Abstract
GroEL can solubilize membrane proteins by binding them in its hydrophobic cavity when detergent is removed by dialysis. The best-studied example is bacteriorhodopsin, which can bind in the GroEL chaperonin at two molecules per tetradecamer. Applying this approach to the holin and antiholin proteins of phage λ, we find that both proteins are solubilized by GroEL, in an ATP-sensitive mode, but to vastly different extents. The antiholin product, S107, saturates the chaperonin at six molecules per tetradecameric complex, whereas the holin, S105, which is missing the two N-terminal residues of S107, forms a hyper-solubilization complex with up to 350 holin molecules per GroEL, or approximately 4 MDa of protein per 0.8 MDa tetradecamer. Gel filtration chromatography and immunoprecipitation experiments confirmed the existence of complexes of the predicted masses for both S105 and S107 solubilization. For S105, negatively stained electron microscopic images show structures consistent with protein shells of the holin assembled around the chaperonin tetradecamer. Importantly, S105 can be delivered rapidly and efficiently to artificial liposomes from these complexes. In these delivery experiments, the holin exhibits efficient membrane-permeabilizing activity. The S107 antiholin can block formation of the hypersolubilization complexes, suggesting that their formation is related to an oligomerization step intrinsic to holin function.
Keywords: membrane proteins, chaperone, holins, liposomes
With the exception of the proteins of the outer membranes of bacteria and related organelles, the transmembrane domains (TMDs) of integral membrane proteins are universally λ-helical and hydrophobic. Studying the structure and function of such proteins is complicated by their insolubility in the absence of detergent. Moreover, the function of these proteins is notoriously difficult to study because of the difficulty in integrating them into defined, pre-formed bilayers in vitro. Recently, we have found that integral membrane proteins can be kept soluble without detergent by binding to the GroEL tetradecameric chaperone (Deaton et al. 2004). Definitive experiments were performed with purified bacteriorhodopsin (BR), an integral membrane protein with seven transmembrane domains. After dialysis of detergent-solubilized BR in the presence of GroEL, it was found that BR-GroEL complexes were formed containing two molecules of BR at saturation. The complexes were sensitive to the presence of ATP, but not AMP. Remarkably, BR retained native conformation in the complexes, which could be used to deliver BR to liposomes efficiently, vectorially, and in functional form.
Our original motive to develop methods for the detergent-free solubilization and delivery of membrane proteins derived from our interest in the function of the bacteriophage λ holin protein. Holins are integral membrane proteins that cause a temporally scheduled permeabilization of the membrane during host cell lysis (Wang et al. 2000). The holin protein, S105, and the antiholin, S107, which binds to and inhibits S105, are both products of the S gene, differing only by the N-terminal Met-Lys extension at the N terminus of S107. Hole formation is thought to depend on oligomerization of the holin, and some mutants defective in lysis, such as S105a52v, appear to be blocked at the dimer stage (Gründling et al. 2000a).
In view of the results with BR, we considered that GroEL might form complexes with purified S105 and S107 and that such complexes might be used to insert the S gene products into artificial membranes. Experiments to test this are reported here. The results are discussed in terms of the nature of the S-GroEL interaction, the fundamental functional properties of holin proteins and the possible role of GroEL in the insertion of membrane proteins in vivo.
Results
Hypersolubilization of the λ holin
To test the ability of GroEL to form a soluble complex with the λ holin, S105 in 1% EBB was mixed with either GroEL, or BSA and dialyzed under conditions where detergent was quantitatively removed within 3 days. Based on results with BR, which has seven TMDs and is bound at two molecules per tetradecameric chaperonin, it was anticipated that four to six molecules of S105, with three TMDs (Fig. 1 ▶), could be solubilized by GroEL. Unexpectedly, the S protein remained quantitatively soluble in the presence of GroEL, at an 80:1 ratio of S105 molecules per GroEL, while completely precipitating in the BSA control (Fig. 2A,B ▶). In similar experiments performed to ascertain the limits of this GroEL-dependent solubilization of S, it was found that S105 could be solubilized at up to 350 molecules per GroEL complex (Table 1; Fig. 3 ▶). At 11.5 kDa per S105 molecule, this level of solubilization, which we refer to as hypersolubilization, would require complexes formed with ~4 MDa of holin protein per GroEL chaperonin, which has a mass of 0.8 MDa. Stored at 4°C, the S105 remains soluble in the presence of GroEL for at least 12 weeks at low S to GroEL ratios and 3–4 weeks at high S to GroEL ratios at 4°C (data not shown).
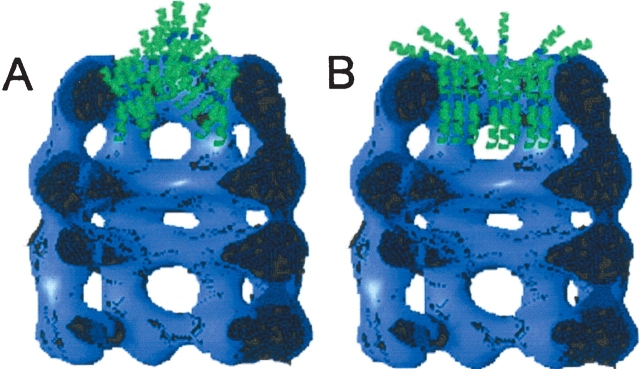
Figure 1.
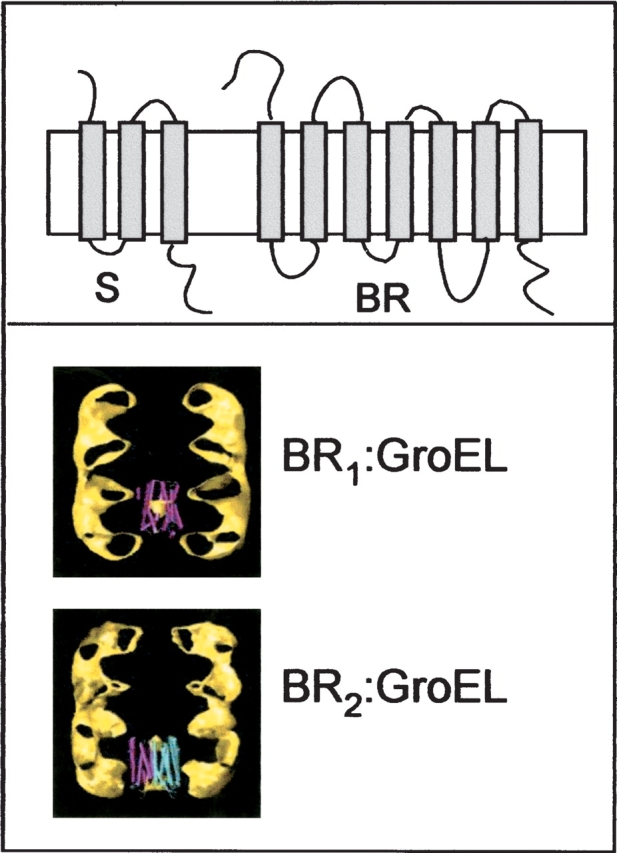
S gene products and BR: topologies and scale compared to GroEL. (Upper panel) Topological model of the S proteins (S105 and S107), with three TMDs, and BR, with seven TMDs. (Lower panel) Modeled images of complexes formed between one or two molecules of BR and the structure of GroEL.
