the X1 dimer in solution.
Two other lectins, 1dbn (MAL) and 1qmo (FRIL), exist as tetramers in crystal structures, while they are dimers in solution. In the crystal structure, 1dbn is a tetramer consisting of type II and X1 interfaces (Imberty et al. 2000), while 1qmo is a tetramer consisting of type II and X2 interfaces (Hamelryck et al. 2000). However, in these cases, there is no conclusive evidence as to the type of dimerization that exists in solution. On the other hand, it has been elucidated that FRIL may exist as a type II dimer in solution rather than X2 dimer, although the possibility of the existence of alternate X2 dimers in solution cannot be completely excluded (Hamelryck et al. 2000). Analysis of pair-wise residue contacts at the intersubunit interfaces and buried surface area of the X2 interface of FRIL had shown that the residue contacts and buried surface area at the X2 interface are much less than what is normally seen in the X2 interface of ConA. These differences in residue contacts and buried surface area may be destabilizing the X2 interface of FRIL, thus preventing it from forming the ConA type tetramer (with type II and X2) in solution. However, this weak X2 interface of FRIL might get stabilized due to its binding with multivalent glyco-conjugates capable of two or three dimensional cross-linking, as seen in the crystal structure. Hence, it may preferably exist as the more stable type II dimer in solution and form II + X2 tetramers under conditions of crystallization (Hamelryck et al. 2000). In the case of MAL, the comparison of the pair-wise residue contacts and the buried surface area at the type II and X1 interfaces, have not yielded any significant differences between each other or from those seen normally in canonical (II) or DB58 (X1) dimers, and hence, there is an uncertainty as to which dimer exists in solution, although both type II and X1 dimers are equally probable (Imberty et al. 2000).
We have analyzed the two interfaces of MAL (II and X1) and FRIL (II and X2) using the strength of the interface cluster as the criterion to understand which one of the dimeric interfaces probably exists in solution. A look at the interface clusters of these proteins shows that both MAL and FRIL have stronger interface clusters at the canonical interface than the X1/X2 interface. The clusters at the canonical interfaces of these two tetramers are formed at higher interaction cutoff than the X1/X2 interface, and at a given interaction cutoff, the canonical interface has more interface cluster-forming residues than X1/X2. Hence, canonical seems to be the stronger interface than X1 or X2 in both these cases. Therefore, it is likely that these proteins exist as canonical dimers rather than X1 or X2 dimers in solution. Nevertheless, the possibility of the alternate X1/X2 dimer cannot be completely excluded.
The latter cases of legume lectins with multiple oligomeric states give an indication that the other tetrameric legume lectins can also exist in multiple oligomeric states, where the dimers and monomers forming the tetramers also exist independent of each other along with the tetramer. There could exist a dynamic equilibrium among all the four oligomeric states, namely, monomer, two different types of dimers, and a tetramer, with preference for any of the four states based on the strengths and stabilities of the different interfaces contributing to the overall stability of the protein. As mentioned earlier, Arcelin-5 remains a monomer, although it is capable of forming the canonical dimer. This shows that the other oligomeric lectins could also exist as monomers, which could be in an equilibrium state with their higher order oligomers. Moreover, the weak higher order oligomers, which are normally not seen in solution (as in the case of FRIL), could be stabilized under suitable conditions as a result of binding with two or three dimensionally cross-linked glycoconjugates as already mentioned by Hamelryck et al. (2000) leading to multiple oligomeric states in these legume lectins. This brings out the striking feature that the state of oligomerization of these legume lectins can be affected by small perturbations in their sequence and environment.
Identification of interface type in Lectins with Unknown Structures (LUS)
One of the motives of this analysis is to predict the oligomerization state and type of a lectin whose structure is yet unknown, from the consensus pattern obtained for the five major interface types using the lectins of known structures. As discussed in the previous sections, the consensus sequence patterns for the five main interface types have been obtained using the lectins with known three-dimensional structures. This is now applied to a set of six lectins with unknown structures (Kouchalakos et al. 1984; Konami et al. 1990, 1992; Kusui et al. 1991; Calvete et al. 1998; K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.). The sequence of each of these six lectins is aligned with the already obtained multiple sequence alignments for each of the five interface types, and then the LUSs are analyzed for the presence of each of the five consensus patterns. A score is given for each of these alignments based on the number of consensus residues present in the LUSs. Figure 2 ▶ shows the alignment of a sample LUS in some interface types, where the conserved consensus residues are highlighted in bold.
