Abstract
The observation of peaks corresponding to both disulfide-bonded and reduced peptides in matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra of disulfides could suggest that the samples are either mixtures prior to analysis or that the measurement process has converted single compounds into mixtures. This is an important distinction when characterizing potentially disulfide-bonded peptides obtained from proteolyzed proteins or from oxidized synthetic peptides. It is well documented that disulfides can undergo in-source decay (ISD) when using a 337-nm laser. However, the mixed matrix 2-(4-hydroxyphenylazo)benzoic acid:α-cyano-4-hydroxycinnamic acid (1:10) not only suppresses the ISD reduction of disulfides to thiols but allows the same low threshold laser power generally used with α-cyano-4-hydroxycinnamic acid to be applied.
Keywords: , Disulfides, peptides, MALDI-TOF-MS, ISD, mixed matrices
The analysis of potentially disulfide-bonded peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) can be obscured by the experimental procedure. It is well documented that MALDI-TOF mass spectra often show the expected peaks corresponding to disulfides accompanied by their reduction products. Patterson and Katta1 and Crimmins et al.2 have offered convincing evidence that disulfides undergo in-source decay (ISD) during the MALDI experiment (337-nm laser) and that increasing the laser power increases the signal intensity for thiols while decreasing the intensity for the disulfide. However, the observation of peaks corresponding to both disulfide-bonded and reduced peptides in MALDI-TOF-MS could suggest that the samples are either mixtures prior to MALDI analysis or that the measurement process has converted single compounds into mixtures. This is an important distinction when characterizing potentially disulfide-bonded peptides obtained from proteolyzed proteins or from oxidized synthetic peptides.
While establishing the disulfide bond assignments in human thrombospondin type 1 domains,3 we searched for a way to use MALDI-TOF-MS to show that our isolated disulfides were not mixtures. Other laboratories have developed methods to characterize disulfide-bond–containing peptides: Jones et al.4 used post-source decay (PSD) to follow disulfide fragmentation; Katta et al.5 showed that ISD could be used for direct sequence analysis of disulfide-containing peptides; Mhatre et al.6 used on-plate reduction of protease digest mixtures to identify disulfide-containing peptides; and Schnaible et al.7 are automating a process that uses an algorithm to look for both disulfides and thiols in ISD spectra followed by fragmentation of the suspected disulfide.
While other laboratories have exploited the reactivity of disulfide bonds for analysis by ISD and PSD MS, our experiments involved changing the matrix and using mixed matrices. This report describes the conditions we found that control the redox state of sulfurcontaining peptides during the MALDI experiment.
MATERIALS AND METHODS
The matrices α-cyano-4-hydroxycinnamic acid (CHCA), 2-(4-hydroxyphenylazo)benzoic acid (HABA), and 2,5-dihydroxybenzoic acid (DHB) were purchased from Sigma (St. Louis, MO). Peptides A and B were prepared from the third type 1 repeat of human thrombospondin-1 that had been expressed in the baculovirus system, purified, and shown to be fully disulfide bonded.3 The peptides were generated by thermolysin digestion at pH 6.5 and separated by high-performance liquid chromatography (HPLC) using a Vydac C18 column (Separations Group, Hesperia, CA). Peptide A contains one disulfide bond and peptide B contains two. MALDI targets were prepared by spotting 0.5 μL of HPLC fraction followed by 0.5 μL of matrix solution. Initially, three matrix solutions were used: CHCA (10 mg/mL in 70% acetonitrile/30% water), HABA (10 mg/mL in 70% acetonitrile/30% water); and DHB (20 mg/mL in 50% ethanol/50% water). While optimizing disulfide-bond analysis, two matrix mixtures were prepared: (1) 1 part HABA, 1 part CHCA; and (2) 1 part HABA, 10 parts CHCA.
