Abstract
Immobilization of proteins and other biological macromolecules on solid supports is a method suitable for purification or screening applications in life science research. Prolinx, Inc. has developed a novel chemical affinity system that can be used for specific immobilization of proteins and other macromolecules via interaction of two small synthetic molecules, phenyldiboronic acid (PDBA) and salicylhydroxamic acid (SHA). This report describes immobilization applications of activated microporous membranes that have been functionalized with SHA derivatives. These SHA-membranes exhibit high capacity and specificity for binding of PDBA-labeled nucleic acids and proteins. Conjugation of active protein with PDBA is performed in solution independent of the immobilization step on SHA membranes. The resulting PDBA–protein conjugate is immobilized directly without purification and retains biological activity. PDBA conjugates may also be released from these SHA-affinity membranes in a controlled manner. Capture and release of PBA-modified oligonucleotides is also demonstrated. SHA-membranes can be used as surfaces for microarrays, and are therefore compatible with high-throughput analyses. These properties make them useful for development of numerous preparative or screening applications.
Keywords: microporous membranes, affinity purification, microarrays
Recent advances in technology have led to methods for generating large sets of genomic and proteomic samples. This has created a need for tools that facilitate rapid screening and analysis of these large sets of samples. Immobilization of proteins and nucleic acids on a solid phase is a useful way to perform separation of biomolecules, screen for enzymatic activity, or interrogate for binding events. Functionalized microporous membranes have been widely used in biological applications, including affinity purification of biomolecules,1,2 biocatalysis,3 and preparation of arrays.4,5 Advantages of microporous membranes include low diffusional resistance, high compressibility and chemical stabilty to harsh conditions of adsorption, elution, and regeneration.1 However, immobilization of macromolecules on a membrane surface with maximum retention of their activity and structural integrity still presents a challenge.6 Direct covalent attachment of protein molecules to a solid support results in a random orientation of the protein and, often, in structural deformation due to multisite immobilization.3 Proteins can also be bound to membranes through noncovalent interactions, such as streptavidin/biotin or recombinant affinity-tagged proteins, but the binding interactions may not be stable, resulting in leaching of protein off the surface.6–8 The streptavidin/biotin linkage is very stable, but requires the use of a large protein, limiting capacity and requiring special handling.7
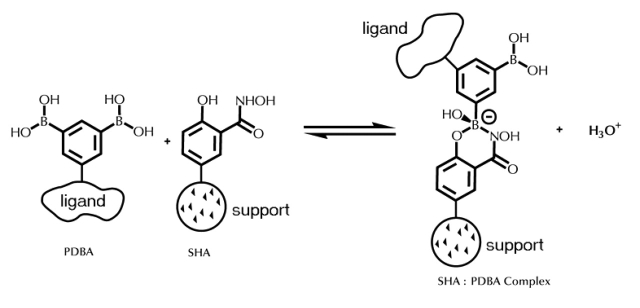
Prolinx, Inc. has developed a novel chemical affinity system for specific solid phase immobilization of proteins and other macromolecules. This chemical affinity system is based on the small molecule affinity pair of phenyldiboronic acid (PDBA) and salicylhydroxamic acid (SHA) (Figure 1).9,10 The use of phenylboronic acids in chromatography has been the subject of many reviews.11–13 Immobilized phenylborates are known to form covalent interactions with cis-diol–containing compounds. The complex between PDBA and SHA has been shown to be stable in aqueous solutions that vary widely with respect to pH, salt, and detergent of denaturant concentration, making it amenable for use in studies of biological macromolecules.9,14,15 Modification of ligand with PDBA can be performed in solution, independent of immobilization to solid phase, giving the PDBA-modifying reagent access to sites throughout the ligand and taking advantage of solution-phase kinetics.16 PDBA conjugates can be immobilized onto SHA-containing surfaces in an oriented manner through attachment of a phenylboronic acid (PBA) group to a defined residue on the surface of the protein. The input ratio of PDBA is optimized empirically to achieve high avidity to the SHA surface while retaining sufficient biological activity of the ligand.9,10 Previous studies have shown that purification of unincorporated PDBA-moieties is not required for efficient binding of PDBA conjugates,16,17 reducing the number of steps required to obtain immobilization. Finally, the formation of the complex between PDBA and SHA during the immobilization step is also compatible with a wide variety of reaction conditions and additives.14 In this study, we present the use of SHA-affinity membranes to achieve highly specific immobilization of proteins or nucleic acids on a membrane surface.