Table 3 gives the presence or absence of the consensus pattern for each of the five dimerization types in all the six LUSs, based on visual inspection. Although the presence of a consensus pattern indicates the possibility of a particular dimerization type, the scores (number of consensus residues present in the LUSs) can be used to rank the dimerization types in case a LUS shows more than one pattern. It is evident from Table 3 that LUS 1 has the pattern required for the type II dimer, LUS 2, 4, and 5 have the patterns required for both type II and X1, and hence, are likely to be tetramers with type II and X1 interfaces (they can also have multiple oligomerization states like DB58, where it exists as an X1 dimer as well as II + X1 tetramer), LUS 6 is an X4 type (can be either dimer or tetramer because there is a possibility of the unusual interface in this case), and LUS 3 does not show any clear demarcation as to which type of interface it prefers. Because LUS 3 does not have any of the patterns required for II, X1, X2, X3, and X4 interface types, we aligned LUS3 sequence with those of PNA and GS1 to check for the presence of the residues involved in the unusual interface types present in these. We find that LUS3 does not have the residues required for both the unusual interfaces of PNA and GS1, indicating that all the seven known interface types are likely to be destabilized in the case of LUS3. In the case of LUS6, where we find that the X4 pattern is conserved, we checked for the presence of residues involved in the unusual interfaces of PNA and GS1, because both these have X4 patterns as well along with their unusual interfaces. We find that the LUS6 sequence does not have the residues required for the unusual interface of PNA. Moreover, it also does not have some residues required for type II canonical interface. Hence, PNA-type quaternary structure (II + X4 + unusual) is highly unlikely in this case. When checked for the presence of the interface cluster forming residues of this unusual interface type of GS1 in LUS6, we found that six out of the eight residues are present in LUS6 as well. Although this is not a conclusive evidence for the formation of this unusual GS1-type interface in LUS6, the possibility cannot be ruled out. The results of this analysis have been compared with the already available experimental results on the state of oligomerization of these LUS (Osawa et al. 1978; Kouchalakos et al. 1984; Young et al. 1984; Konami et al. 1990, 1992; Kusui et al. 1991; Calvete et al. 1998; Cheng et al. 1998; K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.) and the dendrogram obtained from the multiple sequence alignments of all the legume lectins with known and unknown structures (Manoj and Suguna 2001). The results of these comparisons are summarized in Table 4.
Table 3.
Presence of oligomerization-specific sequence motif in lectins with unknown structures
| LUSa (species) \ Interface type | X1 | X2 | X3 | X4 | II |
| LUS1 Onobrychis viciifolia | ×b | × | × | × | ✓c |
| LUS2 Cystisus scoparius | ✓ | × | × | × | ✓ |
| LUS3 Lotus tetragonolobus | × | × | × | × | × |
| LUS4 Vatairea macrocarpa | ✓ | × | × | × | ✓ |
| LUS5 Cicer arietinum | ✓ | × | × | × | ✓ |
| LUS6 Bauhinia purpurea | × | × | × | ✓ | × |
a LUS: lectins with unknown structures.
b×: sequence motif not present.
c ✓: sequence motif present.
Table 4.
Comparison of the predicted interface types with already available data
| Results from current analyses | Experimental resultsc | ||||
| LUSa | State of oligomerization | Type(s) of interface(s) | Results from dendrogramb (type) | State | Reference |
| LUS1 | Dimer | II | II | Dimer | Kouchalakos et al. 1984 |
| LUS2 | Tetramerd | II and X1 | II and X1 | Tetramer | Young et al. 1984 |
| LUS3 | Not conclusive | Not conclusive | Separate node | Tetramer | Cheng et al. 1998 |
| LUS4 | Tetramerd | II and X1 | II and X1 | Tetramer | Calvete et al. 1998 |
| LUS5 | Tetramerd | II and X1 | II and X1 | Unknowne | — |
| LUS6 | Tetramerf | X4 | X4 | Tetramer | Osawa et al. 1978 |
a LUS: lectins with unknown structures.
c Experimental results are based on gel filtration or electrophoresis experiments.
d LUS 2, 4, and 5 have patterns for both II and X1 interfaces. They can either exist as II+X1 tetramers as in DBL, or exist in multiple oligomerization states like in DB58 (X1 dimer and II+X1 tetramer).
e Oligomerization state not known experimentally.
f LUS 6 can be dimeric as in GS4 (X4) or tetrameric as in GS1 (X4+unusual interface) as it has the residues required for X4 and the unusual interface of GS1.
