Abstract
Combinatorial libraries of de novo amino acid sequences can provide a rich source of diversity for the discovery of novel proteins with interesting and important activities. Randomly generated sequences, however, rarely fold into well-ordered proteinlike structures. To enhance the quality of a library, features of rational design must be used to focus sequence diversity into those regions of sequence space that are most likely to yield folded structures. This review describes how focused libraries can be constructed by designing the binary pattern of polar and nonpolar amino acids to favor proteins that contain abundant secondary structure, while simultaneously burying hydrophobic side chains and exposing hydrophilic side chains to solvent. The “binary code” for protein design was used to construct several libraries of de novo proteins, including both α-helical and β-sheet structures. The recently determined solution structure of a binary patterned four-helix bundle is well ordered, thereby demonstrating that sequences that have neither been selected by evolution (in vivo or in vitro) nor designed by computer can form nativelike proteins. Examples are presented demonstrating how binary patterned libraries have successfully produced well-ordered structures, cofactor binding, catalytic activity, self-assembled monolayers, amyloid-like nanofibrils, and protein-based biomaterials.
Keywords: artificial proteins, binary patterning, combinatorial libraries, de novo protein design
Constructing proteins de novo is ultimately about choosing amino acid sequences that fold into structures with desired properties. The number of possible sequences from which to choose is enormous. Even for a relatively small protein, one cannot sample all possibilities. For example, for a chain of 100 residues composed of the 20 naturally occurring amino acids, there are 20100 possibilities. This number is so large that a collection containing one molecule of each sequence would fill a volume larger than Avogadro’s number of universes (Beasley and Hecht 1997). Clearly, it is not practical to explore sequence space by designing and characterizing individual proteins one by one.
Combinatorial methods are far more appealing. However, combinatorial libraries composed of random sequences will rarely yield proteins with desired properties. Therefore, to enhance the likelihood of success, combinatorial collections must be focused into those regions of sequence space most likely to produce well-folded proteins. The numerical power of the combinatorial approach must be tempered with elements of rational design (Beasley and Hecht 1997; Moffet and Hecht 2001).
Designed combinatorial libraries of de novo proteins
To focus a library into the most productive regions of sequence space, which features must be designed explicitly? To delineate these features, we were guided by two unifying themes from natural proteins, which typically fold into structures that (1) contain abundant secondary structure (α-helices and β-sheets) and (2) expose polar side chains to solvent while burying nonpolar side chains in the protein interior. Our strategy for protein design draws on these two features to rationally constrain the diversity of libraries of de novo sequences in ways that favor the formation of folded structures.
Sequences capable of forming abundant secondary structure while simultaneously exposing polar side chains and burying nonpolar side chains can be designed by constraining the periodicity of polar and nonpolar residues to match the structural periodicity of the desired secondary structure (Xiong et al. 1995). Thus, for an α-helical design, the sequence periodicity of polar and nonpolar residues must approximate the structural repeat of 3.6 residue/turn. For example, a sequence of polar (○) and nonpolar (•) residues with the pattern ○•○○••○○•○○••○ has a nonpolar amino acid every three or four positions, consistent with the structural repeat of α-helices. Conversely, for a designed β-strand, polar and nonpolar residues would alternate every other residue. Thus, a designed sequence with the pattern ○•○•○•○ has a sequence periodicity of 2, which matches the structural repeat found in β-strands, with successive side chains pointing up-down-up-down and so forth (Fig. 1 ▶).
Figure 1.
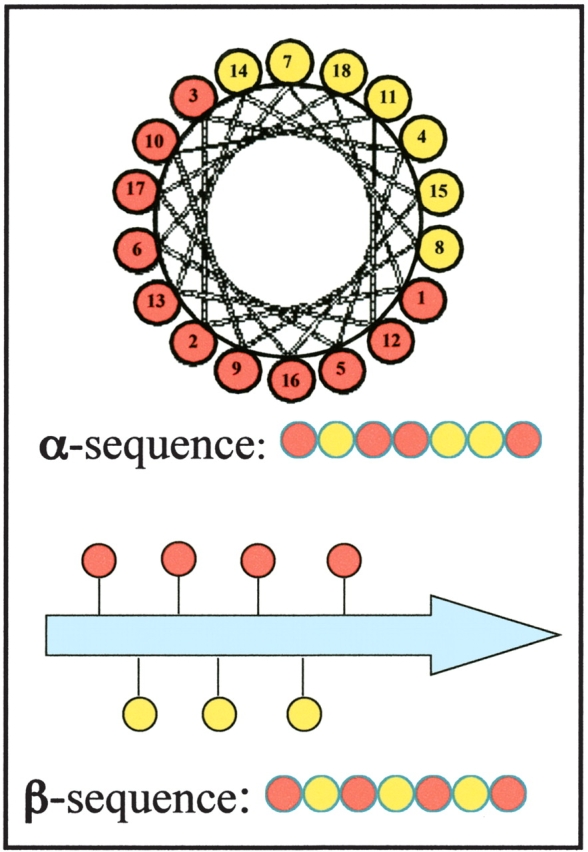
(Top) Helix wheel representation of an amphiphilic α-helix. (Bottom) Representation of an amphiphilic β-strand. Polar and nonpolar residues are shown in red and in yellow, respectively.
We have used this polar/nonpolar patterning as a cornerstone for de novo protein design. Our strategy is based on the premise that the locations of polar and nonpolar residues must be specified explicitly, but their precise identities can be varied extensively. Thus, the strategy uses a “binary code,” which specifies only whether a given position is hydrophobic or hydrophilic. Because the precise identity of each polar or nonpolar residue is not specified, the binary code strategy facilitates the design and construction of libraries with enormous combinatorial diversity.
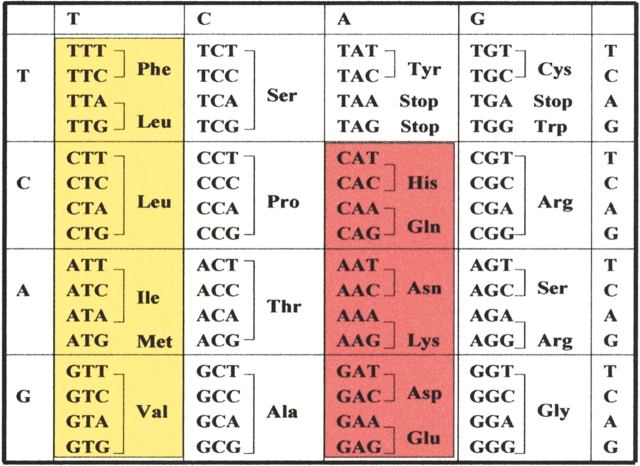
Our initial test of the binary code strategy focused on the design of a library of four-helix bundles (Kamtekar et al. 1993). Each member of the library had a unique amino acid sequence, yet all sequences shared the identical pattern of polar and nonpolar residues (Fig. 2 ▶). Combinatorial diversity was made possible by the organization of the genetic code (Fig. 3 ▶). Six polar residues (Lys, His, Glu, Gln, Asp, Asn) were encoded by the degenerate DNA codon VAN, and five nonpolar residues (Met, Leu, Ile, Val, Phe) were encoded by the degenerate codon NTN (V = A, G, or C; N = A, G, C, or T).
Figure 2.

Ribbon diagram showing the periodicity of polar (red) and nonpolar (yellow) residues in a four-helix bundle. (Reprinted from Kamtekar et al. 1993.)
Figure 3.
The genetic code. Codons for polar residues are highlighted in red. Those for nonpolar residues are highlighted in yellow.
Proteins were produced from a library of synthetic genes expressed in bacteria. Several of the resulting proteins were purified. Biophysical characterization of these proteins demonstrated that the majority of sequences generated by our binary code for α-helical design indeed formed proteins that were soluble, stable, and had the expected α-helical secondary structure (Kamtekar et al. 1993).