Mass spectra were obtained using a Bruker REFLEX II (Billirica, MA) equipped with delayed extraction, a reflectron, and a 337-nm nitrogen laser. Unless noted, spectra were obtained using the reflectron mode and external calibration and are the average of at least 50 shots. Calibrants used included adrenocorticotropic hormone fragment [18–39], angiotensin II, insulin, and matrix. Peptides A and B were reduced on-target with dithiothreitol* as follows: A 0.5-μL aliquot of peptide, which had previously been dried and resuspended in water, was incubated with 0.5 μL of dithiothreitol (20 mM in 8 mM NH3/H2O) for approximately 5 min. The reaction was quenched by the addition of 0.5 μL of matrix solution. Dried spots containing either CHCA or HABA were washed when necessary to remove small water-soluble molecules by placing a drop on target with a pipet and then removing it with a pipet or tissue.
RESULTS AND DISCUSSION
In the MALDI experiment, the disulfide is first mixed with a matrix and then dried. Once in a vacuum, the spot is hit with energy in the form of 337-nm light. Usually the matrix is present in great excess (>1000), and it readily absorbs 337-nm light energy. Two locations where the matrix and analyte are in close contact so that reactions can occur are in the preparation of the spot before the laser fires and in the vacuum just after the laser fires and before the plume disperses. Figure 1 contains the structures of the peptides and matrices used in this report. Figure 2 contains the mass spectra of peptide A in three standard matrices. Peptide A contains one disulfide bond. With CHCA and DHB, the spectra show peaks where the two thiols resulting from reduction of the disulfide bond would be expected. Although the peaks are not large relative to the disulfide peak, they are present. However, with HABA and the same peptide solution used to obtain the CHCA and DHB spectra, only a peak for the disulfide was observed. Why did HABA produce a clean spectrum?
FIGURE 1.
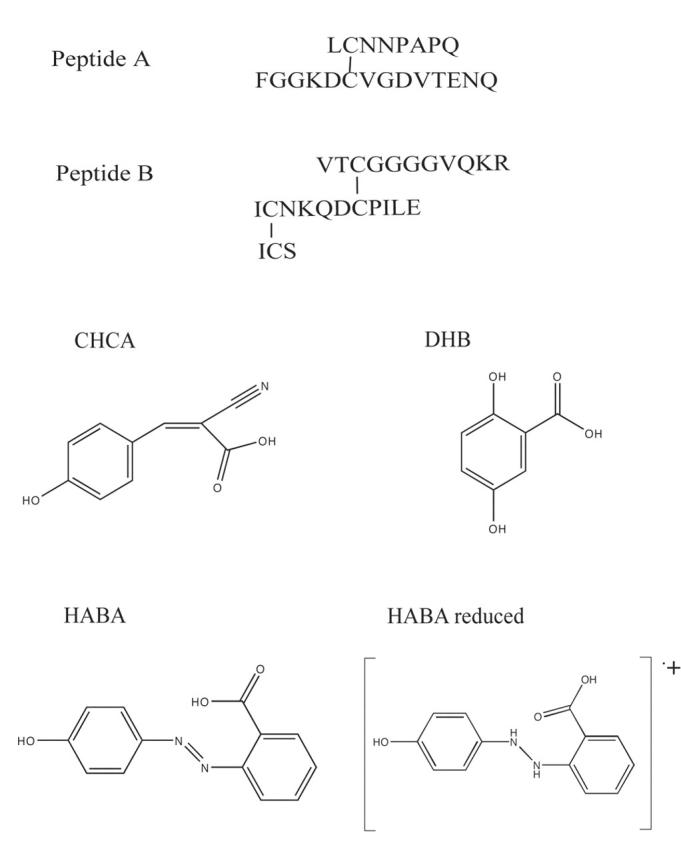
Structures of the peptides and matrices used in this study.
FIGURE 2.
The MALDI-TOF mass spectra of peptide A obtained with (a) CHCA, (b) DHB, and (c) HABA.
When aliquots of peptide B were subjected to on-target reduction with dithiothreitol8,9 mixed with the same three matrices, and subjected to MALDI-TOF-MS, complete reduction of the disulfide bonds was observed. The results are shown in Figure 3. Both CHCA and DHB clearly produced the expected thiols. While peaks corresponding to thiols could be observed using HABA, the spectra were weaker and generally had a more complicated background. Because a lower laser power can be used with CHCA relative to DHB and HABA, the matrix of choice for thiols is CHCA.