Table 4 shows that the results from the present analysis correlate very well with the already available experimental and dendrogram results. LUS1 is a type II dimer from the present analysis as well as from the experimental and dendrogram results. Even the case of LUS 3, which does not score with any of the seven interface types with any clarity, comes up as a separate node in the dendrogram away from all the known lectins, indicating that it is likely to have a quaternary association type that is different from the known legume lectin cases. LUS 2, 4, and 5 are found to be tetramers from experiments; hence, they are likely to be II + X1 tetramers like DBL rather than X1 dimers as in DB58. In the case of LUS 6, we do not have any conclusive evidence as to whether it is a dimer or tetramer from the present analysis, because it has only the X4 pattern conserved in it. Nevertheless, it is likely that LUS 6 forms a GS1-like tetramer consisting of two X4 dimers tetramerizing using an unusual interface because it has most of the residues involved in this unusual type of interface and also because experimentally LUS 6 is known to be a tetramer. The crystal structure of LUS 6 holds conclusive evidence for this, and if LUS 6 does form the unusual interface as seen in GS1 (along with X4), it could also help us get the consensus pattern characteristic of this unusual interface type. Thus, results of the present analysis have correlated well with already existing data. Hence, we have a highly reliable and useful method at hand to correlate the state, nature, and type of quaternary associations in legume lectins, which is highly consistent with the available crystal structures, and has considerable predictive ability for other members of the family where structures are yet to be determined.
Conclusions
Our method of using the clustering algorithm in combination with the traditional multiple sequence alignment methods is a novel way of analyzing quaternary association in proteins, and has provided interesting results in the case of legume lectins. The present analysis is based on a graph-spectral algorithm, which is used to identify clusters of amino acid residues from complex three-dimensional structures of proteins. The algorithm takes into consideration noncovalent tertiary interactions among amino acid residues for the formation of networks of spatially connected residues (clusters) at protein interfaces, and hence, it involves the global topology of the protein and not just the local interactions in the protein structure. One of the main features of the current method is that it can be easily combined with traditional techniques like multiple sequence alignment algorithms, to yield interesting insights into protein structures. This algorithm has, therefore, proven to be very robust and elegant for protein structural analysis, and in this case, has helped us determine the factors responsible for imparting specificities to the nature of quaternary association in the legume lectins. Additionally, the method has proven to have a powerful ability in predicting the nature of quaternary association in lectins with unknown structures. The present analysis has helped us solve the problem of identifying the determinants of the quaternary association in legume lectins, which has been elusive to pair-wise interaction studies and multiple sequence algorithms, and has given a new perspective for looking at such problems that involve the understanding of protein structures. At the fundamental level, it also supports the idea that the state and nature of oligomerization of proteins are also encoded in the primary structure of the protein.
Materials and methods
We have used a combination of the graph-spectral clustering algorithm and multiple sequence alignment to analyze and identify the factors determining the nature and type of interfaces seen in the quaternary structures of legume lectins. The following sections deal in detail with the methodology used for the same.
Data set
The data set for this analysis consists of the crystal structures of all the legume lectins available in the Protein Data Bank that were found to be 28 in number (excluding the redundant structures). The PDB files were obtained from http://www.rcsb.org (Berman et al. 2000). Although lectins mainly form homooligomers in nature, they mainly occur in two different states of oligomerization. These include dimers and tetramers, though monomers are also seen (Arcelin-5). The tetramers are generally dimers of dimers from the structural perspective, thus forming symmetric and identical interfaces in the tetrameric protein. Because our aim is to analyze and characterize the dimeric interfaces of legume lectin oligomers, the first step is to eliminate the identical interfaces within a protein and obtain a nonredundant data set of legume lectin dimers from the available dimers and tetramers. The dimeric legume lectins in this nonredundant data set are then categorized based on the nature and type of their interfaces. The legume lectin dimeric interfaces have thus been categorized as seven different types, namely type II (canonical), X3 (ECorL-type, handshake), X4 (GS4-type, back to back), X1 (DB58-type), X2 (the noncanonical interface of ConA-type), and the unusual interfaces of PNA and GS1. From the 28 legume lectins (pdbs) considered in this data set, we thus obtained 39 dimeric units in total, with 20 type II (canonical), 4 X3 (ECorL-type), 2 X4 (GS4-type), 8 X1 (DB58-type), and 5 X2 (noncanoni-cal interface of ConA-type), as shown in Table 1. The unusual interfaces of PNA and GS1 are considered separately in this analysis because they are the only known examples of their types.