Novel proteins and nativelike structures
The binary code strategy is based on the premise that appropriate patterning of polar and nonpolar residues can drive a polypeptide chain to fold into segments of amphiphilic secondary structure that anneal together to form a desired tertiary structure (Kamtekar et al. 1993; West and Hecht 1995; Xiong et al. 1995). However, because of its combinatorial nature, the binary code strategy does not allow explicit design of specific interresidue interactions. Because unique packing cannot be designed a priori, it is reasonable to question whether “nativelike” structures can nonetheless be isolated from binary code libraries a posteriori.
Well-folded structures versus molten globules
We developed several methods to rapidly screen libraries for sequences that successfully recapitulate the features of well-folded native proteins. These screens were based either on peak dispersion in NMR spectra (Roy et al. 1997a), or protection of hydrogen exchange measured by mass spectrometry (Rosenbaum et al. 1999). Our searches for well-folded proteins in the original 74-residue α-helical library uncovered several proteins that displayed some nativelike characteristics (Roy et al. 1997a,b; Rosenbaum et al. 1999; Roy and Hecht 2000). However, most proteins in the initial collection formed fluctuating structures, reminiscent of molten globule intermediates.
Why did most sequences from the original collection fail to form nativelike structures? One might postulate that fluctuating “molten” structures are exactly what should be expected from a combinatorial strategy that precludes explicit design of specific sequences with predetermined side-chain interactions. However, the alternative result—nativelike structures—would be predicted by numerous studies demonstrating that well-folded proteins can be specified by many different amino acid sequences (Dill 1985; Chothia and Lesk 1987; Bowie et al. 1990; Matthews 1993; Bromberg and Dill 1994; Axe et al. 1996; Gassner et al. 1996; Munson et al. 1996; Riddle et al. 1997). Comparisons of evolutionarily related sequences, theoretical studies using simplified models, and extensive mutagenesis experiments have all led to the realization that protein structures are robust, and explicit design of “jigsaw-puzzle” packing may not be necessary. These considerations led us to question whether the tendency of the original binary code proteins to form fluctuating structures might not be a failure of the binary code strategy per se, but rather a shortcoming of the designed structural scaffold used in its initial implementation.
In particular, we questioned whether the α-helices specified by our original scaffold might simply be too short. Previous workers had shown that for rationally designed α-helical bundles longer helices can enhance stability, and in some cases, favor more nativelike structures (Fairman et al. 1995; Betz and DeGrado 1996). We reasoned that in the context of the binary code strategy, which cannot specify side-chain packing a priori, it might be especially important to use a scaffold that encodes longer α-helices, and hence larger interhelical interfaces.
To test this reasoning, we designed a new structural scaffold to encode longer four-helix bundles (Fig. 4 ▶), and constructed a second-generation library of 102-residue sequences (Wei et al. 2003b). This new library was not constructed from scratch. Instead, to stringently test whether the redesigned features are sufficient to convert a fluctional protein into a well-ordered structure, we chose a molten globule-like protein (sequence #86) from the original 74-residue library (Kamtekar et al. 1993) as the starting point for designing the second-generation elongated library.
Figure 4.
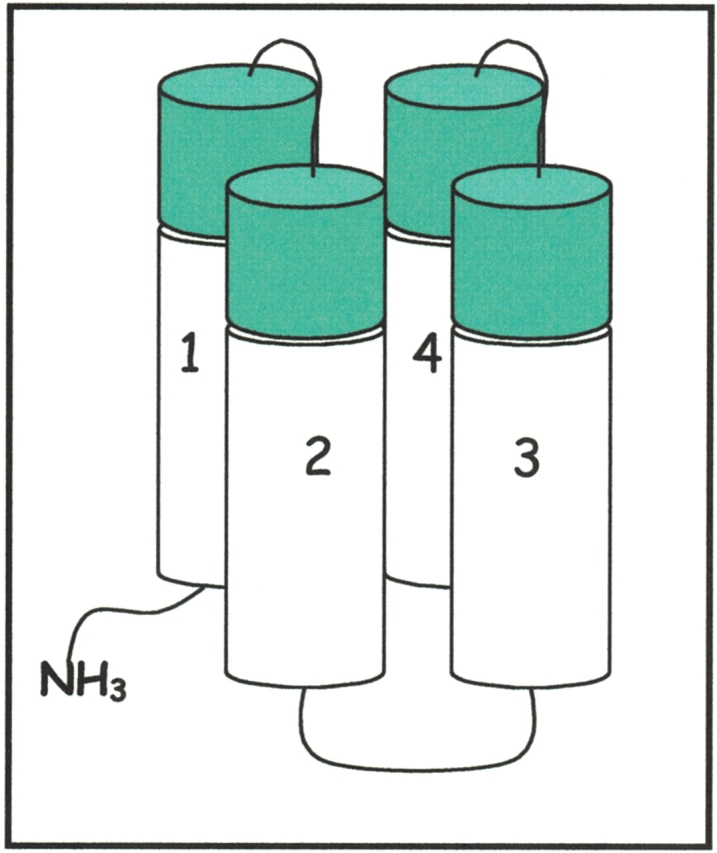
Design of a second-generation library of binary patterned four-helix bundles. Helices were designed to be ~50% longer than in the first library.
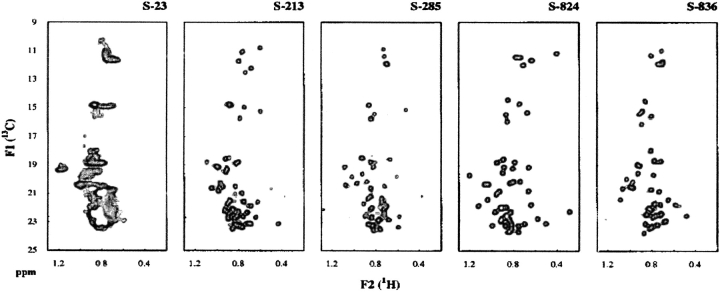
Characterization of five proteins chosen arbitrarily from this new library revealed that all were considerably more stable than the sequences form the initial (74-residue) collection. Most importantly, the second-generation proteins contained an abundance of tertiary interactions indicative of well-ordered, nativelike structures (Wei et al. 2003b). For example, natural abundance 13C HSQC NMR spectra demonstrated that four of the five proteins from this naïve (i.e., unselected and unscreened) second-generation library displayed well-resolved peaks and good dispersion indicative of well-ordered side chains (Fig. 5 ▶). These findings demonstrate that the binary code strategy—if applied to an appropriately designed structural scaffold—can generate large collections of stably folded and well-ordered protein structures.
Figure 5.
Natural abundance 13C,1H-HSQC NMR for five proteins from the unselected second-generation library. γand δ methyl resonances of Ile side chains appear in the 13C dimension at ~11 ppm and ~15 ppm, respectively. Methyl groups from the side chains of Val, Met, Leu, and Thr appear between 18 ppm and 24 ppm. Spectra for four of the five proteins (S-213, S-285, S-824, S-836) display resolved peaks and good dispersion, indicating well-ordered structures. (Reprinted from Wei et al. 2003b.)
Solution structure of a de novo protein from a designed combinatorial library
The ultimate test of whether a design successfully produces the proposed structure is the experimental determination of the actual structure. Attempts to determine the three-dimensional structures of our first-generation α-helical proteins were unsuccessful. This was not surprising given that structural flexibility is known both to disfavor crystal growth and to yield poorly resolved NMR spectra. In contrast with these first-generation sequences, proteins from the second-generation library yield high quality NMR spectra indicative of well-folded (nonfluctuating) structures (Wei et al. 2003b). Therefore the three-dimensional structures of these new proteins are accessible by NMR spectroscopy.