Figure 2.
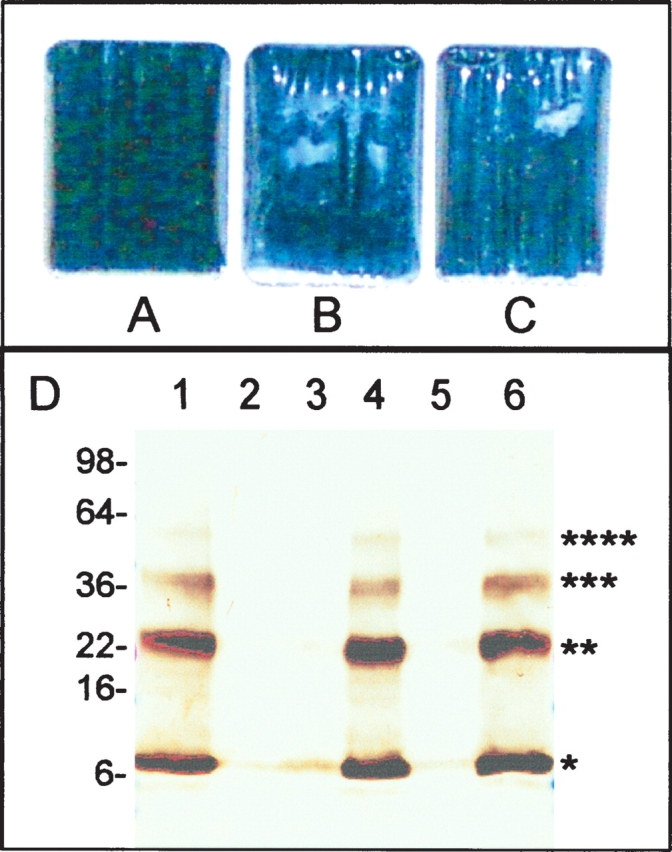
ATP-sensitive solubilization of the λ holin. S105 protein at 2 mg/mL in detergent was placed in dialysis chambers and dialyzed until visible precipitates formed. (A–C) Images of chambers after dialysis. In addition to the detergent-solubilized holin, the dialysis chambers contained (A,C) GroEL (1 mg/mL); (B) BSA (1 mg/mL). In C, the dialysis buffer contained 5 mM ATP. (D) S105 (100 μg/mL) dialyzed with GroEL (lanes 1,2), BSA (lanes 3,4), or GroEL + 5 mM ATP (lanes 5,6). Soluble and insoluble fractions obtained by centrifugation were analyzed by SDS-PAGE and immunoblotting with anti-S antibodies. Lanes 1, 3, 5 = supernatant (soluble fraction); 2, 4, 6 = pellet (insoluble fraction). Molecular masses as determined by mobility of standards are indicated. Monomer, dimer, trimer, and tetramer species of the holin are indicated by asterisks.
Table 1.
Solubilization of the λ holin by GroEL
| Proteina | Input ratiob | % Solublec | Number of experimentsd |
| S105 | 1:10 | 100 | 5 |
| 1:1 | 100 | 50 | |
| 10:1 | 100 | 10 | |
| 50:1 | 100 | 5 | |
| 80:1 | 100 | >100 | |
| 100:1 | 100 | 5 | |
| 150:1 | 100 | 5 | |
| 200:1 | 100 | 5 | |
| 250:1 | 100 | 5 | |
| 300:1 | 100 | 10 | |
| 350:1 | 100 | 10 | |
| 400:1 | 75–80 | 2 | |
| +ATP (5 mM) | 80:1 | 0 | 5 |
| +ADP (5 mM) | 80:1 | Slow precipitation | 2 |
| +AMP (5 mM) | 80:1 | 100 | 2 |
| +AMPPNP (5 mM) | 80:1 | 0 | 1 |
| S105a52v | 1:10 | 100 | 5 |
| 1:1 | 100 | 50 | |
| 10:1 | 100 | 5 | |
| 50:1 | 100 | 5 | |
| 80:1 | 100 | >100 | |
| 100:1 | 100 | 5 | |
| 150:1 | 100 | 5 | |
| 200:1 | 100 | 5 | |
| 250:1 | 100 | 10 | |
| 300:1 | 95 | 10 | |
| 350:1 | 80 | 5 | |
| 400:1 | 50 | 2 | |
| +ATP (5 mM) | 80:1 | 0 | 2 |
| +ADP (5 mM) | 80:1 | Slow precipitation | 2 |
| +AMP (5 mM) | 80:1 | 100 | 2 |
| +AMPPNP (5 mM) | 80:1 | 0 | 1 |
| S107 | 1:10 | 100 | 2 |
| 1:1 | 100 | 10 | |
| 3:1 | 100 | 10 | |
| 5:1 | 100 | 10 | |
| 6:1 | 100 | 10 | |
| 20:1 | <40 | 3 | |
| 50:1 | <15 | 3 | |
| 100:1 | <10 | 2 | |
| S105 and S107 (2:1) | 1:10 | 100 | 2 |
| 1:1 | 100 | 2 | |
| 3:1 | 100 | 2 | |
| 10:1 | <75 | 2 | |
| 100:1 | <15 | 2 |
a Input protein samples in the detergent EBB, as specified in Materials and Methods. GroEL present at 100 μg/mL.
b Ratio of solute protein molecules to GroEL.
c Percent soluble protein after dialysis, estimated from intensity of immunoblot bands, standardized by purified protein.
d Number of trials conducted.
Figure 3.