Classification of legume lectins
Legume lectins are classified on the basis of their quaternary association as follows:
Based on state of oligomerization: State of oligomerization refers to the number or monomeric units present in the oligomer. In the present data set, the legume lectins are in either dimeric or tetrameric state of oligomerization (Table 1). Both the dimeric and tetrameric states are generally homooligomeric in nature except in the case of DB58 and DBL, as discussed earlier.
Based on quaternary structure: Based on the overall quaternary structure, the legume lectins can be classified as Canonical, ECorL-type, GS4-type, GS1-type, DB58-type, DBL-type, ConA-type, PNA-type, and Arcelin-5-type (Table 1). This nomenclature is named after the characteristic representative of each type.
Based on nature of dimeric interfaces: The overall quaternary structure can be considered as a combination of independent dimeric interfaces. Therefore, there is a necessity to differentiate the types of quaternary structures from the types of interfaces. Hence, we have used a different nomenclature for the interface types as given by Manoj and Suguna (2001). Based on the structure and nature of the interfaces, the interfaces have been classified into seven different types. These seven interface types include II (Canonical), X1 (DB58-type), X2 (noncanonical interface of ConA-type), X3 (Handshake, ECorL-type), X4 (back to back, GS4-type), unusual interfaces of PNA, and GS1 (Table 1). These seven interface types differ in their interface structure, compositions, and orientations. The legume lectins can thus be classified based on the type(s) of dimeric inter-face(s) present in them.
Clustering algorithm
A graph-spectral algorithm is used to determine side-chain clusters and their cluster centers at the interface of the dimeric legume lectins in the data set. The method is described in detail in the paper by Kannan and Vishveshwara (1999). In this method, each residue in the protein structure (coordinates obtained from the PDB file) is considered as a node in the graph, and these nodes are connected by edges based on whether or not they satisfy an interaction criterion. This interaction criterion considers the number of atoms from both residues that come within a distance of 4.5 Å and normalizes this value with respect to the size of the residues under consideration. A user-defined cutoff value is fixed for the interaction criterion and any two residues with interaction greater than the cutoff is considered for cluster formation. A cluster is defined as three or more such interacting residues. The interaction cutoff value is generally in the range of 1% (minimum) to 15% (maximum; Kannan and Vishveshwara 1999; Brinda et al. 2002). A lower cutoff value will give huge clusters comprising residues with low interactions, while a high cutoff gives clusters where the interaction among the cluster-forming residues is high. It is important to optimize this interaction cutoff, to obtain significant clusters, distinguishable from the bulk of the protein (Brinda et al. 2002). One of the significant features of the algorithm is that this interaction criterion takes into consideration only the noncovalent spatial interactions between amino acid residues in the three-dimensional structure of the protein. The sequential covalent interactions are eliminated during the evaluation of the interaction criteria. Based on such an interaction criterion, the connection between two nodes in the graph, that is, the contact between two residues in the protein, is evaluated. Such a graph can be represented by an n × n matrix (n being the number of nodes in the graph) called the Laplacian matrix. This matrix is then diagonalized to obtain the eigen spectra of the graph. This eigen spectrum contains a variety of information regarding the graph, including the cluster-forming residues and the residues that form the centers of such clusters (Kannan and Vishveshwara 1999). The cluster-forming residues are those that have come closer to each other in space in the three-dimensional structure of the protein. The vector components of the second lowest eigenvalue have information regarding the cluster-forming residues, while those of the top eigenvalues give us the centers of these clusters (Kannan and Vishveshwara 1999; Vishveshwara et al. 2002). The vector components of the top eigenvalues are very important because they contain significant information regarding the contribution of each node to the stability of the cluster. The higher this magnitude, the more the contribution of the node to the stability of the cluster (Vishveshwara et al. 2002).