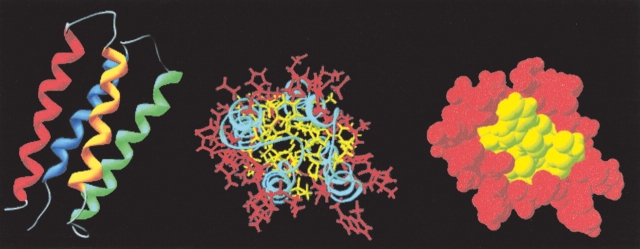
Recently, we solved the solution structure of a protein (S-824) from the second-generation library (Wei et al. 2003a). As shown in Figure 6 ▶, the structure is indeed a four-helix bundle. In accordance with the binary code design, the polar and nonpolar residues segregate on the surface and the interior, respectively (Fig. 6B,C ▶). Moreover, the protein is not a molten globule: As shown in Figure 7 ▶, the interior side chains are well ordered—even by the standards of natural proteins.
Figure 6.
Solution structure of protein S-824 from the second-generation binary patterned library. (Reprinted from Wei et al. 2003a; © 2003 National Academy of Sciences, U.S.A.) (Left) Ribbon diagram of the four-helix bundle. (Center) Head-on view of the four-helix bundle with polar residues colored red and nonpolar residues colored yellow. (Right) Same as center image in space-filling representation.
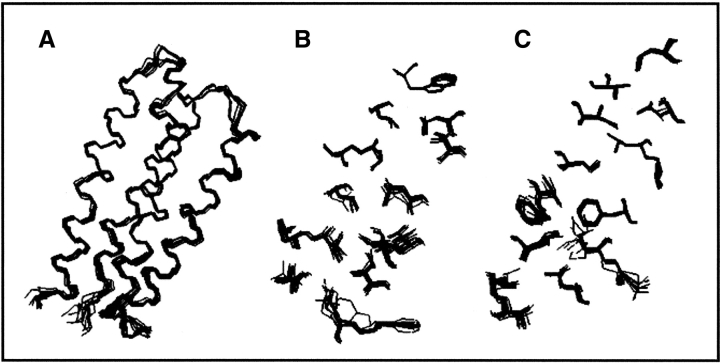
Figure 7.
Overlay of 10 lowest energy structures calculated from the NMR data (Reprinted with permission from Wei et al. 2003a, © 2003 National Academy of Sciences, U.S.A.) (A) Backbone. (B) Nonpolar side chains of α-helices 1 and 2. (C) Nonpolar side chains of α-helices 3 and 4.
It should be noted that only 14 of the 102 positions in the sequence of protein S-824 were designed as specific amino acids (Fig. 8 ▶). These are in the chain termini and in the interhelical turns. The other 88 positions (86% of the total) arose from degenerate DNA codons, which specified the binary pattern, but not the amino acid identities.
Figure 8.
Amino acid sequence of protein S-824 shown in single letter code. Polar and nonpolar residues of the binary patterned α-helices are shown in red and in green, respectively. Turn residues (and the N and C termini) are shown in black. Residues that were encoded by degenerate DNA codons specifying combinatorial mixtures of amino acids are underlined. Of the 102 residues in this sequence, 88 were derived from combinatorially encoded mixtures of amino acids. (Reprinted from Wei et al. 2003a; © 2003 National Academy of Sciences, U.S.A.)
The potential diversity encoded by this binary pattern is enormous. Of the five proteins characterized to date, all are α-helical and four of the five appear well ordered (Wei et al. 2003b). As shown in Figures 6 ▶ and 7 ▶, the first structure determined from this library is a well-folded four-helix bundle similar to that specified by the design. In considering whether well-folded structures are representative of the library as a whole, it is important to emphasize that the five proteins characterized thus far were chosen arbitrarily from a naïve library, which was not subjected to genetic selections or high throughput screens. Therefore, we presume these stably folded protein structures are not “needles in a haystack,” and libraries based on appropriately designed binary patterning can produce vast collections of well-folded de novo proteins.
Libraries of functional de novo proteins
Nature uses two general strategies to endow proteins with function. In the “purist” strategy, nature uses only the protein itself; all the requirements for activity are provided by atoms on the polypeptide chain. However, nature often employs an alternative strategy, and uses nonprotein cofactors. Such cofactors range in size and complexity from a single metal atom to large macrocycles.
In our efforts to devise functionally active proteins we have also used both approaches: We have developed collections of functional proteins both by incorporating cofactors and by pursuing the purist approach.
Functional heme proteins
Cofactors can be thought of as portable activity modules, which are used for different purposes by different proteins. Many cofactors, such as the porphyrin in heme, are preorganized structures. Moreover, the cofactors themselves often possess some level of activity even in the absence of protein. To make functional proteins, nature incorporates these portable activity modules into otherwise inactive proteins.
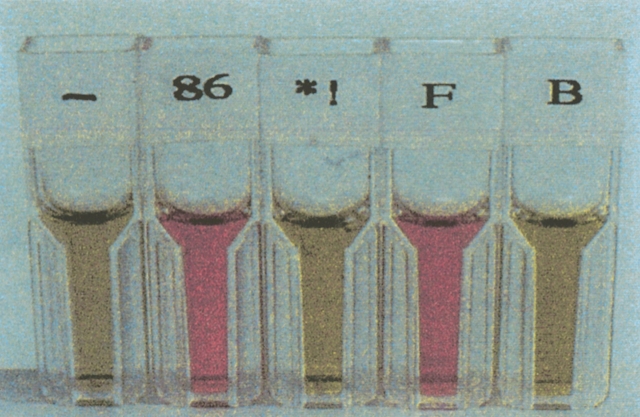
We have followed nature’s lead by using cofactor binding as a step towards the isolation of functional de novo proteins. We screened our binary patterned collections of α-helical proteins for heme binding. In an initial collection of 30 α-helical sequences from the first-generation library (those with 74-residue sequences), we found that 15 bound heme (Rojas et al. 1997). More recent experiments showed that proteins from the second-generation library (102-residue sequences) also bind heme. The novel heme proteins are bright red (Fig. 9 ▶) similar to hemoglobin or cytochrome c. The absorption and resonance Raman spectra of the de novo heme proteins (Rojas et al. 1997) resemble those of natural cytochromes. Because the design of the novel sequences was based solely on global features of polar/nonpolar patterning, our finding that approximately half of them bind heme demonstrates that isolating de novo heme proteins does not require explicit design of a cofactor-binding site. Apparently, heme binding is more permissive and more easily achieved than previously suspected.
Figure 9.
Red color associated with heme binding by proteins from the initial 74-residue α-helical library. Proteins “86” and “F” bind heme. Protein “B” does not. The other cuvettes show negative controls. (Reprinted from Rojas et al. 1997.)
This collection of novel heme proteins provides a unique opportunity for an unbiased assessment of the functional potential of heme proteins that have not been prejudiced either by explicit design or evolutionary selection.
In nature, heme proteins perform a number of functions. Among these are (1) catalysis of redox reactions, (2) binding and transport of small molecules (e.g., oxygen or carbon monoxide), and (3) electron transfer. We have probed our library of de novo heme proteins for all three of these functions (Moffet et al. 2000, 2001, 2003).
The capacity of the binary code proteins for catalytic activity was established by demonstrating that several of the de novo heme proteins are active as peroxidases, capable of catalyzing the two-electron reduction of hydrogen peroxide to water (Moffet et al. 2000). The most active protein in the collection had a turnover number of 17,000 min−1—significantly better that that of microperoxidase (1260 min−1; Adams 1990), and only fourfold lower than the natural enzyme, horseradish peroxidase (60,000 min−1; Hiner at al. 1996). Like natural enzymes, our de novo proteins are inactivated by high concentrations of peroxide. Hence the measured turnover numbers represent a lower limit of the catalytic potential of these proteins.
To assess the abilities of the de novo heme proteins to bind diatomic ligands, we measured affinity for CO, kinetics of CO binding and release, and resonance Raman spectra of the CO complexes for several heme proteins derived from our combinatorial libraries (Moffet et al. 2001). The CO binding affinities for all of the proteins were similar to that of myoglobin, with dissociation constants in the low nanomolar range. Overall, the CO binding properties of the de novo heme proteins span a narrow range of values that falls near the center of the range observed for natural heme proteins.