FIGURE 3.
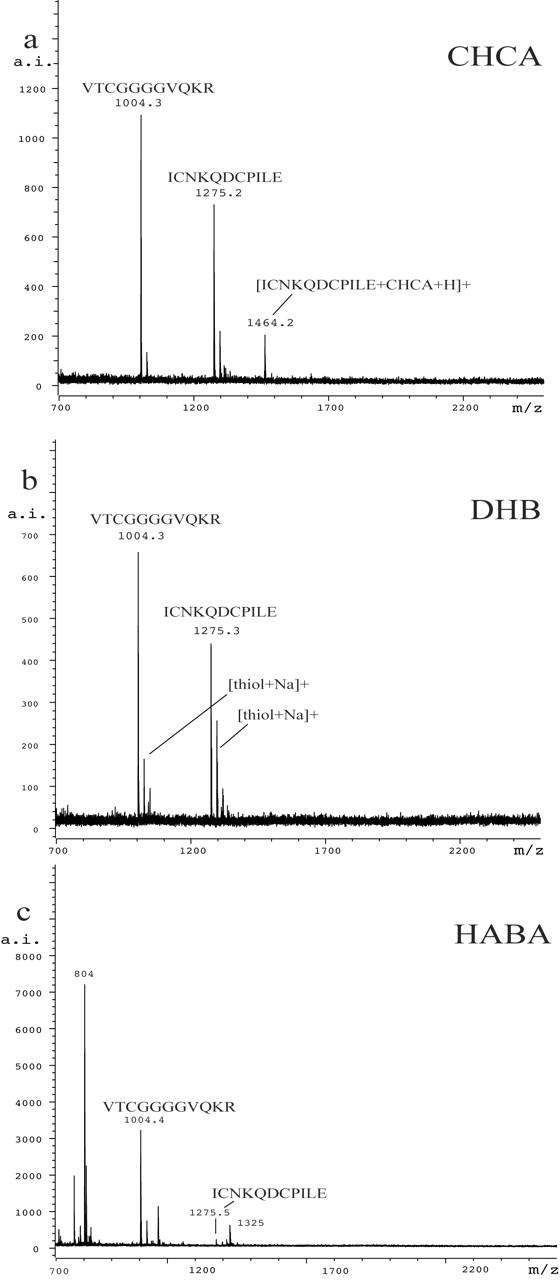
The MALDI-TOF mass spectra of peptide B obtained after on-target reduction was quenched with (a) CHCA, (b) DHB, and (c) HABA.
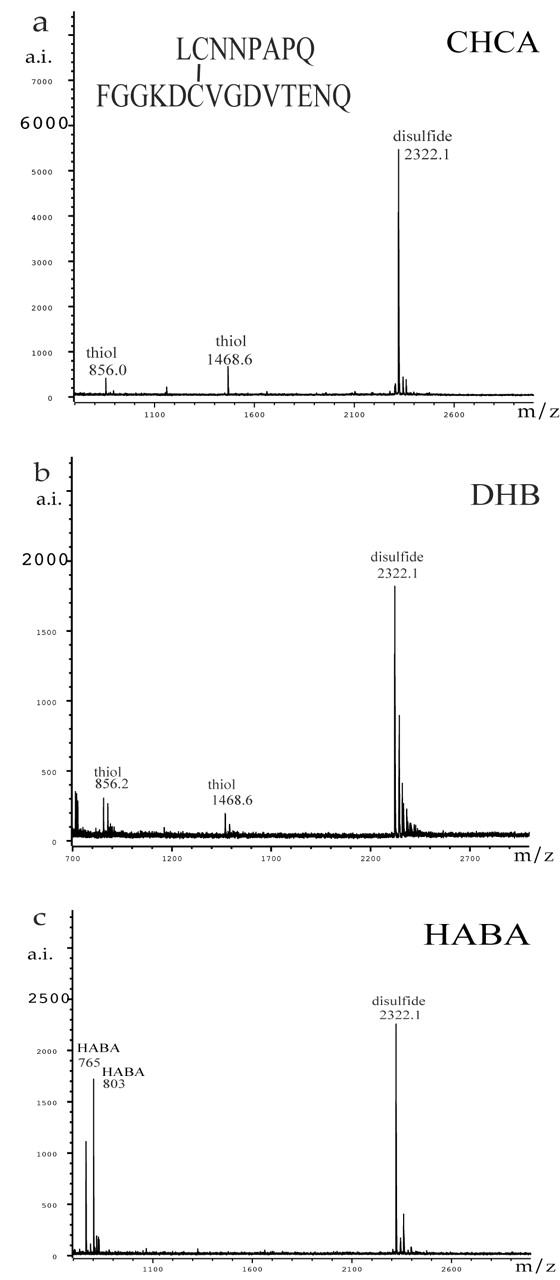
In order to understand and verify the protective effect of HABA for disulfides, we mixed HABA with other matrices. Figure 4 contains three of the best results for peptide A. In Figure 4a, an aliquot of peptide A was put on the target, immediately followed by an aliquot of HABA and then an aliquot of CHCA. Unlike our general practice of pulling the mixture into the pipet for a final mix, no extra mixing was needed. In Figure 4b, one part HABA was mixed with 10 parts CHCA before being mixed with the peptide solution. In Figure 4c, one