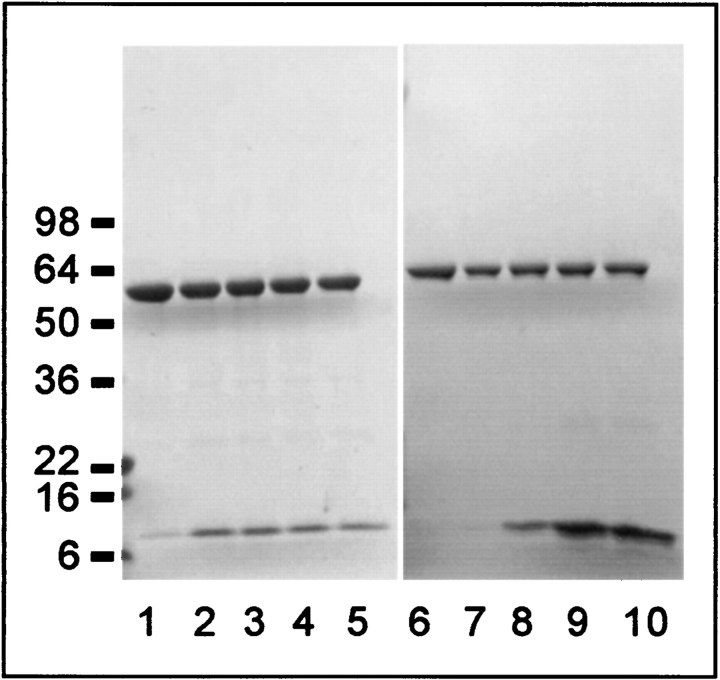
Titration of S105 and S107 into GroEL. GroEL was mixed with EBB-solubilized S105 or S107 at various ratios in 1-mL final volume and then subjected to dialysis to remove detergent, as described in Materials and Methods. Insoluble material was removed by centrifugation and the soluble fraction was concentrated to 0.5 mL before analysis by SDS-PAGE and immunoblotting with anti-S and anti-GroEL antibodies. The amount of solubilized S protein does not increase above 6:1 for S107 (lane 2) but increases continuously through 300:1 (lane 9) for S105. Input solute molecules per GroEL are, for S107 (lanes 1–5): 1, 6, 80, 160, 300. For S105 (lanes 6–10): 1, 6, 80, 300, 400. The position of molecular weight standards are indicated to the left.
The hypersolubilization of the functional S105 holin was also observed with the missense mutant product, S105a52v, which does not permeabilize membranes either in vivo or in vitro (Table 1; Smith et al. 1998; Gründling et al. 2000a). In contrast, the antiholin product of S, S107, was solubilized only at a much lower level, determined by titration experiments to be six molecules per GroEL tetradecamer (Fig. 3 ▶; Table 1), despite the fact that S107 differs from S105 only by the Met-Lys N-terminal dipeptide (Raab et al. 1988). Moreover, the presence of the antiholin prevented the hypersolubilization of the holin (Table 1), mirroring the in vivo dominant-negative character of S107, which binds to and inhibits the lytic function of S105 (Gründling et al. 2000b).
Characterization of the GroEL-S complexes
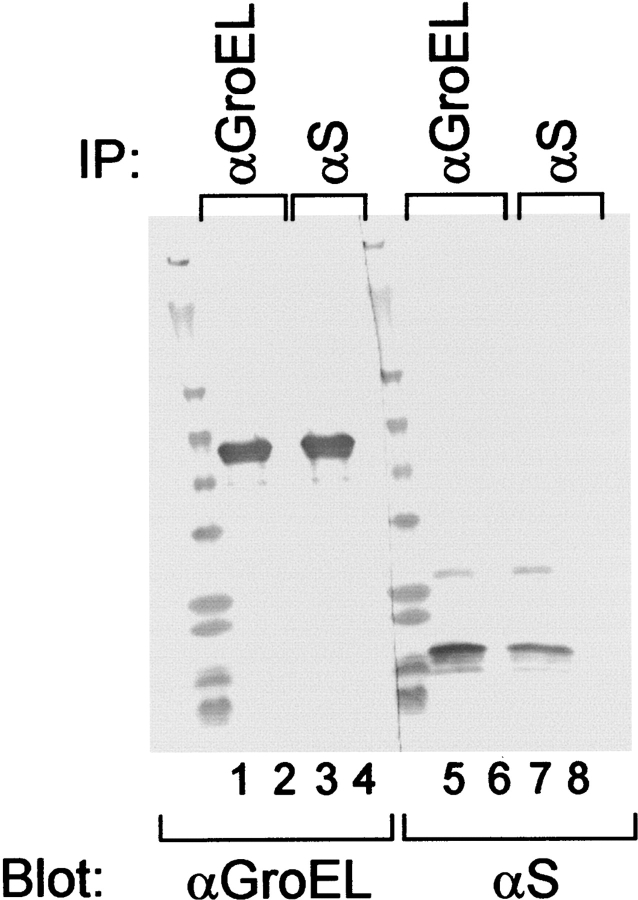
Complexes formed at various S to GroEL ratios were analyzed by gel filtration chromatography. Using the 6:1 S105:GroEL sample, the chaperonin and the holin cochromatographed at a position corresponding to approximately 1 MDa (Fig. 4A ▶), but in the 300:1 sample, complexes containing most of the S protein eluted at a position corresponding to mass of between 2 MDa and 6 MDa (Fig. 4B ▶). Moreover, GroEL and S105 quantitatively coprecipitated with both cognate antibodies, when complexes were formed at 80:1 holin:chaperonin (Fig. 5 ▶). Similar quantitative coprecipitation was observed at 6:1 and 300:1 ratios of S to GroEL (data not shown). Thus, in both the larger and smaller types of complexes, antibodies are able to bind both solute and chaperonin. Moreover, all of the S protein in solution must be complexed with the chaperonin, and all the chaperonin is involved in complexes with S protein. However, the holin S105 and its nonlytic allelic product S105a52v can form soluble complexes with upwards of 300 holin molecules per tetradecamer, which represents more than 3 MDa of solute molecule per 0.8 MDa chaperonin, a solute mass far in excess of anything reported for GroEL. Parallel experiments with S107, which is solubilized at about six molecules per GroEL, showed that the only the smaller ~1-MDa complexes were formed (not shown), consistent with the proposed binding capacity of one chamber of the chaperonin (Ewalt et al. 1997; Song et al. 2003).
Figure 4.
