Abstract
Advanced glycation end products (AGEs), which are composed of various glucose or carbohydrate adducts, are thought to be responsible for several diabetic and age-related complications. However, to date, specific sites on proteins that are modified by AGEs remain largely unknown. We report here the use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to determine the type and localization of several AGEs formed in vitro on human beta-2-microglobulin (β2M), and in vivo on type 2 ryanodine receptor calcium-release channel (RyR2), and sarco(endo)plasmic reticulum (SERCA2a). A PERL script algorithm, developed in-house, makes searching the relatively large amount of data generated by the MALDI-MS more manageable. The outstanding sensitivity of MALDI-TOF-MS coupled with the PERL script algorithm allows such an approach to be a very useful tool in detecting AGEs and other post-translational modifications. We believe that this method could be an important tool when searching for post-translational modifications on proteins.
Keywords: mass spectrometry, post-translational modification, matrix-assisted laser desorption; ionization, advanced-glycation end products
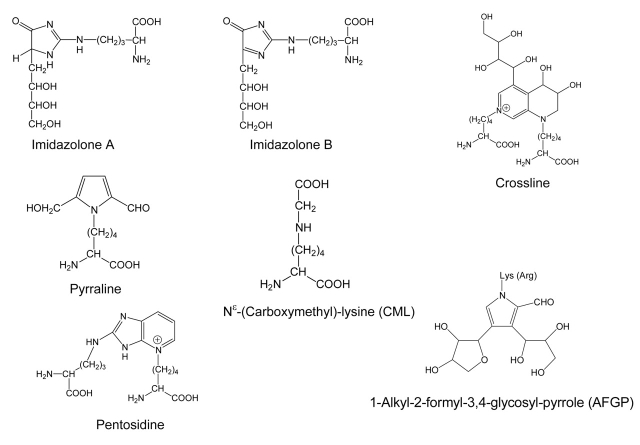
Protein post-translational modifications play important roles in biological activities. It has been a great challenge for biochemists to determine the specific covalent modifications of proteins. A number of analytical methods1–4 have been employed to qualitatively and quantitatively determine identity and location of the modifications; however, mass spectrometry (MS) remains the most powerful method because of its high accuracy and sensitivity.5 Fast atom bombardment mass spectrometry (FAB-MS), liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS), and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) have all been used to study the post-translational modifications of the protein.6 Advanced glycation end products (AGEs) are a heterogeneous group of proteins that have been modified with glucose or carbohydrate adducts, and are thought to be responsible for many complications of diabetes and aging.7 AGEs arise from the nonenzymatic addition of reducing sugars to the side chain of lysine and/or arginine residues of the protein.8 The initial labile Schiff base and Amadori products (AP) undergo a series of rearrangement, dehydration, and fragmentation reactions to produce more complex and irreversibly covalently cross-linked structures.6,8 Several AGE structures, such as imidazolone A and B, Nɛ-(carboxymethyl)lysine (CML), pyrraline, pentosidine, 1-alkyl-2-formyl-3,4-glycosyl-pyrrole (AFGP), and crossline, have been previously identified in association with a number of proteins (Figure 1).6,8 To understand the functional implication of AGEs, structural information is essential. In this study, we describe the use of in-house-developed PERL script algorithm to rapidly determine the identity and localization of AGEs formed in vitro on human beta-2-microglobulin (β2M), and in vivo on type 2 ryanodine receptor calcium-release channels (RyR2) and sarco(endo)plasmic reticulum calcium ATPase (SERCA2a) from a diabetic rat using MALDI-TOF-MS.
FIGURE 1.
Chemical structures of the AGE molecules.
MATERIALS AND METHODS
Isolation and Purification of β2M and β2M-AGEs
β2M was isolated and purified as previously described.9–11 Briefly, ultrafiltrate was collected during hemofiltration with an F-80 polysulfone hollow fiberdialyzer (Fresenius, Ogden, UT) from a chronic (25 yrs) hemodialysis patient with documented β2M amyloidosis. The ultrafiltrate was concentrated, desalted, and separated by isoelectric focusing. The purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was then run over a Toxigel column (Pierce, Rockford, IL) to remove lipopolysaccharide (LPS). The purified β2M was divided into two fractions. One was incubated in glycating buffer (1X PBS, 200 mM d-glucose, 1.5 mM PMSF, 1 mM EDTA) at 37°C for 60 days and represents glycated β2M (β2M-AGEs). The other fraction was incubated with 1.5 mM PMSF and 1 mM EDTA at 4°C for 60 days and represents nonglycated β2M (β2M). At the end of 60 days, the fractions were dialyzed separately to remove glycating buffer and/or preservatives. The proteins were then separated by 10% SDS-PAGE and visualized with standard Coomassie blue dye. The bands corresponding β2M were excised for in-gel trypsin digestion.