part HABA and one part CHCA were mixed before being mixed with the peptide solution. No peaks that could be assigned to disulfide were present in any of the spectra shown in Figure 4. The best signal-to-noise ratio for peptide A was observed with the 1:10 HABA:CHCA mix. Figure 5 displays mass spectra obtained for peptide B. The data collected using just CHCA (Fig. 5a) show strong evidence of decomposition of the disulfide bonds, whereas the data collected using HABA (Fig. 5b) and 1:10 HABA:CHCA (Fig. 5c) show only peptide B.
FIGURE 4.
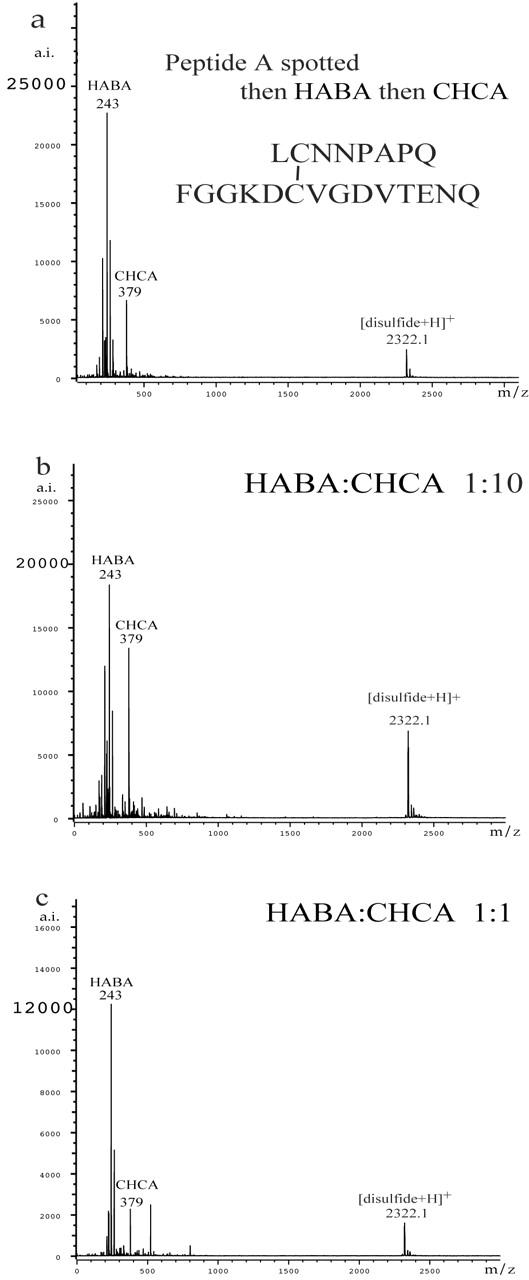
The MALDI-TOF mass spectra of peptide A obtained with (a) HABA then CHCA before drying, (b) 1:10 HABA:CHCA, and (c) 1:1 HABA:CHCA. No peaks assignable to thiols were observed.