FIGURE 1.
Scheme representing the PDBA:SHA complex. The biological ligand is covalently bound to a phenylboronic acid, in this case PDBA. The SHA molecule is incorporated onto a solid support such as a cellulose membrane.
MATERIALS AND METHODS
SHA-Affinity Membranes
SHA-modified regenerated cellulose membranes (0.45 μm pore size) were supplied in sheets by Prolinx, Inc. (Bothell, WA).
Modification of Protein with PDBA
Alkaline phosphatase (AP, Calzyme, San Luis Obispo, CA) was modified using Versalinx Amine Modifying Reagent (PDBA-X-NHS) as described.10 The molar input ratios of Versalinx Amine Modifying Reagent to protein used were determined empirically to yield conjugated protein with good SHA-surface avidity. The molar input ratio of Versalinx Amine Modifying Reagent:protein was 10:1. PDBA-human immunoglobulin G (IgG) was prepared by modifying human IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), with a 15:1 input ratio of Versalinx Amine Modifying Reagent:IgG. Concentrations of protein were determined spectrophotometrically at λ = 280 nm using an HP 845X UV-Visible Chemstation (Hewlett-Packard, Palo Alto, CA).
Immobilization of PDBA-Modified Protein on SHA-Affinity Membranes Inserted into Microcentrifuge Spin Columns
Disks of SHA-membrane 5 mm in diameter were fitted into empty microspin columns (Lida 8500-PX, Nalge Nunc International, Rochester, NY). These membranes were washed with 100 μL phosphate buffered saline (PBS; 140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, pH adjusted to 7.4) by centrifugation in an Eppendorf model 5415C microcentrifuge (Eppendorf, Hamburg, Germany), at 3000 rpm (735 × g) for 1 min. One hundred microliter aliquots of various concentrations of PDBA-AP or unmodified AP in PBS were passed through the membrane by slow centrifugation at 1000 rpm (82 × g) for 5 min. Membranes were then washed as before with 100 μL PBS. AP activity in the flow-through was analyzed using p-nitrophenyl phosphate Sigma-Fast tablets (Sigma-Aldrich, St Louis, MO), one tablet (15 mg) per 15 mL of DEA buffer solution (1 M diethanolamine, 1 mM MgCl2, 0.1 mM ZnSO4, pH 10.2). Absorbance was monitored at λ = 405 nm in a ELX-808 plate reader (Bio-Tek, Winooski, VT). Protein concentrations in the flowthrough were determined by preparing standard curves of AP at 0 μg/mL, 10 μg/mL, 20 μg/mL, 30 μg/mL, and 40 μg/mL, and measuring activity as described above. All experiments were performed in duplicate.
Release of PDBA-Modified Protein Immobilized on SHA-Affinity Membranes
Versalinx Protein Releasing Reagent (100 μL, Prolinx, Bothell, WA) was added to spin columns with immobilized PDBA-AP and AP, and the columns were incubated at 37°C for 30 min. The columns were centrifuged at 1000 rpm (82 × g) for 5 min, and then the membranes were washed as above with 100 μL of PBS. AP activity and protein concentrations for the combined flow-through fractions were determined as described above. Experiments were performed in duplicate.
Immobilization of PDBA-Modified Protein via Dot Blotting
Sheets of SHA-membranes were fitted onto a DHM-96 dot-blotter apparatus (Topac, Hingham, MA). Each well was loaded with 10 μL of PDBA-AP or unmodified AP at various concentrations in PBS. Protein was incubated on membrane for 0 or 40 min, as indicated, at room temperature. Vacuum was applied and wells were washed twice with 100 μL PBS. Protein immobilized on the membranes was visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) using Sigma Fast BCIP/NBT tablets (Sigma-Aldrich, St Louis, MO), one tablet per 10 mL of distilled water, producing a solution of 0.15 mg/mL BCIP, 0.30 mg/mL NBT, 100 mM Tris, 5 mM MgCl2, pH 9.5. Reactions were quenched with methanol after color development to prevent discoloration.