Preparation of RyR2 and SERCA2a from Rat Hearts
Type 2 ryanodine receptor and SERCA2a were isolated from the hearts of 8-week streptozotocin-induced diabetic rats and age-matched controls. All animal procedures were done in accordance with Institutional Animal Care and Use Committees. Briefly, membrane vesicles were prepared simultaneously from control and diabetic rat hearts (2 hearts per preparation × 2 preparations) as previously12 described. Their protein contents were determined using the method of Lowry et al.13 One hundred micrograms of total protein from each vesicle preparation were solubilized in gel dissociation medium containing 10 mg/mL dithiothreitol (DTT) and electrophoresed using 4–20% linear gradient SDS-polyacrylamide gel for 3.5 h at 150 V (one gel per membrane preparation). At the end of the run, the gels were transferred overnight onto polyvinylidene difluoride (PVDF) membranes using a semidry procedure. Next, the membranes were blocked, washed, and incubated with either mouse anti-RyR2 or mouse anti-SERCA2a antibodies for 16 h at 4°C. At the end of this time, the membranes were again washed and incubated for 2 h at room temperature with anti-mouse Ig G-horseradish peroxidase. Membranes were then incubated for 1 min with ECL and autoradiograms were developed after 5 min. After the transfer the polyacrylamide gels were stained with 0.25% Coomassie blue dye for 1 h, destained for 4 h, and then dried between two sheets of cellophane. The protein bands on the membranes that immuno-reacted with RyR2 and SERCA2a antibodies were then located by superimposing the dried gel to the autoradiograms. The identified bands were excised and transferred to 1.5-mL Eppendorf tubes for in-gel trypsin digestion.
In-gel Protein Digestion
The gel slices were cut into small pieces, destained with 50% acetonitrile/50 mM ammonium bicarbonate, reduced with 10 mM DTT (Sigma, St. Louis, MO), and alkylated with 55 mM iodoacetamide (Sigma, St. Louis, MO). After alkylation, the gel pieces were digested with trypsin (6 ng/mL) (Promega, Madison, WI) overnight at 37°C. The resulting peptides were desalted using micro C18 ZipTips. The peptides were eluted from the ZipTips with a 1.8-mL elution solution of 50% acetonitrile (Aldrich, Milwaukee, WI) and 0.1% TFA (Sigma, St. Louis, MO) in water. Alpha-cyano-4-hydroxycinnamic acid (Sigma, St. Louis, MO) was used as matrix. One microliter of matrix (10 mg/mL) and 1 μL of eluted peptides were deposited on a MALDI plate for MALDI-MS analysis.
MALDI-TOF-MS Analysis
Mass spectra were recorded in positive reflectron mode of a MALDI-TOF mass spectrometer (Micromass, Manchester, UK). The time of flight was measured using the following parameters: 3400 V pulse voltage, 15,000 V source voltage, 500 V reflectron voltage, 1950 V MCP voltage, and low mass gate of 500 Da. Internal calibration was performed using auto digestion peaks of bovine trypsin (M + H+, m/z 842.5099 and m/z 2211.1045) in the same series as the samples to be measured.
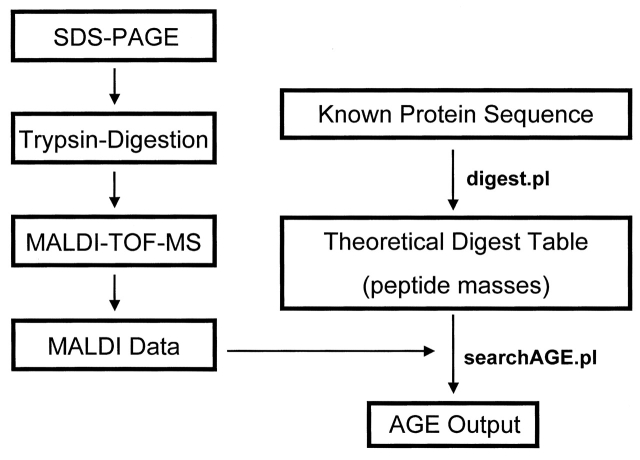
The measured peptide mass profiles were then analyzed using the PERL script algorithm. This algorithm matches any measured mass that corresponds to a theoretical peptide with an AGE modification and sends the result to an output file (Figure 2). The control and experimental spectra were then compared to elucidate the different AGE modifications.