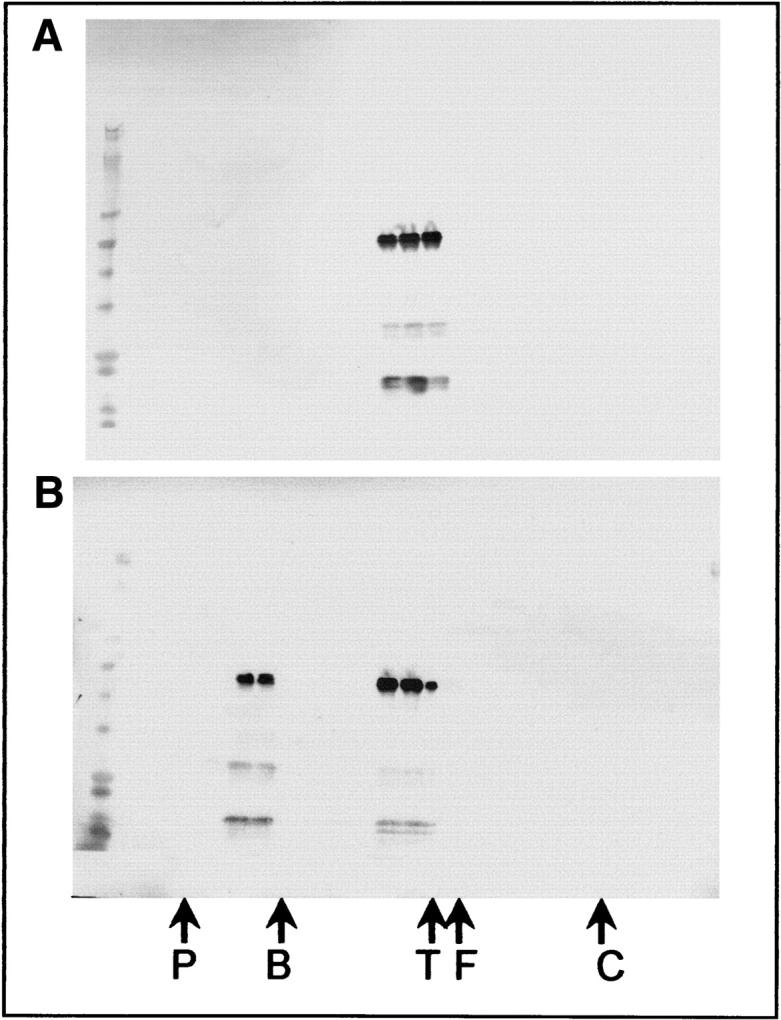
Gel filtration of S-GroEL complexes. S105 samples dialyzed with GroEL at 6:1 (A) and 300:1 (B) were analyzed by gel filtration on a Superose 6GL column. Twenty-four 1-mL fractions were collected, concentrated and analyzed by SDS-PAGE, and immunoblotting with a mixture of anti-S and anti-GroEL antibodies. The peak fractions of gel filtration standards are labeled as follows: P, Polystyrene (6 MDa); B, Blue Dextran 2000 (2 MDa); T, thyroglobulin (660 kDa); F, ferritin (440 kDa); C, chymotrypsinogen (25 kDa). SDS-PAGE standards are shown in the first lane.
Figure 5.
Coimmunoprecipitation of GroEL and solubilized S105. GroEL-solubilized S105 membrane protein samples were subjected to immunoprecipitation and analysis by SDS-PAGE and immunoblotting, using either anti-GroEL or anti-S antibodies as the precipitating (IP) and blotting (Blot) antibodies, as indicated. Lanes 1–4: blot with anti-GroEL; lanes 5–8: blot with anti-S. Shown are immunoprecipitates (odd-numbered lanes) and supernatants (even-numbered lanes) using anti-GroEL (lanes 1,2,5,6) or anti-S (lanes 3,4,7,8).
Effects of nucleotides on S-GroEL complexes
ATP binding by GroEL results in a conformational change that expands the opening of the chaperonin and reduces its affinity for unfolded protein substrates . This change does not require ATP hydrolysis, is much less dramatic with ADP, and does not occur with AMP (Roseman et al. 2001). The addition of ATP to S-GroEL complexes formed at any S to GroEL ratio resulted in the immediate formation of a precipitate that contained all of the S protein present in the sample; the GroEL chaperonin remained soluble. Because both large and small complexes are formed at high S to GroEL ratios, both types must be destabilized by the addition of ATP. AMP-PNP also promoted the release of S from S-GroEL complexes, indicating that this process does not require ATP hydrolysis. However, with AMP-PNP, the release of S occurred more slowly, taking more than 1 h to reach completion. By contrast, ADP caused very slow release of S over the course of several days while AMP had no effect (Table 1). Thus, the effects of nucleotides on S-GroEL complexes were similar to those previously observed with BR-GroEL complexes (Deaton et al. 2004).
Molecular basis of the chaperonin solubilization
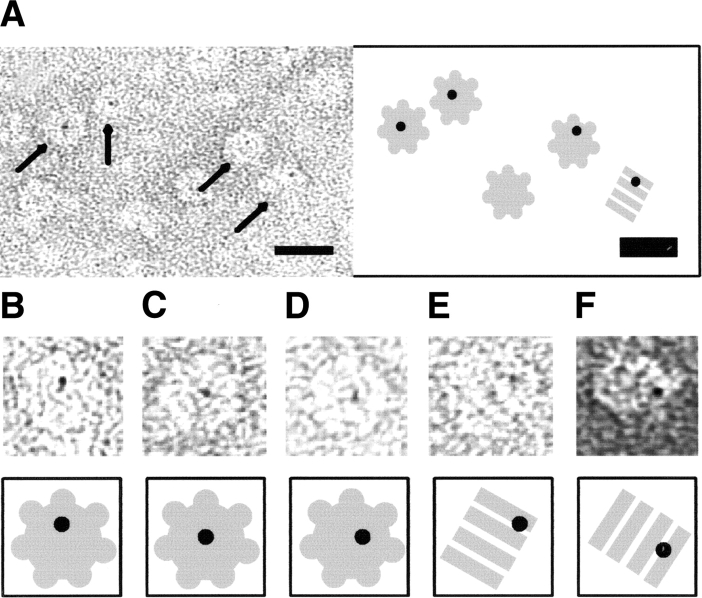
To localize S within the S-GroEL complexes, detergent-solubilized S105 was labeled with amine-reactive Nanogold particles and subjected to dialysis with GroEL. Because of the low yield of Nanogold labeling, the loading ratio of S to GroEL was low, probably less than 1:1. Electron microscopy of GroEL loaded with S105-Nanogold conjugates (Fig. 6 ▶) clearly reveals the association of holin with the GroEL chaperone. Nanogold particles were consistently found within the GroEL lumen, but never around the perimeter of GroEL as deduced from face-on (Fig. 6A–D ▶) in conjunction with side-on projections (Fig. 6E,F ▶). Face-on projections of GroEL are endowed with a sevenfold rotational symmetry, and the positions of the gold particles are close or even coincide with the center of symmetry. With the side-on projections, GroEL displays a point of twofold symmetry separating the two lumenal cavities; in these projections, the positions of gold particles are also indicative for lumenal binding. Nanovan staining (Yang et al. 1994) aided greatly in the detection of labeled molecules. Only occasionally (Fig. 6F ▶) did ammonium molybdate provide a sufficiently low contrast for nanogold to be unambiguously discriminated from protein-related densities. The background in all S105-nanogold conjugate specimens was clean. Control specimens with inactivated gold occasionally showed nanogold aggregates. However, no GroEL was labeled with gold, indicating that nanogold itself does not associate with GroEL unless conjugated to S105. This observation is consistent with the findings of Hainfeld and Furuya (1992), who reported a low affinity of nanogold for proteins in general. Silver-enhanced SDS PAGE revealed that only a fraction of the nanogold particles could be successfully conjugated with S105 (data not shown), resulting in a low labeling efficiency. In electron microscopic analysis, less than 5% of all GroEL molecules were found to be associated with S105-nanogold. Even though the efficiency was low, all identifiable nanogold particles were found in association with GroEL molecules, which means that the labeling, albeit of low yield, is highly specific. The low yield may be due to detergent masking of the reactive lysine groups in S105. These results confirm that, at low S105:GroEL ratios, S105 is bound in the central chamber of the chaperonin.
Figure 6.