All the binary patterned heme proteins characterized thus far bound heme with 1:1 stoichiometry (Moffet et al. 2003). The availability of this collection enabled us to determine “default reduction potentials” for 1:1 heme proteins that have neither been selected by evolution nor explicitly designed for redox activity. We measured the midpoint reduction potentials for five first-generation and three second-generation binary patterned proteins. The potentials ranged from −112 mV to −176 mV (Moffet et al. 2003). These default reduction potentials can be compared with the reduction potentials of (1) heme alone, (2) naturally evolved heme proteins, and (3) rationally designed heme proteins. The midpoint reduction potential for unbound heme is −220 mV. Nature has altered this potential for particular redox functions by evolving heme proteins spanning a wide range of potentials from approximately −400 mV to +400 mV. Attempts to engineer the potentials of either natural proteins (Springs et al. 2000, 2002) or novel protein maquettes (Shifman et al. 2000) have produced narrower ranges.
Functional proteins without cofactors
Many proteins in nature are fully active without the aid of cofactors. Therefore we were also interested in probing the ability of the binary code strategy to generate functional proteins without cofactors.
Previous attempts by other groups to generate proteins with enzymelike activities have relied on a number of different approaches (Pollack et al. 1986; Tramontano et al. 1986; Stewart et al. 1994; Pinto et al. 1997; Broo et al. 1998; Benson et al. 2000; Hilvert 2000; Bolon and Mayo 2001; Keefe and Szostak 2001; Yamauchi et al. 2002; Looger et al. 2003). Some groups used rational design and/or computational methods to engineer active sites and catalytic function into either natural or novel protein sequences. Other groups selected or screened for desired activities by using the mammalian immune system (e.g., for catalytic antibodies), evolution in vitro, or other methods.
Recently, we probed whether any of our de novo α-helical proteins might catalyze ester hydrolysis (Wei and Hecht 2004). For our initial probes of catalytic activity, we focused on hydrolysis of p-nitrophenyl esters. We chose this activity because (1) hydrolysis of p-nitrophenyl esters is relatively easy to achieve (Menger and Ladika 1987), (2) it is straightforward to assay, and (3) there are precedents of novel esterases being isolated both from catalytic antibodies and from rational designs (Pollack et al. 1986; Tramontano et al. 1986; Menger and Ladika 1987; Stewart et al. 1994; Broo et al. 1998; Bolon and Mayo 2001; Yamauchi et al. 2002).
We measured the esterase activity of S-824, the binary patterned protein whose structure was solved at high resolution (see the section “Solution structure of a de novo protein from a designed combinatorial library” and Figs. 6 ▶ and 7 ▶). Protein S-824 displayed a rate enhancement (kcat/kuncat) of 8700. The observed activity is similar to or better than that observed for several esterases designed previously using rational design and/or automated computational methods (Broo et al. 1998; Bolon and Mayo 2001). Moreover, the observed activity rivals those of the first catalytic antibodies (Pollack et al. 1986; Tramontano et al. 1986).
Because hydrolysis of p-nitrophenyl esters by protein S-824 is presumed to involve a histidine nucleophile (Wei and Hecht 2004), it was important to establish that the rate catalyzed by protein S-824 is above that catalyzed by free imidazole. At pH 7, protein S-824 hydrolyzed p-nitrophenyl acetate ~100-fold faster than does 4-methylimidazole (Wei and Hecht 2004). Even after correcting for the 11 histidines in the sequence of S-824, it is clear that the de novo protein catalyzes ester hydrolysis more effectively than a simple imidazole-based catalyst. Like natural enzymes, S-824 catalyzed multiple turnovers and remained active after prolonged exposure to excess substrate.
To assess whether the activity of S-824 is representative of other proteins in our binary patterned libraries, we measured the esterase activity of six additional binary patterned proteins. These proteins were from two libraries: the original library of 74-residue sequences and the second-generation library of 102-residue sequences described above. Both libraries were naïve in that they were neither designed to bind substrate nor subjected to high throughput screens for activity. All six of the additional proteins displayed esterase activity significantly above background (Wei and Hecht 2004).
These findings suggest that although the exquisite levels of activity and specificity typical of natural enzymes may have required eons of evolutionary selection, proteins with moderate levels of activity are surprisingly common in libraries of de novo sequences designed by binary patterning. The activities of these unselected proteins provide a reference state for the levels of activity that have been reported for proteins obtained by selection and/or computational design.
Amyloid-like fibrils from binary patterned libraries
The deposition of insoluble amyloid plaque is associated with several neurodegenerative diseases including Alzheimer’s disease and spongiform encephalopathies (e.g., mad cow disease; Prusiner 1997; Koo et al. 1999; Selkoe 2001; Dobson 2002). The primary component of amyloid differs from one disease to another: In Alzheimer’s disease, it is the 42-residue Aβ peptide, whereas in spongiform encephalopathies, it is the prion protein. Despite substantial differences in both sequence and length, these diverse proteins assemble into amyloid structures that are remarkably similar to one another. They all form fibrils composed of β-strands running perpendicular to the fibril axis (cross-β structure). The similarity among the structures of the different amyloids suggests the various amyloids may share unifying structural determinants. However, the extreme dissimilarity between the various amyloidogenic sequences, coupled with the unavailability of a high-resolution structure, has limited understanding of the sequence determinants of amyloidogenesis (Wood et al. 1995; Selkoe and Podlisny 2002; Wurth et al. 2002; Williams et al. 2004).
What are the molecular determinants of amyloidogenesis? Which features of an amino acid sequence cause it to assemble into amyloid? These questions can be addressed by two very different approaches: (1) One can probe the sequence determinants of natural amyloid proteins by screening randomly generated amino acid substitutions (mutations) to identify those that prevent amyloidogenesis or (2) one can test hypotheses about possible sequence determinants by using them as the basis for the design of de novo amyloid-like proteins.
We have used both approaches. Our work probing the sequence determinants of amyloidogenesis in a natural system (the Alzheimer’s Aβ peptide) has been described previously (Wurth et al. 2002) and will not be reviewed here. Our design of libraries of de novo amyloid-like fibrils is described in this section.
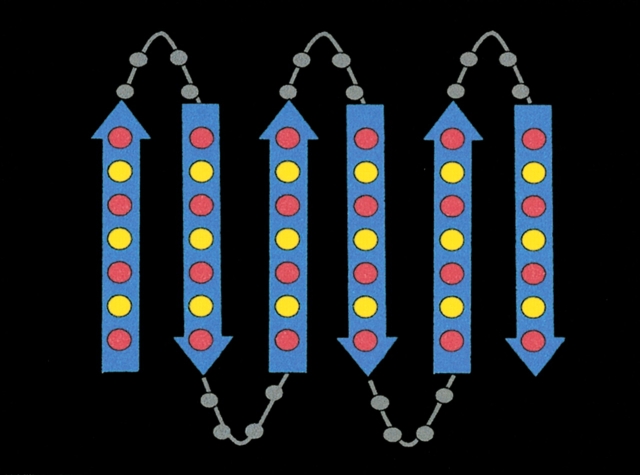
To probe the sequence determinants of amyloidogenesis by de novo protein design, we once again used the binary code strategy. We designed a binary patterned combinatorial library of de novo proteins (West et al. 1999) using the β-strand pattern shown in Figure 1 ▶. All sequences in the library were constrained by the ○•○•○•○ alternating pattern consistent with amphiphilic β-structure. The precise identities of the side chains were not constrained and were varied combinatorially. Polar sites were allowed to be His, Lys, Asn, Asp, Gln, or Glu and nonpolar residues were allowed to be Leu, Ile, Val, or Phe. A schematic diagram of the binary pattern for proteins containing six β-strands punctuated by turns is shown in Figure 10 ▶. The amino acid sequences of proteins from this library are shown in Figure 11 ▶.
Figure 10.
Binary pattern for a library of six-stranded β-sheet proteins. Arrows designate β-strands. Red circles represent positions occupied by polar amino acids, and yellow circles represent those occupied by nonpolar amino acids.
Figure 11.