FIGURE 2.
Experimental flow-chart using PERL script algorithm. The proteins are separated by SDS-PAGE and digested by trypsin. Mass spectral data are obtained by MALDI-TOF-MS. The digest.pl script algorithm takes the Known Protein Sequence and generates a list of theoretical peptide masses including modifications and single miscleavage that may occur in the system. The list of masses with corresponding peptide sequences is output as a Theoretical Digest Table. The searchAGE.pl script algorithm is simply a comparison tool for the Theoretical Digest Table and the experimental MALDI Data. All masses measured by MALDI that match to a theoretical mass from the Theoretical Digest Table within a tolerable error range are output as AGE Output. These results are presented in Table 2.
RESULTS AND DISCUSSION
The identification of the AGEs reported here relies on the relative mass changes (▵mass) that result from formation of the AGEs (Table 1). After MALDI-TOF-MS spectra were obtained, the peak list was generated as the text file and uploaded into the in-house-developed PERL script algorithm, and all ▵mass corresponding to the AGEs were used to search against the experimental peak list. Two restrictions were employed when search was performed: (1) only a single modification was allowed within any fragment; (2) the peptide must contain a missed cleavage site (lysine/arginine). Using these criteria, the m/z that corresponds to a specific AGE formation was then carefully examined in the spectra from both control and treated samples. This mass spectrometric method enables one to search the ▵m/z with high accuracy and speed.
TABLE 1.
Theoretical Delta Mass (▵M) for AGEs
| AGE | Delta mass (Da) |
| Pentosidine | +58.03 |
| Nɛ-(carboxymethyl)-lysine (CML) | +58.02 |
| Pyrraline | +108.02 |
| Imidazolone A | +144.03 |
| Imidazolone B | +142.03 |
| Schiff base | +162.02 |
| Amadori product (AP) | +162.02 |
| Crossline | +252.11 |
| 1-Alkyl-2-formyl-3,4-glycosyl-pyrrole (AFGP) | +270.07 |
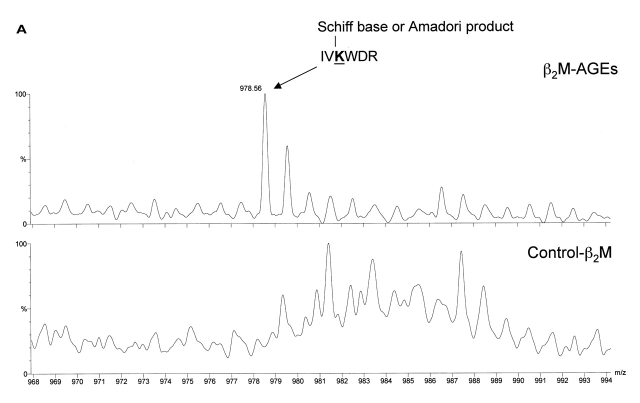
Figure 3A shows the presence of the m/z 978.5 in β2M-AGEs sample but its absence in normal β2M sample. The mass of 978.5 Da corresponds to the peptide IVKWDR (a.a. 112–117) plus a Schiff base or AP (+162) which is the precursor of AGEs. The lysine residue in this peptide becomes resistant to tryptic cleavage after AGE modification, resulting in a miscleaved peptide. However, MALDI-TOF-MS cannot distinguish between structural isomers such as Schiff base and AP. We also detected the m/z 2130.1 peak from the β2M-AGEs sample with a total ion current (TIC) of 260, but it was absent or nearly absent in the control β2M (TIC of 108). The tryptic β2M peptide DEYACRVNHVTLSQPK (a.a. 96–111) plus an AFGP modification (+270 Da) would result the m/z 2129.9, giving the measured mass of 2130.1, an error of 103 ppm. The PERL script searchAGE.pl process also revealed other AGEs such as crossline (m/z 2159.2) and pentosidine (m/z 2371.2) (see Table 2) at lower abundance.