Immobilization of PBA-Modified Oligonucleotides on SHA-Affinity Membranes
PBA-modified oligonucleotides were synthesized at Prolinx and contain the M13 forward primer sequence (5′-AGCGGATAACAATTTCACACAGGA-3′), modified at the 5′ end with four PBA moieties using four additional cycles with PBA phosphoramidite (Prolinx). Unmodified oligonucleotide of the same sequence (M13/pUC -48 reverse sequencing primer) was obtained from New England Biolabs (Beverly, MA). Oligonucleotides were end-labeled with 32P using the 3′-End-Labeling Kit (Roche Applied Science, Indianapolis, IN) and α32P-cordycepin 5-triphosphate (Perkin-Elmer Life Sciences, Boston, MA). Disks of SHA-affinity membranes 5 mm in diameter were produced using a hole punch. Three layers of disks were laid flat in the bottom of a microcentrifuge spin column containing a 0.2 μm cellulose acetate frit (PGC Scientific, Frederick, MD). Nonradiolabeled oligonucleotides were resuspended to a final concentration of 1000 nM and spiked with 0.3% (w/w) of the corresponding radiolabeled oligonucleotide in 30 μL 10X SSC (1.5 M NaCl, 150 mM sodium citrate, pH 7) and incubated with membranes for 15 min at room temperature. After binding, membranes were washed once with 1 volume of 1X SSC then once with 1 volume of 0.1X SSC. Bound oligonucleotides were released by adding 60 μL 1 mM Versalinx Nucleic Acid Releasing Reagent (Prolinx) in distilled, deionized water and incubating at 37°C for 10 min. The quantities of oligonucleotides bound to membranes and released into the flow-through were determined. The amount of radioactivity in each fraction was determined by addition of 2 mL Opti-Fluor scintillation cocktail (PerkinElmer, Life Sciences, Wellesley, MA). Counting was performed in a Beckman LS-6500 Multi-Purpose Scintillation Counter (Beckman-Coulter, Fullerton, CA). Experiments were performed in duplicate. Percentages of bound and released oligonucleotides relative to input quantity of radiolabeled were calculated.
Preparation of Protein Microarrays on SHA-Affinity Membranes
Small pieces (approximately 5 × 10 mm) of membrane were affixed to the surface of 1 × 3-inch glass microscope slides using Jacquard Gutta Resist Clear polymer (Rupert, Gibbon and Spider, Healdsburg, CA) to glue the outer edges of the membrane to the glass surface. PDBA-human IgG or unmodified human IgG (50 μg/mL) were spotted in Versalinx Spotting Solution (Prolinx) in a 4 × 30 microarray format, 300 pL/spot, 500 μm pitch, using a Packard BioChip Arrayer (PerkinElmer Life Sciences) according to manufacturer’s specifications. The array was incubated at room temperature for 1 h, washed briefly with Versalinx Wash Buffer #1 (Prolinx), and then developed with 0.1 μg/mL Cy3-goat anti-human IgG (Jackson ImmunoResearch Laboratories) in Versalinx Wash Buffer #2 (Prolinx, Inc.) for 30 min at room temperature. The array was washed for 5 min in PBS; dried briefly in air; and visualized on a ScanArray Lite scanner (Packard BioScience, Meriden, CT) using a laser power of 75%, PMT gain of 65%, and spatial resolution of 10 μm.