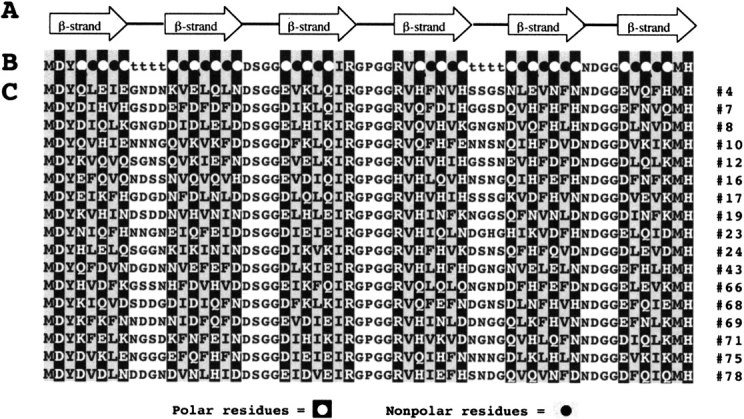
(A) Schematic illustration of the design of a combinatorial library of de novo proteins containing six β-strands (arrows) punctuated by turns. (B) Designed binary sequence pattern. Alternating pattern in the β-strands is indicated with polar residues (○) as open circles on a black background and nonpolar residues (•) as closed circles on a gray background. Combinatorial diversity is incorporated at positions marked ○, •, and t (turn). Fixed residues are incorporated at the termini and in some of the turns. (C) Amino acid sequences (single letter code) of 17 de novo proteins from the combinatorial library. (Reprinted from West et al. 1999, © 1999 National Academy of Sciences, U.S.A.)
The designed proteins were expressed from a library of synthetic genes and several were purified and characterized. Circular dichroism spectroscopy confirmed that the proteins formed β-structure. Electron microscopy demonstrated that they self-assembled into large oligomers resembling amyloid-like fibrils (Fig. 12 ▶).
Figure 12.

Transmission electron micrograph of a 63-residue binary patterned de novo protein designed to form amphiphilic β-strands. The observed fibrillar structures resemble those seen for natural amyloid proteins. (Reprinted from West et al. 1999; © 1999 National Academy of Sciences, U.S.A.)
These libraries of de novo amyloid proteins can be compared with our earlier work designing α-helical proteins (see above). In both cases, combinatorial libraries were designed by specifying the binary pattern of polar and non-polar residues. Yet the resulting proteins display dramatically different properties: In the previous work, the sequences formed α-helices that folded intramolecularly into small globular domains. In contrast, the sequences described here form β-strands and self-assemble intermolecularly into high-order oligomers that assume fibrillar structures. What causes these dramatically different structures? The lengths of the sequences are not dramatically different, nor are their overall compositions. We propose that the determining difference is the binary patterning itself. In the earlier work, the library was constrained by the pattern ○•○○••○○•○○••○, consistent with the periodicity of α-helical structure. In contrast, this library is constrained by the pattern ○•○•○•○, consistent with the periodicity of amphiphilic β-strands.
If it is indeed true that alternating patterns of polar and nonpolar residues have an inherent propensity to form amyloid fibrils, then one might expect that these binary patterns would be disfavored by natural selection. We tested this possibility with a bioinformatics study: We analyzed a database of 250,514 natural protein sequences comprising 79,708,024 residues and calculated the frequencies of alternating patterns relative to other patterns with similar compositions. The results of this search revealed that alternating patterns occur in nature significantly less often than other patterns with similar compositions (Broome and Hecht 2000). The underrepresentation of alternating binary patterns in natural proteins, coupled with the observation that such patterns promote amyloid-like structures in de novo proteins, suggests that sequences of alternating polar and nonpolar amino acids are inherently prone to form amyloid-like structures and consequently have been disfavored by evolutionary selection.
Monomeric β-sheet proteins
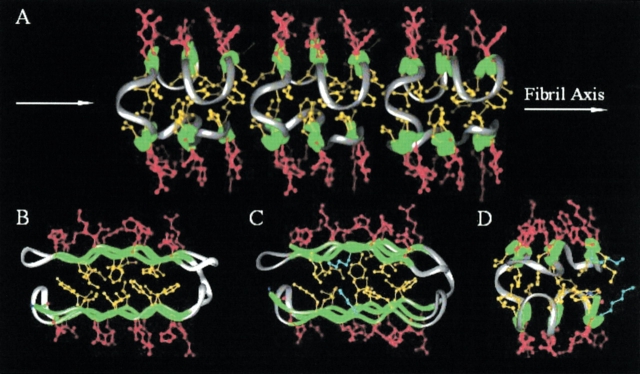
The binary patterned β-sheet proteins described in the previous section were designed to encode proteins containing six amphiphilic β-strands separated by turns. Each β-strand was designed to be seven residues long, with polar and nonpolar amino acids arranged with an alternating periodicity (Figs. 10 ▶, 11 ▶). The initial design specified the identical polar/nonpolar pattern (○•○•○•○) for all of the β-strands: No strand was explicitly designated to form the edges of the β-sheets. With all β-strands preferring to occupy interior (as opposed to edge) locations, intermolecular oligomerization was favored, and the proteins assembled into amyloid-like fibrils (shown in Fig. 12 ▶ and modeled in Fig. 13A ▶).
Figure 13.
(A) Schematic representation of a fibril formed by open-ended oligomerization of a six-stranded binary patterned β-sheet protein. β-strands are shown in green and turns in silver. Polar side chains are shown in red, and nonpolar side chains, in yellow. (B) Model of a monomeric six-stranded β-sandwich (rotated 90° relative to A). In the monomer, the hydrophobic side chains (yellow) of the edge stands are accessible to water. For simplicity, flat β-sheets are depicted. In reality, a six-stranded β-sandwich would be twisted. (C) Model of a monomeric six-stranded β-sandwich in which lysine side chains (blue) are substituted in place of Ile 5 in the N-terminal β-strand and Val 60 in the C-terminal β-strand of sequence 17 (see Fig. 3 ▶). In the monomeric structure, the charged ends of the lysine side chains on the edge strands would be exposed to solvent. (D) Same as panel C, rotated by 90°. (Reprinted from Wang and Hecht 2002, © 2002 National Academy of Sciences, U.S.A.)
To assess whether explicit design of edge-favoring strands might tip the balance in favor of monomeric β-sheet proteins, we used a strategy suggested by Richardson and Richardson (2002) to redesign the first and/or last β-strands of several sequences from the original library. In the redesigned β-strands, the binary pattern was changed from ○•○•○•○ to ○•○K○•○ (where K denotes lysine). The presence of a lysine on the nonpolar face of a β-strand should disfavor fibrils because such structures would bury an uncompensated charge. The nonpolar-to-lysine mutations, therefore, would be expected to favor monomeric structures in which the ○•○K○•○ sequences form edge strands with the charged lysine side chain accessible to solvent (Fig. 13C,D ▶). To test this hypothesis, we constructed several mutant sequences in which the central nonpolar residue of either the N-terminal β-strand, the C-terminal β-strand, or both was changed to lysine. Characterization of the redesigned proteins showed that they indeed formed monomeric β-sheet proteins (Wang and Hecht 2002).
Protein-based biomaterials
Self-assembled monolayers
The amyloid-like fibrils described above and shown in Figure 12 ▶ are self-assembled one-dimensional structures. To explore the possibility of fabricating two-dimensional protein layers, we studied the ability of our de novo proteins to self-assemble into monolayers at an air/water interface (Xu et al. 2001).