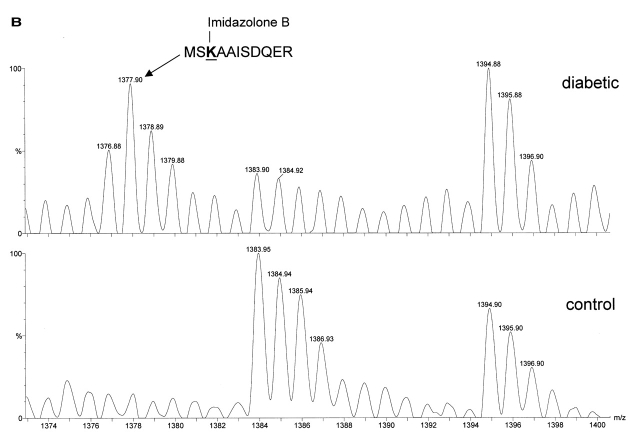
FIGURE 3A, B.
Comparison of MALDI-TOF mass spectra from tryptic digests of proteins. A: Human β2-microglobulin: Lys-114 in this peptide is modified with either a Schiff base or an Amadori product. B: Rat RyR2: Lys-3441 in this peptide is modified with imidazolone B. Peak m/z 1383.95 represents peptide (3365)DLYAFYPLLIR(3375.
TABLE 2.
AGE-modified Peptides Observed by MALDI-TOF-MSa
| Peptide observed (residue nos.) | Modification | Theoretical mass MH+ (m/z) | Measured mass MH+ (m/z) | ▵M (ppm) | Protein source |
| IVKWDR (112-117) | Schiff base or AP | 978.49 | 978.56 | −70 | Human β2M |
| TPKIQVYSR (24-32) | Schiff base or AP | 1253.64 | 1253.76 | −120 | Human β2M |
| DEYACRVNHVTLSQPK (96-111) | AFGP | 2129.98 | 2130.13 | −150 | Human β2M |
| IVKWDR (112-117) & TPKIQVYSR (24-32) | Crossline | 2159.19 | 2159.20 | −10 | Human β2M |
| IVKWDR (112-117) & IQVYSRHPAENGK (27-39) | Pentosidine | 2371.27 | 2371.27 | 0 | Human β2M |
| KAVWHK (3584-3589) | CML | 826.45 | 826.13 | 320 | Rat RyR2 |
| VDVSRISER (1607-1615) | Imidazolone B | 1202.60 | 1202.71 | −110 | Rat RyR2 |
| MSKAAISDQER (3439-3449) | Imidazolone B | 1377.60 | 1377.90 | −300 | Rat RyR2 |
| AALDFSDAREK (4677-4687) | Pyrraline | 1330.63 | 1330.82 | −190 | Rat RyR2 |
| ANACNSVIKQLMK (468-480) | AFGP | 1689.75 | 1689.81 | −60 | Rat SERCA2a |
| KEFTLEFSR (481-489) | AFGP | 1426.60 | 1426.72 | −120 | Rat SERCA2a |
| KSVQR (135-139) & VPADIRLTSIK (159-169) | Pentosidine | 1887.10 | 1886.86 | 240 | Rat SERCA2a |
| KSVQR (135-139) & AFTGREFDELSPSAQR (651-666) | Pentosidine | 2485.24 | 2485.14 | 100 | Rat SERCA2a |
aModified residues in bold and underline.
In RyR2 samples, the peptide MSKAAISDQER (a.a. 3439-3449) plus imidazolone B modification on lysine residue (Fig. 3b, m/z 1377.9) was detected in the sample prepared from the diabetic rat hearts. This mass was barely detected in RyR2 from the control animals. Other AGEs such as CML and pyrraline were also detected in RyR2 sample from the diabetic rat hearts (Table 2).
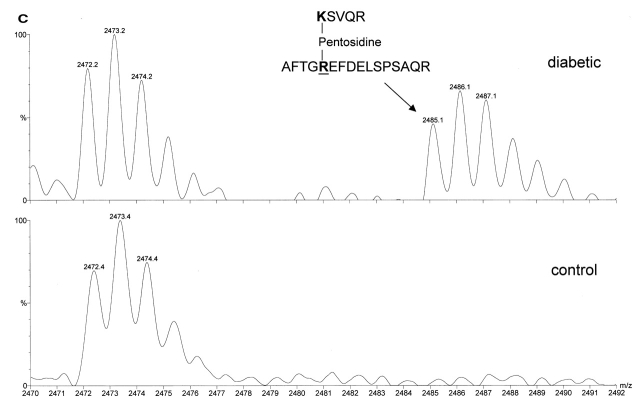
Similarly, several AGE-modified peptides were detected only in SERCA2a from the diabetic rat hearts. For example, KEFTLEFSR (a.a. 481–489) was modified with an AFGP (m/z 1426.7) on Lys-481 (Table 2). Interestingly, pentosidine, which requires intramolecular crosslinking, was detected with the m/z of 2485.1. The PERL script algorithm analysis revealed two involved peptides, KSVQR (a.a. 135-139) and VPADIRLTSIK (a.a. 159–169) (Figure 3C). Other modified peptides are shown in Table 2.