FIGURE 5.
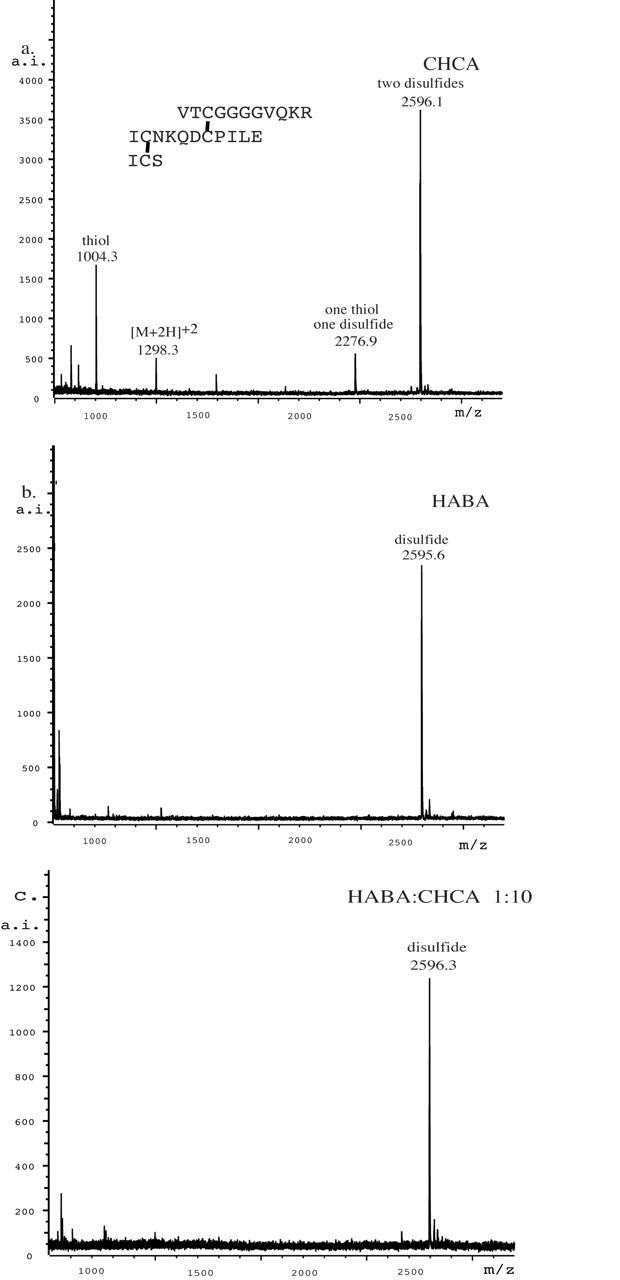
The MALDI-TOF mass spectra of peptide B obtained with (a) CHCA, (b) HABA, and (c) 1:10 HABA:CHCA.