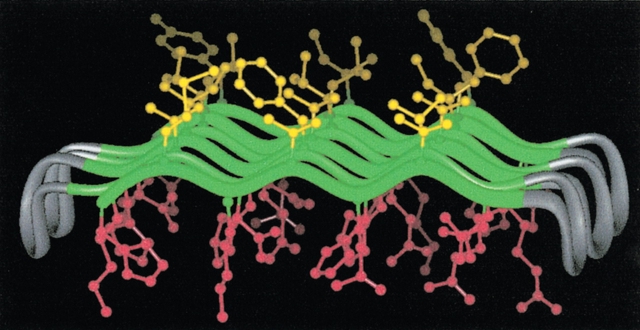
Several proteins were probed for their abilities to form amphiphilic monolayers. The proteins were chosen from the binary patterned library described above, which was designed to form six β-strands punctuated by reverse turns (Figs. 10 ▶, 11 ▶). We used Langmuir–Blodgett techniques to study the properties of the monolayers at the air/water interface, and spectroscopic methods to assess secondary structure. The results of these studies demonstrated that (1) the proteins self-assembled into monolayers at an air/water interface; (2) the monolayers were dominated by β-sheet secondary structure, as shown by both circular dichroism and infrared spectroscopies; (3) the measured area per protein molecule was approximately 500–600 Å2. This matches the area expected for a model of an amphiphilic β-sheet shown in Figure 14 ▶. If the polar turns project down into the aqueous solvent, as shown in the figure, then the measured area would comprise only the 42 residues in the six β-strands, which would indicate an area of approximately 12–14 Å2 per residue (Xu et al. 2001).
Figure 14.
Model of a binary patterned β-sheet protein at an air/water interface. The model shows six antiparallel β-strands. Each strand contains seven residues: four polar (red) and three nonpolar (yellow). The modeled conformation shows a facial amphiphile with a hydrophobic face (toward air) and a hydrophilic face (toward water). (Reprinted from Xu et al. 2001, © 2001 National Academy of Sciences, U.S.A.)
Our finding that distinctly different sequences from our library assemble into very similar structures suggests that β-sheet monolayers can be encoded by the designed pattern of polar and nonpolar amino acids. Moreover, because the designed pattern is compatible with a wide variety of different sequences, it may be possible to fabricate β-sheet monolayers using combinations of side chains that are explicitly designed for particular applications of novel biomaterials.
Template-directed assembly of de novo designed proteins
A number of biological materials owe their unusual structural characteristics and mechanical properties to long-range order induced by the lamination of proteins between layers of inorganic mineral (Lowenstam and Weiner 1989; Heuer et al. 1992; Sarikaya and Aksay 1995; Aksay et al. 1996; Belcher et al. 1996; Falini et al. 1996; Weiner and Addadi 1997). A well-studied example of protein/mineral layering occurs in the nacre of mollusk shells (mother of pearl). These laminated structures are composed of alternating layers of a protein-rich matrix and aragonite, a crystal form of calcium carbonate. The proteins in the protein-rich layer are dominated by β-sheet secondary structure. In such composites, both the protein layer and the mineral layer adopt structures different from those they assume in isolation. Interactions between such layers and the ordered structures that result from these interactions enable nature to produce biomaterials that are simultaneously hard, strong, and tough.
With the long-term goal of constructing artificial biomaterials with laminated structures, we developed a biomimetic system using an ordered surface to template the assembly of a de novo designed β-sheet protein. The protein used in this study was chosen from the same combinatorial library used above (Fig. 11 ▶) to form amyloid-like fibrils in a homogeneous aqueous environment or β-sheet monolayers at an air/water interface. Formation of facial amphiphiles is favored at an interface between polar and nonpolar phases. In the previous example, monolayers formed by self-assembly at the interface between water and air. In contrast, to study template-directed assembly, we chose the nonpolar phase to be the highly ordered surface of pyrolytic graphite. The graphite lattice is hexagonal; therefore structures templated by the graphite surface would be expected to show threefold symmetry.
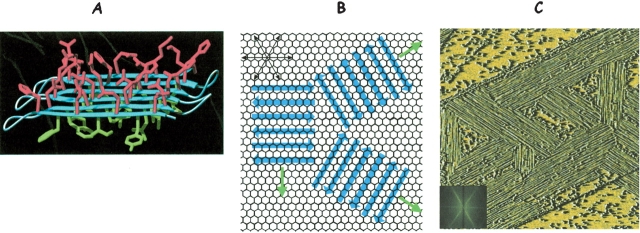
To probe the ability of our de novo proteins to undergo template-directed assembly, we deposited protein onto a graphite surface and used atomic force microscopy (AFM) to image the resulting assemblies (Brown et al. 2002). As shown in Figure 15C ▶, the AFM images demonstrate that the protein assembles on the graphite surface into ordered fibers aligned in three orientations at 120° to each other. This symmetry indicates that the hexagonal lattice of graphite (Fig. 15B ▶) directs nucleation of fibers on the surface. The straightness of the fibers and their persistent length of several microns suggest that the template also influences the addition of protein monomers onto the growing fiber. The size of these structures indicates that the hexagonal lattice of the graphite surface templates assembly of millions of protein molecules into a highly ordered structure, reminiscent of those found in natural biomaterials.
Figure 15.
(A) A binary patterned de novo protein modeled as a flat six-stranded amphiphilic β-sheet with polar residues (red) projecting up and nonpolar residues (green) projecting down toward the hydrophobic graphite surface. (B) Schematic representation of the six-stranded β-sheet protein assembled on a graphite surface. β-strands are shown as blue arrows. The threefold symmetry of the graphite template is recapitulated in the assembly of the protein. The long axis of the fibers is shown perpendicular to the β-strands and is indicated with green arrows. (C) AFM image of a binary patterned protein deposited on graphite. (Inset) A Fourier transform of this image. The threefold symmetry is apparent both in the AFM image and in its Fourier transform. (Reprinted from Brown et al. 2002 with permission from J. Am. Chem. Soc., © 2002 Am. Chem. Soc.)
Conclusions and prospects for future work
The experiments described in the preceding sections demonstrate that binary patterning of polar and nonpolar amino acids can focus libraries of de novo sequences into productive regions of sequence space. The libraries were designed without using either residue-by-residue rational design or computational methods. Moreover, neither genetic selections in vivo nor high throughput methods in vitro were used to screen the libraries. Nonetheless, appropriately designed binary patterned libraries produced α-helical proteins, β-sheet proteins, well-ordered structures, cofactor binding proteins, catalytically active enzymes, self-assembled monolayers, amyloid-like nanofibrils, and template-directed assemblies of two-dimensional biomaterials.
These initial successes notwithstanding, thus far the potential of the binary code for protein design has been explored only cursorily. To date, we have completed detailed studies of only several representative proteins from each collection. Although characterization of these few proteins has provided a proof-of-principle demonstration that the binary code strategy is an effective method for discovering novel proteins with interesting properties (e.g., well-ordered structures, cofactor binding, enzyme-like activity, etc.), the range of properties that might ultimately be uncovered will require detailed studies of many more proteins. For example, the first binary patterned protein whose three-dimensional structure was solved formed a four-helix bundle that is well ordered and displays nativelike interior packing (Figs. 6 ▶, 7 ▶). Will this be true for all members of this library? Or will there be a range of behaviors with some structures being less well packed? If it turns out that the majority of sequences from this library form highly ordered structures with nativelike packing (as suggested by Fig. 5 ▶ and described by Wei et al. 2003b), then this would imply that even when packing is not explicitly designed, protein sequences nonetheless “find a way” to achieve good packing. Such a result would have significant implications both for de novo protein design and for biological evolution. Our ability to address this question, however, will require detailed structural and dynamic studies of many more proteins form our libraries. Such studies are currently underway.
Future work will aim to merge the binary code strategy with high throughput screens and selections. In recent years, a number of powerful methods have been developed to facilitate the isolation of rare “winners” from vast collections of inactive candidates (Smith 1985; Hanes and Plückthun 1997; Chen et al. 2001; Keefe and Szostak 2001; Lin and Cornish 2002). These methods are often applied to libraries of randomly generated sequences. However, because the vast majority of sequence space is not likely to encode well-folded functionally active proteins (Mandecki 1990; Davidson and Sauer 1994; Davidson et al. 1995; Prijambada et al. 1996), application of these methods to randomly generated sequences typically requires very large libraries and yields “hits” only very rarely (Keefe and Szostak 2001). In contrast, application of screens or selections to focused libraries is likely to produce successful hits far more frequently. As described in this review, the binary code strategy can focus libraries of de novo sequences into regions of sequence space that favor folded protein structures. Therefore, we anticipate that application of high throughput screens and selections to binary patterned libraries will provide a rich source of novel proteins with interesting and useful activities.