The clustering algorithm has been used to identify the side-chain clusters in the present data set of legume lectins using cutoff criterion above 4%. This cutoff value has been optimized for the present data set to give distinct interface clusters. The interface clusters in these dimers are those that have contributions from both chains of the dimer. The interface cluster-forming residues in these lectin dimers have been used for all further analyses. These interface cluster-forming residues in each dimer are compared with the others within the same dimerization type, to identify those residues that are structurally conserved at the interface of the particular dimerization type. The interface clusters in the unusual interfaces of PNA and GS1 have also been obtained in the same manner.
Multiple sequence alignment using ClustalW
The amino acid sequence of the monomer chain of all the legume lectins in the data set have been obtained from the Swiss-Prot sequence database (Boeckmann et al. 2003). Some lectins have circularly permuted sequences. These have been rearranged for the sake of multiple sequence alignments. Those sequences belonging to the same interface type have been grouped together to obtain five sets of sequences corresponding to each of the five major dimerization types seen in legume lectins. Multiple sequence alignment of each of the five sequence sets has been carried out using the ClustalW algorithm (Higgins et al. 1994) using default input parameters. From each of these multiple sequence alignments, we then identify the residues that are conserved in sequence (either completely or partially conserved or conservatively mutated) within the same interface type. This multiple sequence alignment could not be carried out for the two unusual interface types of PNA and GS1 because there is only one example in each of these interface types.
Identification of patterns specific for each interface type
The structurally conserved interface cluster-forming residues were mapped on to the multiple sequence alignments of each dimerization type so as to obtain the set of residues that are conserved both in the sequence and in the interface clusters. The consensus of the two, that is, residues conserved in the sequence and in the interface clusters, sums up the residues important for the formation of the particular dimerization type and thus gives the sequence motif specific for a particular type of dimerization. Another significant factor used in this method is that, in obtaining the final consensus pattern responsible for a particular type of interface, we have considered all forms of conservation in sequence, namely, complete conservation, conservative mutation, and partial conservation of residues. Any residue that fits into any of the three types of sequential conservation and is also conserved in the interface cluster of the legume lectins belonging to a particular category is taken into consideration for obtaining the final consensus sequence pattern responsible for quaternary association. As a confirmative step, we have also compared the consensus pattern of each interface type with that of the others, to determine whether the pattern is present in the other interface types or whether it is exclusive to the particular type. This would not only help us identify the deterministic sequence patterns for each dimerization type but would also help us rationalize why some lectins are monomeric while the others are dimeric or tetrameric. The tetrameric ones are likely to have more than one consensus pattern, because they have to form more than one type of interface with the other monomers. The consensus patterns have been obtained only for the five interface types that have more than one example in each (II, X1, X2, X3, and X4). These patterns could not be obtained for the unusual interfaces of PNA and GS1 because of nonavailability of more structures in these cases. Nevertheless, the interface cluster-forming residues in these unusual interfaces have been identified and analyzed.
Analysis of legume Lectins with Unknown Structures (LUS)
Because one of our aims is to predict the nature of quaternary association in legume lectins given the sequence, we have taken a set of six sequences that are known to be lectins by sequence similarity and other biochemical experiments, but whose structures are not yet available (Osawa et al. 1978; Kouchalakos et al. 1984; Young et al. 1984; Konami et al. 1990, 1992; Kusui et al. 1991; Calvete et al. 1998; Cheng et al. 1998; K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.). Also known is the ligand specificity and state of quaternary association of these lectins from these experimental data. We have tried to predict the nature and types of interfaces that these lectins can form from the consensus patterns determined using the lectins with known structures. The set of six legume Lectins with Unknown Structures (LUS) is obtained from their respective sequence characterization papers (Kouchalakos et al. 1984; Konami et al. 1990, 1992; Kusui et al. 1991; Calvete et al. 1998; K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.). The Swissprot (Boeckmann et al. 2003) accession numbers of these sequences are P02874 (Onobrychis viciifolia; Kouchalakos et al. 1984), P29257 (Cytisus scoparius; Konami et al. 1992), P19664 (Lotus tetragonolobus; Konami et al. 1990), P81371 (Vatairea macrocarpa; Calvete et al. 1998), and P16030 (Bauhinia purpurea; Kusui et al. 1991). The Cicer arietinum (K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.) lectin sequence is an unpublished work, and its Gene Bank accession number is AAO62538.