RESULTS AND DISCUSSION
Retention of PDBA Protein on SHA-Affinity Membranes
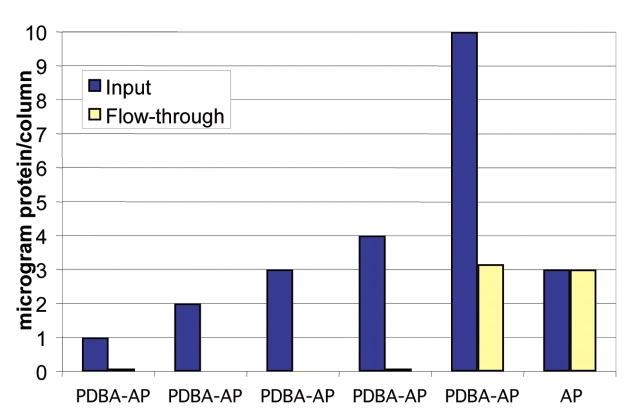
SHA-affinity membranes efficiently remove PDBA-AP from solution; 5-mm disks (0.196 cm2) of membrane capture up to 6 μg of PDBA-AP during a 5-min centrifugation (Figure 2). The capture is specific to PDBA-modified protein: When unmodified AP was centrifuged through SHA-affinity membranes, all of this protein was recovered in the flow-through, indicating that there was essentially no nonspecific binding (Figure 2). Based on these results, the capacity of these membranes is calculated to be at least 30 μg/cm2 for AP, a dimer of 140,000 Da. This is comparable to the capacity of nonaffinity membranes with active aldehyde groups for nonspecific interactions with immunoglobulin (approximately 150,000 Da), reported to be up to 30 μg/cm2 (Sartorius AG, Goettingen, Germany). The high capacity of SHA affinity membranes suggests that the SHA groups are readily accessible for interacting with PDBA. Moreover, the immobilization of PDBA-modified protein attains this capacity after a 5-min centrifugation, while maximal capacity from the nonspecific aldehyde–immunoglobulin interaction requires 1 h of incubation, so SHA affinity membranes enable rapid and efficient binding of specific conjugates.
FIGURE 2.
Immobilization of PDBA-modified protein on SHA-affinity membranes inserted into microcentrifuge spin columns. Disks of SHA-membranes, 5 mm in diameter, were overlayed with 100 μL aliquots of PDBA-AP or unmodified AP in PBS. The protein solutions were passed through the membranes by slow centrifugation. AP activity in the flow-through was analyzed using pNPP (para-nitrophenyl phosphate) and monitoring absorbance at l = 405 nm. Protein concentrations were determined by preparing standard curves of AP at 0 μg/mL, 10 μg/mL, 20 μg/mL, 30 μg/mL, and 40 μg/mL.
The PDBA:SHA complex is stable to conditions that usually disrupt antibody/antigen interactions, enabling the use of PDBA:SHA chemistry for affinity purification using antibodies,10,18 such as purification of His-tagged proteins using PDBA-modified a-His Tag antibodies.19 We have previously shown that immobilization using PDBA:SHA affords higher capacity and higher specific activity that some commercially available affinity chromatography methods, such as biotin-streptavidin.10,16
Release of PDBA-Protein from SHA-Affinity Membranes
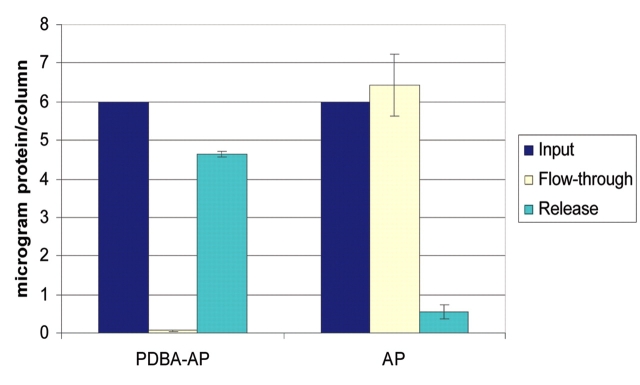
Release of PDBA-modified proteins from SHA-affinity membranes is achieved under mild conditions using a competitive releasing reagent. SHA-affinity membranes with immobilized PDBA-AP were incubated with Versalinx Protein Releasing Reagent at 37°C for 30 min. Greater than 75% of the immobilized PDBA-AP was released (Figure 3). SHA-affinity membranes that had been presented with unmodified AP were also incubated with the releasing reagent. These membranes showed essentially no activity representing released AP, as would be expected because of low nonspecific binding on these surfaces. The Versalinx Protein Releasing Reagent is composed of a proprietary mixture of inert small molecules that are not expected to interfere with downstream applications of released protein. If desired, residual releasing reagent can be removed from the recovered protein by dialysis or size exclusion. This releasing reagent facilitates disruption of the highly stable PDBA:SHA complex without the use of harsh conditions, enabling recovery of a significant amount of immobilized PDBA modified ligand (or ligand–analyte complex) away from the SHA-affinity membrane. Such an application is useful if the modified ligand is available in limiting amounts or if the intention is to maintain the intact ligand:analyte complex.