Electron micrographs of negatively stained S105-nanogold conjugates in the presence of GroEL. GroEL molecules labeled with S105-Nanogold can be clearly discerned, and are highlighted by arrows in the overview (A) and individually boxed molecules (B–F). Note that (A–E) were stained with methylamine vanadate (Nanovan); (F), with ammonium molybdate. The scale bar corresponds to 20 nm. A cartoon interpretation of each panel is provided to highlight top-down and side-on GroEL molecules.
Uranyl-acetate staining of S105 solubilized at a ratio of 80:1 S105:GroEL revealed unexpected morphologies. Protein shells appear to be formed around 15% to 30% of the chaperonin complexes, suggesting a molecular basis for the hypersolubilization of the S105 holin (Fig. 7 ▶). Most of these shells were approximately 30 nm in diameter, or about twice the diameter of GroEL. Assuming the protein shells consist of oligomers of S105, they could contain S105 protein at about 400 molecules per tetradecamer, which would account for the overall 80:1 ratio if 20% of the GroEL molecules are involved and the rest carry S105 at about six molecules per tetradecamer, as in the case of S107. When S105 was loaded into GroEL at 1:1 or 2:1, shells were not observed (not shown). Also, chains of GroEL molecules were occasionally observed at the higher input ratios of S105 to GroEL, but not at low input ratios (not shown), suggesting that S105 molecules could form mixed oligomers with the tetradecamer.
Figure 7.
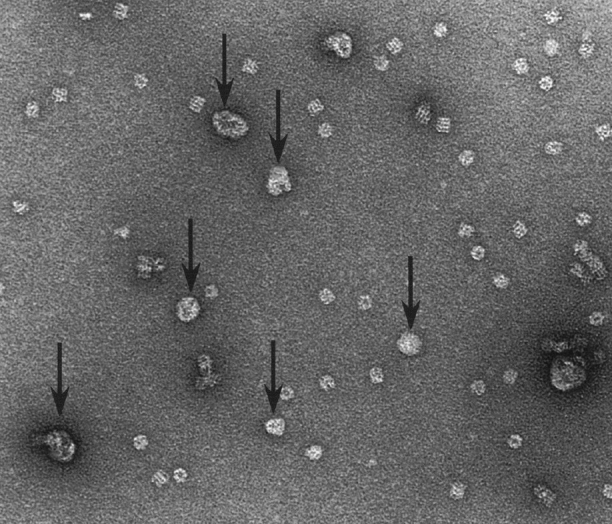
S105 forms shells around GroEL molecules. Arrows indicate structures found in S105 and GroEL mixtures, dialyzed from detergent at an S105:GroEL ratio of 80:1. Free GroEL molecules are readily discerned.
S105 delivered from GroEL complexes is functional irrespective of the loading ratio
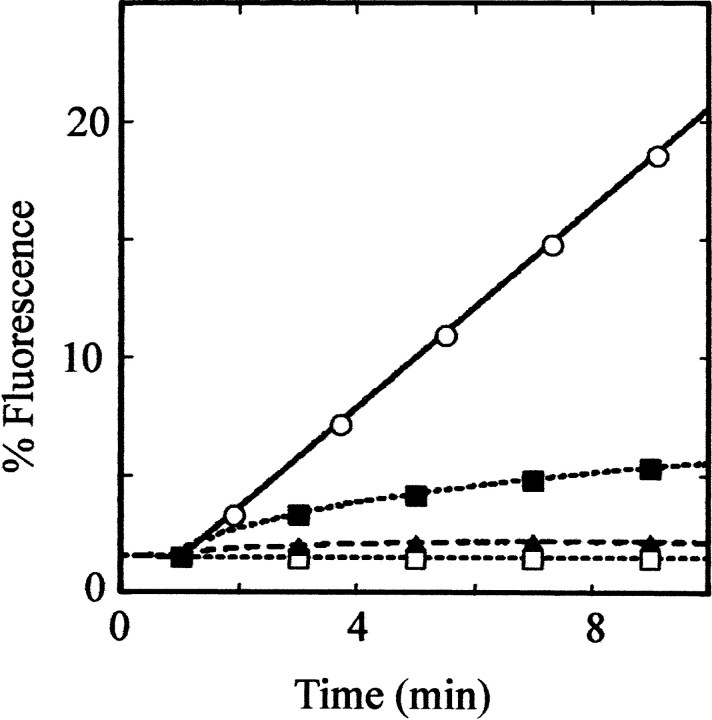
BR solubilized by forming complexes with GroEL was shown to retain its normal conformation, as judged by its spectroscopic characteristics, and could be delivered efficiently and in functional form to artificial membranes, as judged by the ability to energize the membranes upon exposure to light (Deaton et al. 2004). Although there is no independent measure of the conformational state of the S protein in complexes with GroEL, it is possible to assess its function by using a membrane permeabilization assay (Smith et al. 1998). GroEL-S complexes were prepared at both high and low ratios of S to GroEL, and the large ~4-MDa complexes were purified by gel filtration, as shown in Figure 4 ▶. Both types of S-GroEL complexes were shown to effect dye release from liposomes, whereas in controls done with GroEL alone or GroEL loaded with the S105a52v nonlytic allelic product there was no significant dye release above background (Fig. 8 ▶). Moreover, the rate of permeabilization of liposomes is clearly much higher for the large complexes than for the smaller complexes, with comparable levels of S105. Thus the large complexes, although clearly carrying S protein far in excess of the binding capacity of the chaperonin chambers, contain S protein capable of inserting efficiently into artificial membranes and then proceeding to form holin lesions.
Figure 8.
S105 delivered from GroEL complexes is functional for membrane permeabilization. λ Holin protein solubilized at low (6:1) or high (300:1) ratios of S gene product to GroEL was added to pre-formed liposomes loaded with calcein, and the release of dye monitored by fluorescence. The large complexes were purified by size exclusion chromatography. Protein added at t = 1 min. One hundred percent fluorescence corresponds to the value after addition of detergent. (Open circles) S105, 300:1 (7 μg/mL S105, 1.7 μg/mL GroEL); (solid squares) S105, 6:1 (5 μg/mL S105, 50 μg/mL GroEL); (solid triangles) S105a52v, 6:1 (10 μg/mL S105a52v, 100 μg/mL GroEL); (open squares) GroEL only (200 μg/mL).
Discussion
Hypersolubilization of the holin S105 and its application to the study of holin function
In a previous report, we demonstrated that BR could be complexed with GroEL and then transferred efficiently to pre-formed membranes (Deaton et al. 2004), providing a new approach for the study of integral membrane proteins. This method is particularly attractive for the study of bacteriophage holins, which accumulate harmlessly in the host membrane until suddenly triggering and disrupting the membrane, an event that terminates the viral life cycle. Clearly this “hole formation” event cannot be studied if proteoliposomes containing holins are formed by traditional methods involving removal of detergent from mixtures of solubilized lipid and protein. Thus, it was gratifying that the protein products of the λ holin gene S were found to retain solubility after removal of detergent in the presence of GroEL. Surprisingly, however, the amount of the lethal holin S105 that could be solubilized was in excess of 300 molecules of S105 per GroEL. In contrast, titration experiments showed that the antiholin S107, differing from S105 only by the N-terminal two residues Met-Lys, was solubilized up to a limit of six molecules per GroEL, approximately what would be expected from the estimated capacity of the chaperonin chamber (Ewalt et al. 1997; Song et al. 2003). S protein solubilized by GroEL in this way is stable for weeks at 4°C, irrespective of the loading ratio. Moreover, the allelic state of the S105 protein was irrelevant, because the S105a52v product, which is nonlytic in vivo, was also hypersolubilized by GroEL. Finally, the hypersolubilized material is at least as effective, on a per-S molecule basis, as the complexes formed at low ratios, in terms of delivery to and permeabilization of liposomes.