Each of these six lectins whose structures are unknown is aligned separately with each of the five sets of lectin sequences with known structures, belonging to the five major interface types. Figure 2 ▶ shows an example of the alignment of an LUS in some of the interface types. These alignments are then analyzed for the presence of the consensus pattern of the particular type of interface in the unknown lectin sequence by visual inspection. The alignment of each of the LUS with all the five sets is then scored by manually counting the number of consensus residues of the particular interface type present in the LUS. While evaluating these scores, all types of conservation of residues, namely complete, partial, and conservative mutation, are taken into consideration. Wherever there is a possibility of formation of the unusual interface of PNA-type or GS1-type based on whether II + X4 or X4 (respectively) are already present, we have checked for the presence of the interface cluster forming residues of these two unusual interface types in the LUS sequence.
The presence or absence of a consensus sequence pattern can be clearly seen just by visual inspection, and this gives the information regarding the nature, type, and state of oligomerization of the LUS. The use of a simple scoring scheme (the number of consensus residues present) helps us in ranking each interface type for all the unknowns. This also enables us to identify whether the lectin would be a dimer or a tetramer based on the scores, because tetramers would score high in more than one category. The results thus obtained are then compared with the already existing results obtained from experiments and pure sequence alignment algorithms.
There are already some experimental data available on some of these LUS (Osawa et al. 1978; Kouchalakos et al. 1984; Young et al. 1984; Konami et al. 1990, 1992; Kusui et al. 1991; Calvete et al. 1998; Cheng et al. 1998; K.R. Koundal, I.A. Qureshi, R. Kansal, and P.K. Dash, unpubl.), which give us an idea about the state of oligomerization in these unknowns. But they do not give any information regarding the nature and type of oligomerization. Also, Manoj and Suguna (2001) have carried out multiple sequence alignments of all these LUS along with all those legume lectins whose structures are known and obtained a phylogenetic tree (dendrogram) from the same. Based on where the LUS occur in the dendrogram, they have tried to predict which type of interface it forms. The LUS will most likely take up the interface type similar to the one closest in position to it in the dendrogram. We have also compared the results of the present analysis with these already available results from the dendrogram. The results obtained from the dendrogram are indirect, and are not conclusive because these legume lectins have very high sequence similarity within and across the different dimerization types. The identification of the interface type in the present method is carried out by specifically matching the signature of the type of interface. Moreover, combining the clustering algorithm along with the sequence alignment algorithms is likely to yield better results because this method takes into consideration contributions from both sequence and structure.
Acknowledgments
A.S. thanks the Department of Biotechnology (DBT), India, for funding this project. S.V. acknowledges the computational genomics initiative at IISc, funded by the Department of Biotechnology, India, for support. K.V.B. thanks the Council of Scientific and Industrial Research, India, for the fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04651004.
References
- Banerjee, R., Mande, S.C., Ganesh, V., Das, K., Dhanaraj, V., Mahanta, S.K., Suguna, K., Surolia, A., and Vijayan, M. 1994. Crystal structure of peanut lectin, a protein with an unusual quaternary structure. Proc. Natl. Acad. Sci. 91 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.-C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O’Donovan, C., Phan, I., et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinda, K.V., Kannan, N., and Vishveshwara, S. 2002. Analysis of homodimeric protein interfaces by graph-spectral methods. Protein Eng. 4 265–277. [DOI] [PubMed] [Google Scholar]