Acknowledgments
This review article is based on a lecture presented by Michael Hecht on July 29, 2003, on the occasion of his receiving the Emil T. Kaiser Award at the 17th Annual Symposium of The Protein Society. He expresses enormous gratitude to the many students (graduate and undergraduate) and postdoctoral associates who have been part of his research group over the past 15 years. Without their efforts, the research described in this review article would not have occurred. Recent work was supported by NIH grant R01 GM062869.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04690804.
References
- Adams, P.A. 1990. The peroxidasic activity of the haem octapeptide microperoxidase-8 (MP-8): The kinetic mechanism of the catalytic reduction of H2O2 by MP-8 using 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulphonate)(ABTS) as reducing substrate. J. Chem. Soc. Perkin Trans. 2 8 1407–1414. [Google Scholar]
- Aksay, I.A., Trau, M., Manne, S., Honma, I., Yao, N., Zhou, L., Fenter, P., Eisenberger, P.M., and Gruner, S.M. 1996. Biomimetic pathways for assembling inorganic thin films. Science 273 892–898. [DOI] [PubMed] [Google Scholar]
- Axe, D.D., Foster, N.W., and Fersht, A.R. 1996. Active barnase variants with completely random hydrophobic cores. Proc. Natl. Acad. Sci. 93 5590–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley, J.R. and Hecht, M.H. 1997. Protein design: The choice of de novo sequences. J. Biol. Chem. 272 2031–2034. [DOI] [PubMed] [Google Scholar]
- Belcher, A.M., Wu, X.H., Christensen, R.J., Hansma, P.K., Stucky, G.D., and Morse, D.E. 1996. Control of crystal phase and orientation by soluble mollusk-shell proteins. Nature 381 56–58. [Google Scholar]
- Benson, D.E., Wisz, M.S., and Hellinga, H.W. 2000. Rational design of nascent metalloenzymes. Proc. Natl. Acad. Sci. 97 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz, S.F. and DeGrado, W.F. 1996. Controlling topology and native-like behavior of de novo designed peptides: Design and characterization of anti-parallel four-stranded coiled coils. Biochemistry 35 6955–6962. [DOI] [PubMed] [Google Scholar]
- Bolon, D.N. and Mayo, S.L. 2001. Enzyme-like proteins by computational design. Proc. Natl. Acad. Sci. 98 14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, J.U., Reidhaar-Olson, J.F., Lim, W.A., and Sauer, R.T. 1990. Deciphering the message in protein sequences: Tolerance to amino acid substitutions. Science 247 1306–1310. [DOI] [PubMed] [Google Scholar]
- Bromberg, S. and Dill, K.A. 1994. Side-chain entropy and packing in proteins. Protein Sci. 3 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broo, K.S., Nilsson, H., Nilsson, J., and Baltzer, L. 1998. Substrate recognition and saturation kinetics in de novo designed histidine-based four-helix bundle catalysts. J. Am. Chem. Soc. 120 10287–10295. [Google Scholar]
- Broome, B.M. and Hecht, M.H. 2000. Nature disfavors sequences of alternating polar and nonpolar amino acids: Implications for amyloidogenesis. J. Mol. Biol. 296 961–968. [DOI] [PubMed] [Google Scholar]
- Brown, C.L., Aksay, I.A., Saville, D.A., and Hecht, M.H. 2002. Template-directed assembly of a de novo designed protein. J. Am. Chem. Soc. 124 6846–6848. [DOI] [PubMed] [Google Scholar]
- Chen, G., Hayhurst, A., Thomas, J.G., Harvey, B.R., Iverson, B.L., and Georgiou, G. 2001. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nature Biotechnol. 19 537–542. [DOI] [PubMed] [Google Scholar]
- Chothia, C. and Lesk, A.M. 1987. The evolution of protein structures. Cold Spring Harbor Symp. Quant. Biol. 52 399. [DOI] [PubMed] [Google Scholar]
- Davidson, A.R. and Sauer, R.T. 1994. Folded proteins occur frequently in libraries of random amino acid sequences. Proc. Natl. Acad. Sci. 91 2146–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, A.R., Lumb, K.J., and Sauer, R.T. 1995. Cooperatively folded proteins in random sequence libraries. Nat. Struct. Biol. 2 856–864. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. 1985. Theory for the folding and stability of globular proteins. Biochemistry 24 1501–1509. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 2002. Getting out of shape. Nature 418 729–730. [DOI] [PubMed] [Google Scholar]
- Fairman, R.T., Chao, H.-G., Mueller, L., Lavoie, T.B., Shen, L., Novotny, J., and Matsueda, G.R. 1995. Characterization of a new four-chain coiled coil: Influence of chain length on stability. Protein Sci. 4 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini, G., Albeck, S., Weiner, S., and Addadi, L. 1996. Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271 67–69. [Google Scholar]
- Gassner, N.C., Baase, W.A., and Matthews, B.W. 1996. A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc. Natl. Acad. Sci. 93 12155–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes, J. and Plückthun, A. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. 94 4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer, A.H., Fink, D.J., Laraia, V.J., Arias, J.L., Calvert, P.D., Kendall, K., Messing, G.L., Blackwell, J., Rieke, P.C., Thompson, D.H., et al. 1992. Innovative materials processing strategies: A biomimetic approach. Science 255 1098–1105. [DOI] [PubMed] [Google Scholar]
- Hilvert, D. 2000. Critical analysis of antibody catalysis. Annu. Rev. Biochem. 69 751–793. [DOI] [PubMed] [Google Scholar]
- Hiner, A.N.P., Hernandex-Ruiz, J., Arnao, M.B., Garcia-Canovas, F., and Acosta, M. 1996. A comparative study of the purity, enzyme activity, and inactivation by hydrogen peroxide of commercially available horseradish peroxidase isoenzymes A and C. Biotech. Bioeng. 50 655–662. [DOI] [PubMed] [Google Scholar]
- Kamtekar, S., Schiffer, J.M., Xiong, H., Babik, J.M., and Hecht, M.H. 1993. Protein design by binary patterning of polar and non-polar amino acids. Science 262 1680–1685. [DOI] [PubMed] [Google Scholar]
- Keefe, A.D. and Szostak, J.W. 2001. Functional proteins from a random-sequence library. Nature 410 715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, E.H., Lansbury Jr., P.T., and Kelly, J.W. 1999. Amyloid diseases: Abnormal protein aggregation in neurodegeneration. Proc. Natl. Acad. Sci. 96 9989–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. and Cornish, V.W. 2002. Screening and selection methods for large-scale analysis of protein function. Angew Chem. Int. Ed. Engl. 41 4402–4425. [DOI] [PubMed] [Google Scholar]
- Looger, L.L., Dwyer, M.A., Smith, J.J., and Hellinga, H.W. 2003. Computational design of receptor and sensor proteins with novel functions. Nature 423 185–190. [DOI] [PubMed] [Google Scholar]
- Lowenstam, H.A. and Weiner, S. 1989. On biomineralization. Oxford University Press, Oxford, UK.