FIGURE 3.
Release of PDBA-modified protein immobilized on SHA-affinity membranes. Versalinx Protein Releasing Reagent (100 μL) was added to spin columns containing immobilized PDBA-AP, and AP and the columns were incubated at 37°C for 30 min. The releasing reagent was passed through the membranes by slow centrifugation. AP activity in the flow-through was analyzed using pNPP and monitoring absorbance at l = 405 nm. Protein concentrations were determined by preparing standard curves of AP at 0 μg/mL, 10 μg/mL, 20 μg/mL, 30 μg/mL and 40 μg/mL.
Biological Activity of PDBA-Protein Immobilized on SHA-Affinity Membranes
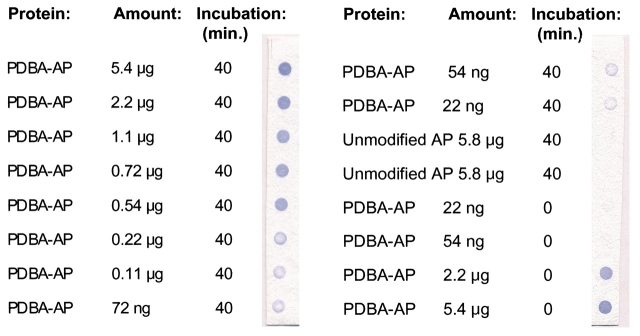
Activity of PDBA-AP immobilized on SHA-affinity membranes is shown in Figure 4. Immobilized proteins were presented with the AP substrate BCIP in the presence of NBT. This substrate produces a purple color when dephosphorylated by AP. Enzyme activity was detected from as little as 22 ng of input PDBA-AP. Immobilization of PDBA-AP via dot-blotting is rapid, as demonstrated by samples that were developed immediately, with no incubation time (Figure 4). Time–course studies showed that immobilization via dot-blotting is largely complete in less than 10 min (data not shown). For areas of the SHA-affinity membrane to which unmodified AP had been presented, no BCIP/NBT activity was detected, indicating that unmodified AP did not immobilize on the membrane. These results also show that the SHA-affinity membranes do not themselves react with BCIP/NBT substrate.
FIGURE 4.
Immobilization of PDBA-modified protein via dot blotting. SHA-membranes were fitted onto a dot-blotter apparatus and each well was loaded with 10 μL of PDBA-AP or unmodified AP at concentrations indicated. Protein solutions were incubated on membranes for 0 or 40 min, as indicated, at room temperature. Vacuum was applied and wells were washed twice with 100 μL PBS. Membranes were visualized using BCIP/NBT.
The retention of AP biological activity is consistent with earlier studies showing that proteins modified with PDBA retain a significant portion of their biological activity.9,10 The molar input ratio of PDBA modifying reagent to protein can be optimized to yield conjugated protein with good SHA-surface avidity. Such conjugated proteins retain more than 80% enzymatic activity.9,18 This suggests that PDBA-modification introduces only minimal structural changes into the protein molecule, and that such proteins can be used for solid phase enzymatic assays or for affinity capture of analytes.