Two types of ATP-sensitive complexes formed between S protein and GroEL
Gel filtration analysis of the S105-GroEL solutions formed at high S105-to-GroEL ratios revealed that most of the holin was in complexes of ~4 MDa, the largest GroEL–protein complexes yet reported. In contrast, complexes formed at low ratios of S105, or between S107 and GroEL, eluted at the same position as GroEL alone, as expected because the molecular weight of S105 or S107 is only about 11.5 K, compared to the 840 K of the chaperonin. Visualization of the S105–GroEL complexes by electron microscopy revealed that, in the samples formed at high ratio, the chaperonin appeared to be completely covered with protein, presumably a shell of S105 molecules. In contrast, complexes formed at low levels of S protein per chaperonin contain S only in the chamber of the tetradecamer, as judged by nanogold staining. Both types of complexes are sensitive to ATP. A reasonable model, then, for these complexes is that the hydrophobic surface of the apical domain of the GroEL subunits is required for the binding and solubilization of the S protein solute. In the current model for the GroEL–GroES cycle for refolding of denatured proteins, this surface is exposed alternately on each heptameric subcomplex of the tetradecamer (Roseman et al. 2001). The fact that ATP binding instantly releases the bound membrane protein solute is strong evidence that the restriction to single cavity loading reflects the conformational dynamics of GroEL. ATP binding is known to cause a conformational change, which is thought to cause internal rotation of the apical domain for all of the subunits of one heptamer. The fact that the shell complexes are also sensitive to ATP suggests that the foundation of the shell is a complex with S protein bound in the chamber of GroEL, and that there are interactions between the chamber-bound and external S proteins that are necessary for the stability of the complex.
In both complexes, GroEL and S are accessible to their respective antibodies. This suggests that the S105 shells are not rigid assemblies that would block access of antibodies to GroEL. A substantial number of GroEL molecules lacking shells are present even at the maximum S105 to GroEL loading, as judged by gel filtration analysis and electron microscopy. The shells may be in dynamic equilibrium, with complexes exchanging S105 protein with one another, and occasionally segregating chaperonins lacking the protein shell. Nevertheless, the immunoprecipitation experiments show that all of the GroEL molecules contain S105 under these conditions, even if not all of them carry S105 shells.
The S gene is expressed at relatively low levels in vivo, such that on the order of 103 molecules of total S gene product are present per cell at the time of lysis (Chang et al. 1995). Considering the much larger numbers of GroEL molecules in the cytosol, the large complexes formed at high input ratio of S105 to the chaperonin are not indicative of complexes that might form in vivo. Nevertheless, in view of the fact that the hypersolubilization effect occurs with S105 but not with S107, clues to the physical differences between the holin and antiholin might be deduced from consideration of models for the structure of the large complexes. The gross dimensions and apparent molecular mass suggests that upwards of 300 S105 molecules polymerize in shells around S105-saturated GroEL tetradecamers. Because the large complexes can be destroyed by the addition of ATP, it is likely that these shells are connected to the chamber-bound S105 molecules. Models of the growth of the shell from complexes with S proteins bound in the chamber are shown in Figure 9 ▶.
Figure 9.
Models for formation of S-GroEL complexes. (A) The TMDs of the S105 protein associate with each other within the GroEL chamber. The oligomers that form are oriented so that continued polymerization beyond the GroEL chamber is possible via TMD-TMD interaction. (B) The hydrophobic faces of one or more of the S105 TMDs bind to the hydrophobic surface of the apical domain of GroEL. The hydrophilic surface of TMDs 1 and/or 3 are exposed to the opening of the GroEL chamber and serve as nucleation sites for the polymerization of S105 around the outside of the GroEL molecule.
Whether there is S105 in both chambers is unknown, although preliminary results from negative-stain single particle analysis suggests that extra density is only found in one chamber, not only for S105 but also for other integral membrane proteins like BR and LacY (J. Sun, C.G. Savva, J.F. Deaton, H.R. Kaback, M. Svrakic, R. Young, and A. Holzenburg, in prep.). Irrespective of whether both chambers are loaded, more than 300 S molecules must be present in the external shell. Assuming 300 S105 molecules are aligned by TMD–TMD interactions to make up the surface of a 30-nm large complex, the density of S105 would be ~0.4 molecules/nm2, approximately what would be expected from tight helical packing of the S105 molecules, each with three TMDs. S105 oligomerizes to a high degree as part of its function in the timing of host lysis (Gründling et al. 2000a), so this extraordinary mostly two-dimensional polymer might reflect the ability of GroEL to maintain S105 in a polymerization-proficient conformation similar to that it assumes in the bilayer. Indeed, the S105 protein in these complexes is even more efficient at permeabilization of liposomes than S105 bound at low ratios to GroEL (Fig. 8 ▶). The fact that the S105a52v protein also forms the large complexes might be taken as evidence against this model. However, the two-dimensional concentration of S protein in vivo is never more than 10−4 molecules/nm2 of membrane, more than 103-fold less than in these putative shells. Thus, with the A52V missense change, a TMD–TMD interaction defect that would block polymerization in vivo by changing affinity by up to two orders of magnitude would not be relevant at the high loading ratios in vitro.
In contrast, the limit of six molecules of the antiholin S107 per chaperonin approximates the putative binding capacity of the GroEL cavity. Significantly, mixtures of S107 and S105 also form only the small complexes, indicating that binding is restricted to the cavity. This dominant-negative effect parallels the lysis-inhibiting function of the S107, and suggests that the antiholin may function by blocking high-level polymerization in vivo.
GroEL as a vector for delivery of integral proteins to bilayers
With the results presented here and previously (Deaton et al. 2004), GroEL has been shown to have the ability to deliver BR and both the holin and antiholin products of the λ S gene to liposomes rapidly, efficiently, and in functional form. These findings support previous suggestions that GroEL may have a role in the posttranslational localization of integral membrane proteins in vivo (Bochkareva et al. 1996). In any case, the important practical implications for the study of membrane proteins in their native environment is, alone, sufficient justification for further study of this phenomenon. In particular, the fact that neither ATP nor the cochaperonin GroES is required for release of the cargo into the bilayer of the liposome is intriguing. The C-terminal region of each GroEL subunit has affinity for membranes in vitro (Torok et al. 1997). The binding of GroEL loaded with a membrane protein to the surface of the bilayer may be associated with conformational changes analogous to those observed during the GroES-ATP binding cycle for GroEL loaded with denatured soluble proteins; the liposome system described here should be suitable for investigating this possibility. In any case, given the ability to deliver both BR, with its light-dependent proton-pumping ability, and S gene products to artificial membranes efficiently, it is now feasible to study the function of holins in the membrane environment and, importantly, the dependency of hole-formation on the energy state of the membrane. Attempts at reconstitution of the entire holin-mediated lysis triggering event, including the function of the antiholin, are now underway.