- Mandecki, W. 1990. A method for construction of long randomized open reading frames and polypeptides. Protein Eng. 3 221–226. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. 1993. Structural and genetic analysis of protein stability. Annu. Rev. Biochem. 62 139–160. [DOI] [PubMed] [Google Scholar]
- Menger, F.M. and Ladika, M. 1987. Origin of rate accelerations in an enzyme model: The p-nitrophenyl ester syndrome. J. Am. Chem. Soc. 109 3145–3146. [Google Scholar]
- Moffet, D.A. and Hecht, M.H. 2001. De novo proteins from combinatorial libraries. Chem. Rev. 101 3191–3204. [DOI] [PubMed] [Google Scholar]
- Moffet, D.A., Certain, L.K., Smith, A.J., Kessel, A.J., Beckwith, K.A., and Hecht, M.H. 2000. Peroxidase activity in heme proteins derived from a designed combinatorial library. J. Am. Chem. Soc. 122 7612–7613. [Google Scholar]
- Moffet, D.A., Case, M.A., House, J.C., Vogel, K., Williams, R., Spiro, T.G., McLendon, G.L., and Hecht, M.H. 2001. Carbon monoxide binding by de novo heme proteins from a designed combinatorial library. J. Am. Chem. Soc. 123 2109–2115. [DOI] [PubMed] [Google Scholar]
- Moffet, D.A., Foley, J., and Hecht, M.H. 2003. Midpoint reduction potentials and heme binding stoichiometry of de novo proteins from designed combinatorial libraries. Biophys. Chem. 105 231–239. [DOI] [PubMed] [Google Scholar]
- Munson, M., Balasubramanian, S., Fleming, K.G., Nagi, A.D., O’Brien, R., Sturtevant, J.M., and Regan, L. 1996. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 5 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, A.L., Hellinga, H.W., and Caradonna, J.P. 1997. Construction of a catalytically active iron superoxide dismutase by rational protein design. Proc. Natl. Acad. Sci. 94 5562–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack, S.J., Jacobs, J.W., and Schultz, P.G. 1986. Selective chemical catalysis by an antibody. Science 234 1570–1573. [DOI] [PubMed] [Google Scholar]
- Prijambada, I.D., Yomo, T., Tanaka, F., Kawama, T., Yamamoto, K., Hasegawa, A., Shima, Y., Negoro, S., and Urabe, I. 1996. Solubility of artificial proteins with random sequences. FEBS Lett. 382 21–25. [DOI] [PubMed] [Google Scholar]
- Prusiner, S.B. 1997. Prion diseases and the BSE crisis. Science 278 245–251. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. and Richardson, D.C. 2002. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. 99 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D.S., Santiago, J.V., Bray-Hall, S.T., Doshi, N., Grantcharova, V.P., Yi, Q., and Baker, D. 1997. Functional rapidly folding proteins from simplified amino acid sequences. Nat. Struct. Biol. 4 805–809. [DOI] [PubMed] [Google Scholar]
- Rojas, N.R., Kamtekar, S., Simons, C.T., McLean, J.E., Vogel, K.M., Spiro, T.G., Farid, R.S., and Hecht, M.H. 1997. De novo heme proteins from designed combinatorial libraries. Protein Sci. 6 2512–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, D.M., Roy, S., and Hecht, M.H. 1999. Screening combinatorial libraries of de novo proteins by hydrogen-deuterium exchange and electro-spray mass spectrometry. J. Am. Chem. Soc. 121 9509–9513. [Google Scholar]
- Roy, S. and Hecht, M.H. 2000. Cooperative thermal denaturation of proteins designed by binary patterning of polar and nonpolar amino acids. Biochemistry 39 4603–4607. [DOI] [PubMed] [Google Scholar]
- Roy, S., Helmer, K.J., and Hecht, M.H. 1997a. Detecting native-like properties in combinatorial libraries of de novo proteins. Fold. Des. 2 89–92. [DOI] [PubMed] [Google Scholar]
- Roy, S., Ratnaswamy, G., Boice, J.A., Fairman, R., McLendon, G., and Hecht, M.H. 1997b. A protein designed by binary patterning of polar and nonpolar amino acids displays native-like properties. J. Am. Chem. Soc. 119 5302–5306. [Google Scholar]
- Sarikaya, M. and Aksay, I.A. 1995. Biomimetics: Design and processing of materials. AIP Press, Woodbury, NY.
- Selkoe, D.J. 2001. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 81 741–766. [DOI] [PubMed] [Google Scholar]
- Selkoe, D.J. and Podlisny, M.B. 2002. Deciphering the genetic basis of Alzheimer disease. Annu. Rev. Genom. Hum. Genet. 3 67–99. [DOI] [PubMed] [Google Scholar]
- Shifman, J.M., Gibney, B.R., Sharp, R.E., and Dutton, P.L. 2000. Heme redox potential control in de novo designed four α-helical bundle proteins. Biochemistry 39 14813–14821. [DOI] [PubMed] [Google Scholar]
- Smith, G.P. 1985. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228 1315–1317. [DOI] [PubMed] [Google Scholar]
- Springs, S.L., Bass, S.E., and McLendon, G.L. 2000. Cytochrome b562 variants: A library for examining redox potential evolution. Biochemistry 39 6075–6082. [DOI] [PubMed] [Google Scholar]
- Springs, S.L., Bass, S.E., Bowman, G., Nodelman, I., Schutt, C.E., and McLendon, G.L. 2002. A multigeneration analysis of cytochrome b562 redox variants: Evolutionary strategies for modulating redox potential revealed using a library approach. Biochemistry 41 4321–4328. [DOI] [PubMed] [Google Scholar]
- Stewart, J.D., Krebs, J.F., Siuzdak, G., Berdis, A.J., Smithrud, D.B., and Benkovic, S.J. 1994. Dissection of an antibody-catalyzed reaction. Proc. Natl. Acad. Sci. 91 7404–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontano, A., Janda, K.D., and Lerner, R.A. 1986. Catalytic antibodies. Science 234 1566–1570. [DOI] [PubMed] [Google Scholar]
- Wang, W. and Hecht, M.H. 2002. Rationally designed mutations convert de novo amyloid-like fibrils into soluble monomeric β-sheet proteins. Proc. Natl. Acad. Sci. 99 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. and Hecht, M.H. 2004. Enzyme-like proteins from an unselected library of designed amino acid sequences. Protein Eng. Des. Sel. 17 67–75. [DOI] [PubMed] [Google Scholar]
- Wei, Y., Kim, S., Fela, D., Baum, J., and Hecht, M.H. 2003a. Solution structure of a de novo protein from a designed combinatorial library. Proc. Natl. Acad. Sci. 100 13270–13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Liu, T., Sazinsky, S.L., Moffet, D.A., Pelczer, I., and Hecht, M.H. 2003b. Stably folded de novo proteins from a designed combinatorial library. Protein Sci. 12 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, S. and Addadi, L. 1997. Design strategies in mineralized biological materials. J. Mater. Chem. 7 689–702. [Google Scholar]
- West, M.W. and Hecht, M.H. 1995. Binary patterning of polar and nonpolar amino acids in the sequences and structures of native proteins. Protein Sci. 42032–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M.W., Wang, W., Patterson, J., Mancias, J.D., Beasley J.R., and Hecht, M.H. 1999. De novo amyloid proteins from designed combinatorial libraries. Proc. Natl. Acad. Sci. 96 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A.D., Portelius, E., Kheterpal, I., Guo, J.T., Cook, K.D., Xu, Y., and Wetzel, R. 2004. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 335 833–842. [DOI] [PubMed] [Google Scholar]
- Wood, J., Wetzel, R., Martin, J.D., and Hurle, M.R. 1995. Prolines and amyloidogenicity in fragments of the Alzheimer’s peptide β/A4. Biochemistry 34 724–730. [DOI] [PubMed] [Google Scholar]
- Wurth, C., Guimard, N.K., and Hecht, M.H. 2002. Mutations that reduce aggregation of the Alzheimer’s Aβ42 peptide: An unbiased search for the sequence determinants of Aβ amyloidogenesis. J. Mol. Biol. 319 1279–1290. [DOI] [PubMed] [Google Scholar]
- Xiong, H., Buckwalter, B.L., Shieh, H.M., and Hecht, M.H. 1995. Periodicity of polar and non-polar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. Proc. Natl. Acad. Sci. 92 6349–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G., Wang, W., Groves, J.T., and Hecht, M.H. 2001. Self-assembled monolayers from a designed combinatorial library of de novo β-sheet proteins. Proc. Natl Acad. Sci. 98 3652–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, A., Nakashima, T., Tokuriki, N., Hosokawa, M., Nogami, H., Arioka, S., Urabe, I., and Yomo, T. 2002. Evolvability of random polypeptides through functional selection within a small library. Protein Eng. 15 619–626. [DOI] [PubMed] [Google Scholar]