- Buts, L., Dao-Thi, M.H., Loris, R., Wyns, L., Etzler, M., and Hamelryck, T. 2001. Weak protein–protein interactions in lectins: The crystal structure of a vegetative lectin from the legume Dolichos biflorus. J. Mol. Biol. 309 193–201. [DOI] [PubMed] [Google Scholar]
- Calvete, J.J., Santos, C.F., Mann, K., Grangeiro, T.B., Nimtz, M., Urbanke, C., and Sousa-Cavada, B. 1998. Amino acid sequence, glycan structure, and proteolytic processing of the lectin of Vatairea macrocarpa seeds. FEBS Lett. 425 286–292. [DOI] [PubMed] [Google Scholar]
- Cheng, W., Bullitt, E., Bhattacharyya, L., Brewer, C.F., and Makowski, L. 1998. Electron microscopy and x-ray diffraction studies of Lotus tetragonolobus A isolectin cross-linked with a divalent Lewisx oligosaccharide, an oncofetal antigen. J. Biol. Chem. 273 35016–35022. [DOI] [PubMed] [Google Scholar]
- Hamelryck, T.W., Poortmans, F., Goossens, A., Angenon, G., Van Montagu, M., Wyns, L., and Loris, R. 1996. Crystal structure of Arcelin-5, a Lectin-like defense protein from Phaseolus vulgaris. J. Biol. Chem. 271 32796–32802. [DOI] [PubMed] [Google Scholar]
- Hamelryck, T.W., Moore, J.G., Chrispeels, M.J., Loris, R., and Wyns, L. 2000. The role of weak protein–protein interactions in multivalent lectin-carbohydrate binding: Crystal structure of cross-linked FRIL. J. Mol. Biol. 299 875–883. [DOI] [PubMed] [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty, A., Gautier, C., Lescar, J., Perez, S., Wyns, L., and Loris, R. 2000. An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides. J. Biol. Chem. 275 17541–17548. [DOI] [PubMed] [Google Scholar]
- Kannan, N. and Vishveshwara, S. 1999. Identification of side-chain clusters in protein structures by a graph spectral method. J. Mol. Biol. 292 441–464. [DOI] [PubMed] [Google Scholar]
- Konami, Y., Yamamoto, K., and Osawa, T. 1990. The primary structure of the Lotus tetragonolobus seed lectin. FEBS Lett. 268 281–286. [DOI] [PubMed] [Google Scholar]
- Konami, Y., Yamamoto, K., Osawa, T., and Irimura, T. 1992. The primary structure of the Cytisus scoparius seed lectin and a carbohydrate-binding peptide. J. Biochem. 112 366–375. [DOI] [PubMed] [Google Scholar]
- Kouchalakos, R.N., Bates, O.J., Bradshaw, R.A., and Hapner, K.D. 1984. Lectin from sainfoin (Onobrychis viciifolia scop.). Complete amino acid sequence. Biochemistry 23 1824–1830. [DOI] [PubMed] [Google Scholar]
- Kusui, K., Yamamoto, K., Konami, Y., and Osawa, T. 1991. cDNA cloning and expression of Bauhinia purpurea lectin. J. Biochem. 109 899–903. [DOI] [PubMed] [Google Scholar]
- Liener, I.E., Sharon, N., and Goldstein, I.J. 1986. The lectins: Properties, functions and applications in biology and medicine. Academic Press, New York.
- Loris, R., Hamelryck, T., Boukaert, J., and Wyns, L. 1998. Legume lectin structure. Biochim. Biophys. Acta 1383 9–36. [DOI] [PubMed] [Google Scholar]
- Manoj, N. and Suguna, K. 2001. Signature of quaternary structure in the sequences of legume lectins. Protein Eng. 10 735–745. [DOI] [PubMed] [Google Scholar]
- Osawa, T., Irimura, T., and Kawaguchi, T. 1978. Bauhinia purpurea agglutinin. Methods Enzymol. 50 367–372. [DOI] [PubMed] [Google Scholar]
- Srinivas, V.R., Reddy, G.B., Ahmad, N., Swaminathan, C.P., Mitra, N., and Surolia, A. 2001. Legume lectin family, the “natural mutants of the quaternary state,” provide insights into the relationship between protein stability and oligomerization. Biochim. Biophys. Acta 1527 102–111. [DOI] [PubMed] [Google Scholar]
- Svensson, C., Teneberg, S., Nilsson, C.L., Kjellberg, A., Schwarz, F.P., Sharon, N., and Krengel, U. 2002. High-resolution crystal structures of Erythrina cristagalli lectin in complex with lactose and 2′-α-L-fucosyllactose and correlation with thermodynamic binding data. J. Mol. Biol. 321 69–83. [DOI] [PubMed] [Google Scholar]
- Tempel, W., Tschampel, S., and Woods, R.J. 2002. The Xenograft antigen bound to Griffonia simplicifolia lectin 1-B4. J. Biol. Chem. 277 6615–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan, M. and Chandra, N. 1999. Lectins. Curr. Opin. Struct. Biol. 9 707–714. [DOI] [PubMed] [Google Scholar]
- Vishveshwara, S., Brinda, K.V., and Kannan, N. 2002. Protein structure: Insights from graph theory. J. Th. Comp. Chem. 1 187–211. [Google Scholar]
- Young, N.M., Watson, D.C., and Williams, R.E. 1984. Structural differences between two lectins from Cytisus scoparius, both specific for D-galactose and N-acetyl-D-galactosamine. Biochem. J. 222 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]