Specific Capture and Release of Oligonucleotides by SHA-Affinity Membranes
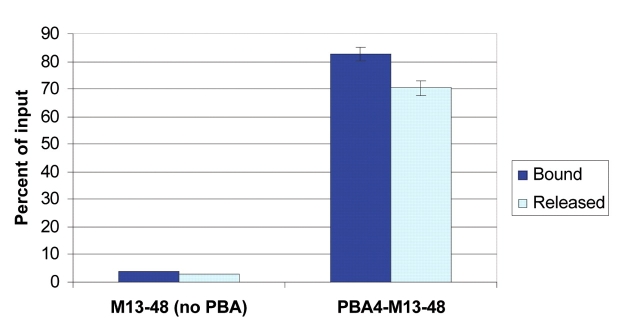
Radiolabeled PBA-modified oligonucleotides were introduced to SHA-affinity membranes and incubated for 15 min. Bound and unbound oligonucleotides were assayed by scintillation (Figure 5). The membranes bound greater than 80% of the 30-pmol input PBA-modified oligonucleotides. By contrast, less than 10% of the same quantity of unmodified oligonucleotides (30 pmol) were retained by the membranes, indicating that nonspecific binding is very low. Release of the specifically bound oligonucleotides was accomplished using Versalinx Nucleic Acid Releasing Reagent. Approximately 70% of captured PBA-modified oligonucleotides were released by this method (Figure 5). These results suggest that these membranes can be used for nucleic acid-based applications.
FIGURE 5.
Immobilization of PBA-modified oligonucleotides on SHA-affinity membranes. Radiolabeled PBA-modified or -unmodified oligonucleotides (1000 nM in 30 μL 10× SSC) were bound to SHA-affinity membranes for 15 min at room temperature. After binding, membranes were washed, and bound oligonucleotides were released by adding 60 μL 1 mM Versalinx Nucleic Acid Releasing Reagent in distilled water and incubating at 37°C for 10 min. Bound and released oligonucleotides were determined by scintillation counting of membrane, flow-through, and input fractions, and are expressed as a percentage of input fractions.
SHA-Affinity Membranes Can Be Used as Microarray Substrates
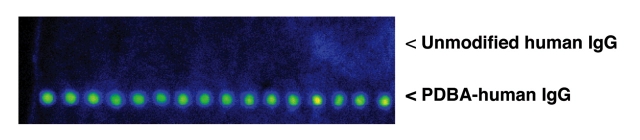
SHA-affinity membranes were used as substrates for protein microarrays (Figure 6). PDBA-modified human IgG protein was immobilized to the membrane using an arrayer according to manufacturer’s specifications. Unmodified human IgG was spotted on the same membrane. Detection of immobilized protein was performed using Cy3-labeled goat anti-human IgG. The spots corresponding to PDBA-modified protein showed bright fluorescence, while the spots corresponding to unmodified protein did not fluoresce. The detection of PDBA-modified protein confirms that the protein was in fact immobilized on the membrane and had retained its ability to be recognized by a specific antibody. We have previously reported the preparation of protein microarrays on glass slides using PDBA:SHA chemistry.17 This method is more rapid than traditional protein immobilization techniques and results in good reproducibility of binding properties across the array.17 SHA-modified membranes provide the researcher with another convenient tool for immobilization of PDBA conjugates with retention of biological activity.
FIGURE 6.
SHA-affinity membranes as microarray surfaces. PDBA-human IgG or unmodified human IgG (50 μg/mL) were spotted onto SHA-affinity membranes in a 4 × 30 microarray format using standard arrayer protocol. The membrane was incubated at room temperature for 1 h, washed with Versalinx Wash Buffer #1, and then developed with 0.1 μg/mL Cy3-goat anti-human IgG for 30 min at room temperature. The array was visualized on a ScanArray Lite at a laser power of 75%, PMT gain of 65%, and spatial resolution of 10 μm.
SHA-functionalized membranes represent a new platform for the specific solid phase immobilization of proteins and other macromolecules using PDBA complex formation. Specific binding of macromolecules to SHA-affinity membranes is rapid, efficient, and convenient. These membranes can be used for development of affinity purification systems, protein microarrays, and other applications to facilitate rapid screening and analysis of large sets of samples.