FIGURE 3C.
C: Rat SERCA2a: Lys-135 and Arg-655 are intramolecularly crosslinked with pentosidine.
Taken together, this study demonstrates a rapid MALDI-MS procedure for identity and location of AGEs on protein samples. The outstanding sensitivity of MALDI-TOF-MS makes this technique a very useful tool for detection of post-translational modifications such as AGEs, and could be used for the analysis of different forms of naturally occurring AGEs in many human diseases. This program can be downloaded from our web site (http://proteomics.biochemistry.iu.edu).
Acknowledgments
We would like to thank Indiana Genomics Initiative (INGEN) for funding the purchase of the reflectron MALDI-TOF mass spectrometer, Drs. N. X. Chen and S. M. Moe of Indiana University School of Medicine for providing β2-microglobulin samples. This work was supported in part by a grant from the National Institutes of Health (HL66898).
REFERENCES
- 1.Jadoul M, Ueda Y, Yasuda Y, et al. Influence of hemodialysis membrane type on pentosidine plasma level, a marker of “carbonyl stress.” Kidney Int 1999;55: 2487–2492. [DOI] [PubMed] [Google Scholar]
- 2.Pongor S, Ulrich PC, Bencsath FA, Cerami A. Aging of proteins: Isolation and identification of a fluorescent chromophore from the reaction of polypeptides with glucose. Proc Natl Acad Sci U SA 1984;81:2684–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Abed Y, Bucala R. Structure of a synthetic glucose derived advanced glycation end product that is immunologically cross-reactive with its naturally occurring counterparts. Bioconjug Chem 2000;11:39–45. [DOI] [PubMed] [Google Scholar]
- 4.Miyata T, Ueda Y, Shinzato T, et al. Accumulation of albumin-linked and free-form pentosidine in the circulation of uremic patients with end-stage renal failure: Renal implications in the pathophysiology of pentosidine. J Am Soc Nephrol 1996;7:1198–1206. [DOI] [PubMed] [Google Scholar]
- 6.Niwa T. Mass spectrometry in the search for uremic toxins. Mass Spectrom Rev 1997;16:307– 332. [DOI] [PubMed] [Google Scholar]
- 7.Vlassara H. Recent progress on the biologic and clinical significance of advanced glycosylation end products. J Lab Clin Med 1994;124:19–30. [PubMed] [Google Scholar]
- 8.Yim MB, Yim HS, Lee C, Kang SO, Chock PB. Protein glycation: Creation of catalytic sites for free radical generation. Ann N Y Acad Sci 2001;928:48–53. [PubMed] [Google Scholar]
- 9.Chen NX, O’Neill KD, Niwa T, Moe SM. Signal transduction of beta2m-induced expression of VCAM-1 and COX-2 in synovial fibroblasts. Kidney Int 2002;61:414–424. [DOI] [PubMed] [Google Scholar]
- 10.Jaradat MI, Schnizlein-Bick CT, Singh GK, Moe SM. Beta(2)-microglobulin increases the expression of vascular cell adhesion molecule on human synovial fibroblasts. Kidney Int 2001;59:1951–1959. [DOI] [PubMed] [Google Scholar]
- 11.Moe SM, Singh GK, Bailey AM. Beta-2-microglobulin induces MMP-1 but not TIMP-1 expression in human synovial fibroblasts. Kidney Int 2000;57:2023–2034. [DOI] [PubMed] [Google Scholar]
- 12.Bidasee KR, Dincer UD, Besch HR, Jr. Ryanodine receptor dysfunction in hearts of streptozotocin-induced diabetic rats. Mol Pharmacol 2001;60: 1356–1364. [DOI] [PubMed] [Google Scholar]
- 13.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–275. [PubMed] [Google Scholar]