Materials and methods
Reagents and general procedures
Calbiochem was the source for all detergents, Calbiosorb Biobeads, and ATP. Bovine serum albumin (BSA), AMP, ADP, and AMP-PNP were obtained from Sigma. BES was obtained from USB. Detergents were used at 1% final concentration. When necessary, samples were concentrated using a Microcon centrifugal filter unit (Amicon), with a molecular weight cutoff of 3000, according to the manufacturer’s instructions. To concentrate protein for SDS-PAGE, 100 μL samples were precipitated by addition of 2 mL of cold 95% EtOH; pellets were recovered by centrifugation for 10 min at 1000g and dissolved in sample buffer. SDS-PAGE and immunoblotting have been described (Smith et al. 1998). The antibodies recognizing S gene products have been described (Chang et al. 1995). Antibodies against GroEL were obtained from StressGen. Polyclonal chicken antibodies against BR were produced by Aves, Inc. against purified BR. Modifications to standard immunoblotting and immunoprecipitation protocols necessary to use the chicken antibodies were done according to manufacturer’s specifications. For immunoprecipitation experiments, primary antibodies were added to the sample at a dilution of 1:2000 and incubated on a roller drum for 1 h at room temperature. Metal-decorated Magnabind secondary antibodies (Pierce) were added to the sample according to the manufacturer’s instructions, and again incubated for 1 h on a roller drum at room temperature. For each precipitation, the Magnabind antibodies and complexes were pulled to the side using a magnet-lined tube rack and washed twice; this procedure prevents contamination of the immune complexes with proteins that spontaneously precipitate out of solution. The magnetically separated complexes were analyzed by SDS-PAGE and immunoblotting.
Gel filtration chromatography
Gel filtration analyses were performed on an AKTA (Amersham Pharmacia) workstation. Samples were filtered through a 0.22-μm pore-size sterilization filter, and concentrated to 1 mL. Samples were chromatographed on a Superose 6 10/300 GL column and collected in 1-mL fractions. The column was calibrated with the following molecular weight standards: polystyrene (~6 MDa); Blue Dextran 2000 (~2 MDa); thyroglobulin (670 kDa); ferritin (440 kDa); and chymotrypsinogen (25 kDa), according to the manufacturer’s instructions. For analytical purposes, eluted fractions were concentrated by cold ethanol precipitation and analyzed by SDS-PAGE and immunoblotting. For liposome permeabilization assays, selected fractions were concentrated and used without further manipulation.
Protein preparations
Purified GroEL was initially obtained from Stressgene. Subsequent experiments were done with GroEL purified from an over-expression system, as described in Kamireddi et al. (1997) The two preparations behaved identically in our hands.
Oligohistidine-tagged holin and antiholin proteins were purified as described (Smith et al. 1998), with some modifications. Membranes from cells producing these proteins were subjected to differential solubilization in 1% EBB, 10% glycerol, 20 mM BES, 0.5 M NaCl, 35 mM MgCl2 (pH 8.0) for 2 to 18 h at 37°C with shaking. Insoluble material was removed by centrifugation at 100,000g for 45 min and the soluble extract was filtered through a 22-μm syringe filter (Genemate Bioexpress) and loaded onto a Hitrap Chelating HP nitrilotriacetic acid column (1 mL) charged with CoCl2 and equilibrated with 1% EBB, 20 mM BES, 0.5 M NaCl, 10% glycerol (pH 7.6). After loading, the column was washed with 1% EBB, 20 mM BES (pH 7.6) and eluted at a flow rate of 0.5 mL/min with a pH gradient from pH 7.6 to pH 2.5, using 1% EBB, 20 mM sodium acetate, 0.5 M NaCl as the limit buffer. Oligohistidine-tagged proteins were eluted with a pH gradient Eluted protein fractions were neutralized with 0.1 M NaOH.
GroEL solubilization of holins
To form complexes between S gene products and GroEL, 800 μL of 1% EBB, 20 mM BES, 0.5 M NaCl (pH 7.6) was placed in a tube and 100 μL of a GroEL solution in the same buffer was added. Finally, 100 μL of S protein in the same buffer was added. At each step, the solution was mixed by pipet. For most experiments, the final concentration of GroEL was 100 μg/mL, and the concentration of S protein was adjusted to achieve the desired molar ratio to the tetradecameric chaperonin. For the purpose of visualization of the precipitate, some experiments contained a final concentration of 2 mg/mL S105 and 1 mg/mL GroEL or BSA. For titration experiments, the final concentration of GroEL was 50 μg/mL for S105 and 75 μg/mL for S107. The 1-mL solution containing detergent-solubilized S protein and GroELwas placed into a dialysis bag and dialyzed against 500 mL of 20 mM BES, 0.5 M NaCl (pH 7.6), supplemented with Calbiosorb Bio-Beads, according to the manufacturer’s instructions. Buffer and Bio-Beads were changed every 8 h. Dialysis was continued until there was quantitative precipitation in a control sample containing the subject protein but with GroEL replaced by an equal mass of BSA. In these experiments, the efficacy of detergent removal was assessed using calcein-loaded liposomes as previously described (Smith et al. 1998). In all cases, detergent was reduced to less than 10% of its critical micellar concentration.
To test the effects of nucleotides on the solubilization process, 5 mM ATP, ADP, or AMP was added to the dialysis solution. To assess the effects of nucleotides on the stability of the complexes between the membrane proteins and GroEL, the same nucleotides or AMP-PNP were added directly, at 5 mM final concentration.
Electron microscopy of Nanogold-labeled S-GroEL complexes
Mono-NHS-Nanogold (6 nM; Nanoprobes) was dissolved in 100 μL 10% DMSO. Labeling was initiated by adding 80 μL of S105 (2.6 mg/mL in 1% EBB, 0.5 M NaCl, 20 mM BES buffer [pH.7.8]) to 40 μL of Nanogold. After incubation at 4°C overnight, the mixture was subjected to gel filtration through a Sephadex 75 column (1.5 × 25 cm) on an AKTA FPLC (Pharmacia). Fractions (1 mL) were collected and analyzed by SDS-PAGE, rendered nonreducing by omitting β-mercaptoethanol from the sample buffer. Duplicate gels were stained with Coomassie Brilliant Blue and LI-Silver (Nanoprobes) for detection of protein and nanogold, respectively. The fractions containing the conjugate were pooled and concentrated to 200 μL. The concentrated conjugates were then incubated with 75 μL of GroEL (1 mg/mL in 10 mM KCl, 10 mM MgCl2, 10 mM Tris [pH 7.6]) and dialyzed as described. Control samples were prepared by adding equivalent amounts of GroEL and deactivated nanogold, incubated for 3 days in 200 mM Tris-HCl (pH 7.5).