This research was funded through private investment, not through grants, and the authors have no conflicts of interest with the work.
REFERENCES
- 1.Charcosset C. Purification of proteins by membrane chromatography. J Chem Technol Biotechnol 1998; 71:95–110. [Google Scholar]
- 2.Klein E. Affinity membranes: A 10-year review. J Membr Sci 2000;179:1–27. [Google Scholar]
- 3.Butterfield DA, Bhattacharyya D, Daunert S, et al. Catalytic biofunctional membranes containing site-specifically immobilized enzyme arrays: A review. J Membrane Sci 2001;181: 29–37 [Google Scholar]
- 4.Kramer A, Schuster A, Reineke U, et al. Combinatorial cellulose-bound peptide libraries: Screening tools for the identification of peptides that bind ligands with predefined specificity. Methods: Companion Methods Enzymol 1994;6:388–395. [Google Scholar]
- 5.Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol 2001;12:59–64. [DOI] [PubMed] [Google Scholar]
- 6.Rao SV, Anderson KW, Bachas LG. Oriented immobilization of proteins. Mikrochim Acta 1998;128:127–143. [Google Scholar]
- 7.Hodneland CD, Lee Y-S, Min D-H, et al. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proc Natl Acad Sci USA 2001; 99:5048–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes KA, Booth LR, Kaiser R J, et al. Optimization of a protein microarray platform based on a small-molecule chemical affinity system. In: Kambhampati D (ed): Protein Microarray Technology. New York: Wiley. 2003:in press.
- 9.Stolowitz ML, Ahlem C, Hughes KA, et al. Phenylboronic acid-salicylhydroxamic acid bioconjugates 1. A novel boronic acid complex for protein immobilization. Bioconjug Chem 2001;12:229–239. [DOI] [PubMed] [Google Scholar]
- 10.Wiley JP, Hughes KA, Kaiser RJ, et al. Phenylboronic acid-salicylhydroxamic acid bioconjugates 2. Polyvalent immobilization of protein ligands for affinity chromatography. Bioconjug Chem 2001;12:240–250. [DOI] [PubMed] [Google Scholar]
- 11.Bergold A, Scouten WH. Borate chromatography. In: Scouten WH (ed): Solid Phase Chromatography. New York: Wiley. 1983:149–187.
- 12.Mazzeo JR, Krull IS. Immobilized boronates for the isolation and separation of bioanalytes. Biochromatography 1989;4:124–130. [Google Scholar]
- 13.Singhal RP, DeSilva SSM. Boronate affinity chromatography. Adv Chromatogr 1992;31:293–335. [PubMed] [Google Scholar]
- 14.Hughes KA, Wiley JP. Use of small-molecule affinity-based matrices for rapid protein purification. In: Vaillancourt PE (ed): E. Coli Gene Expression Protocols; Methods in Molecular Biology. Totowa, NJ: Humana Press. 2002; 205: 215–223. [DOI] [PubMed] [Google Scholar]
- 15.Sienkiewicz PA, Roberts DC. Chemical affinity systems-I. PH dependence of boronic acid-diol affinity in aqueous solution. J Inorg Nucl Chem 1980;42:1559–1575 [Google Scholar]
- 16.Hughes KA, Lucas DD, Stolowitz ML, et al. Novel affinity tools for protein immobilization: Implications for proteomics. Am Biotech Lab 2001;1:36–38. [Google Scholar]
- 17.Booth LR, Clary ST, Gall AS, et al. Fabrication of peptide microarrays utilizing small-molecule affinity technology. Am Biotech Lab 2002;20:76–78 [Google Scholar]
- 18.Wiley JP, Booth LR, Hughes KA, et al. A rapid and efficient chemical affinity system for protein capture and release. Am Genomic/Proteomic Tech 2002;2 :21–25. [Google Scholar]
- 19.Bergseid M, Baytan AR, Wiley JP, et al. Use of a small-molecule based chemical affinity system for the purification of proteins. Biotechniques 2000;29:1126–1133. [DOI] [PubMed] [Google Scholar]