S105-Nanogold/GroEL and control samples were prepared for electron microscopy by adding 4 μL of sample onto formvar-carbon coated copper grids (400 mesh) that had previously been rendered hydrophilic by glow-discharging. After incubation for 60 sec, the samples were blotted to remove excess solution and stained with Nanovan (Nanoprobes). Duplicate grids were prepared similarly but stained with 2% uranyl acetate and 2% ammonium molybdate. Samples were examined using a Zeiss 10C electron microscope operating at 80 kV. Micrographs were taken at a calibrated magnification of 55,400.
In vitro membrane permeabilization assays
Liposomes were made by mixing 245 μL of dioleyl-L-α-phospha-tidylcholine (20 mg/mL), 210 μL of dioleylphosphatidylglycerol (10 mg/mL), and 143 μL of cholesterol (7 mg/mL) in chloroform (Avanti Polar Lipids Inc.). The mixture was dried under a stream of nitrogen and suspended in 1-mL solution containing 0.15 M of the self-quenching fluorophore calcein (in 10 mM Tris and 150 mM NaCl; pH adjusted to approximately 7.6 with 2 M NaOH). The resulting lipid suspension was extruded 40 times through a 200-nm polycarbonate membrane in a Liposofast extrusion device (Avestin Inc.). The mainly unilamellar liposomes loaded with calcein were separated from unincorporated calcein by gel filtration chromatography on a G-50 Sephadex column (1.5 × 25 cm) equilibrated in TBS (10 mM Tris [pH 7.6], 150 mM NaCl). Fractions at the void volume containing the liposomes were clearly visible and were collected manually.
The permeabilization of the calcein-loaded liposomes was measured using a SLM 8100 spectrofluorometer (Spectronic Instruments, Inc.) with excitation and emission set at 490 nm and 520 nm, respectively. The reaction mixture contained 40 μL of the liposome suspension (28 μg lipid, ~1012 liposomes) in a final volume of 2 mL of TBS. The assay was initiated by the addition of the indicated protein sample in 20 μL, after which fluorescence was followed as a function of time. Baseline calcein release was defined as that occurring after the addition of TBS instead of the protein sample, while complete release was obtained by the addition of 10 μL 10% Triton X-100.
Acknowledgments
The authors are grateful to Prasad Reddy for his generous gift of a groEL overexpression strain, to Jim Sacchettini for useful suggestions and materials, to the rest of the Young and Holzenburg Laboratories for general support and encouragement, and to Norma Teetes for her logistical assistance in the publication of this manuscript. Work reported here was supported by PHS Grant GM27099 to R.Y. and by funding for the Program for Membrane Structure and Function from the Life Science Task Force at Texas A&M University to R.Y. and A.H. A.H. gratefully acknowledges support by the Office of the Vice President for Research at Texas A&M University.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BR, bacteriorhodopsin
BES, N,N′-bis(2-hydroxyethyl)-2-2aminoethanesulfonic acid
BSA, bovine serum albumin
DMSO, dimethylsulfoxide
EBB, Empigen BB
SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis
TBS, Tris-buffered saline
TMD, transmembrane domain.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04735104.
References
- Bochkareva, E., Seluanov, A., Bibi, E., and Girshovich, A. 1996. Chaperonin-promoted post-translational membrane insertion of a multispanning membrane protein lactose permease. J. Biol. Chem. 271 22256–22261. [DOI] [PubMed] [Google Scholar]
- Chang, C.-Y., Nam, K., and Young, R. 1995. S gene expression and the timing of lysis by bacteriophage λ. J. Bacteriol. 177 3283–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton, J., Sun, J., Holzenburg, A., Struck, D.K., Berry, J., and Young, R. 2004. Functional bacteriorhodopsin is efficiently solubilized and delivered to membranes by the chaperonin GroEL. Proc. Natl. Acad. Sci. 101 2281–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewalt, K.L., Hendrick, J.P., Houry, W.A., and Hartl, F.U. 1997. In vivo observation of polypeptide flux through the bacterial chaperonin system 4. Cell 90 491–500. [DOI] [PubMed] [Google Scholar]
- Gründling, A., Bläsi, U., and Young, R. 2000a. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 182 6082–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling, A., Smith, D.L., Bläsi, U., and Young, R. 2000b. Dimerization between the holin and holin inhibitor of phage λ. J. Bacteriol. 182 6075–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainfeld, J.F. and Furuya, F.R. 1992. A 1.4-nm gold cluster covalently attached to antibodies improves immunolabeling. J. Histochem. Cytochem. 40 177–184. [DOI] [PubMed] [Google Scholar]
- Kamireddi, M., Eisenstein, E., and Reddy, P. 1997. Stable expression and rapid purification of Escherichia coli GroEL and GroES chaperonins. Protein Expr. Purif. 11 47–52. [DOI] [PubMed] [Google Scholar]
- Raab, R., Neal, G., Sohaskey, C., Smith, J., and Young, R. 1988. Dominance in λ S mutations and evidence for translational control. J. Mol. Biol. 199 95–105. [DOI] [PubMed] [Google Scholar]
- Roseman, A.M., Ranson, N.A., Gowen, B., Fuller, S.D., and Saibil, H.R. 2001. Structures of unliganded and ATP-bound states of the Escherichia coli chaperonin GroEL by cryoelectron microscopy. J. Struct. Biol. 135 115–125. [DOI] [PubMed] [Google Scholar]
- Smith, D.L., Struck, D.K., Scholtz, J.M., and Young, R. 1998. Purification and biochemical characterization of the λ holin. J. Bacteriol. 180 2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.L., Li, J., Huang, Y.S., and Chuang, D.T. 2003. Encapsulation of an 86-kDa assembly intermediate inside the cavities of GroEL and its single-ring variant SR1 by GroES. J. Biol. Chem. 278 2515–2521. [DOI] [PubMed] [Google Scholar]
- Torok, Z., Horvath, I., Goloubinoff, P., Kovacs, E., Glatz, A., Balogh, G., and Vigh, L. 1997. Evidence for a lipochaperonin: Association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc. Natl. Acad. Sci. 94 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.-N., Smith, D.L., and Young, R. 2000. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54 799–825. [DOI] [PubMed] [Google Scholar]
- Yang, Y.S., Datta, A., Hainfeld, J.F., Furuya, F.R., Wall, J.S., and Frey, P.A. 1994. Mapping the lipoyl groups of the pyruvate dehydrogenase complex by use of gold cluster labels and scanning transmission electron microscopy. Biochemistry 33 9428–9437. [DOI] [PubMed] [Google Scholar]