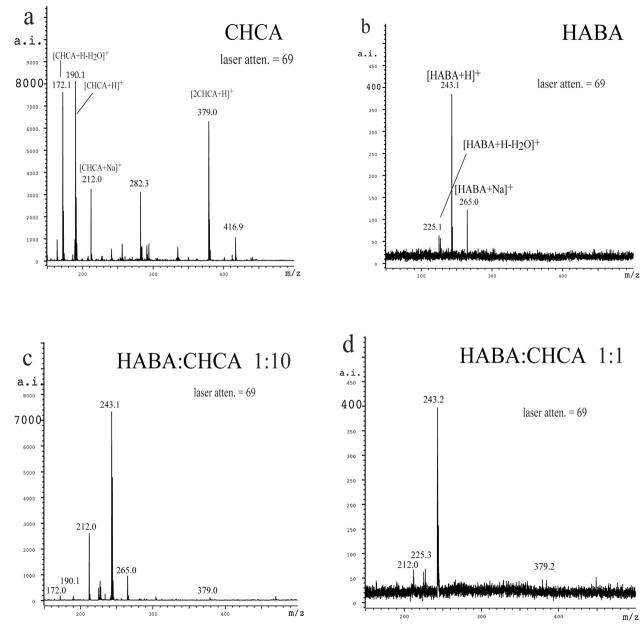
Several notable points were revealed upon examination of the matrix regions for CHCA, HABA, the 1:10 HABA:CHCA mixture, and the 1:1 HABA:CHCA mixture, obtained at the same laser attenuation and number of laser pulses (Fig. 6). First, whenever HABA is present in combination with CHCA, the signals for HABA ions dominate. Second, lower signal intensity is obtained for HABA relative to CHCA. To achieve the same signal intensity for HABA, lower laser attenuation (higher power) is required (data not shown). With the 1:10 and 1:1 HABA:CHCA mixtures, HABA ions dominate. Interestingly, with the 1:10 mix the HABA ions appear at the same laser attenuation used for CHCA alone. So while CHCA ions do not dominate, the presence of CHCA allows the MALDI experiment to be run at same high-laser attenuation (low power) settings used for CHCA. This advantage is not evident in the 1:1 mixture, in which the HABA ions still dominate but the laser power advantage of CHCA has disappeared.
FIGURE 6.
A comparison of the matrix regions of (a) CHCA, (b) 1:10 HABA:CHCA, (c) 1:1 HABA:CHCA, and (d) HABA.
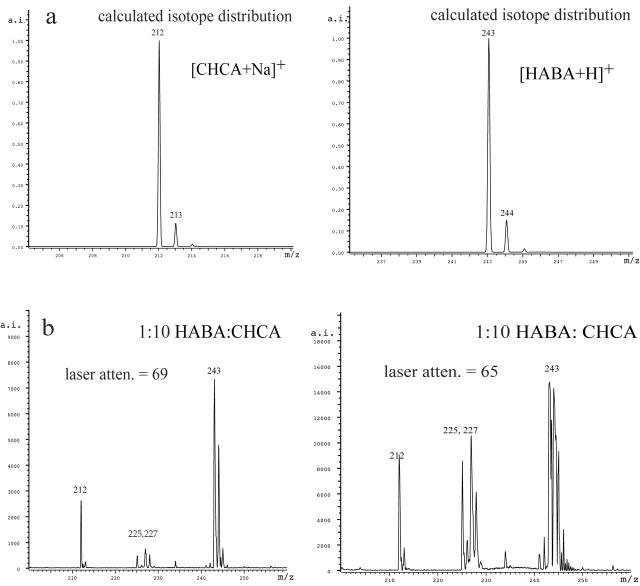
Looking carefully at Figure 6, it can be seen that, in the presence of CHCA, the isotope pattern for the [HABA+H]+ ion does not match the predicted pattern. The intensity of the peak at m/z 244 is too high relative to m/z 243, indicating that more than one species is responsible for the pattern. The elemental formula for [HABA+H]+ is [C13H11N2O3]+ and has a predicted monoisotopic mass of 243. Using an isotope pattern calculator—such as IsoPro 3.1,10 the University of Sheffield’s ChemPuter website,11 or one that is part of a mass spectrometer’s software—the intensity for m/z 244 should be 15% of that for m/z 243 (Fig. 7). In the 1:10 mix and the 1:1 mix, the relative intensity is much greater than predicted (Fig. 7). If the observed excess intensity at m/z 244 corresponds to reduced HABA (radical cation with m/z 244), then the protective effect HABA has for disulfides can be explained by the reduction of HABA in the MALDI experiment prior to the disulfides. Looking at the isotopic clusters assignable to [CHCA+H]+ at m/z 190, and the cluster assignable to [CHCA+Na]+ at m/z 212, the patterns in the 1:10 mix are near normal (Fig. 7). However, when only CHCA is present, the amount of m/z 191 is too large, again indicating that more than one species is producing the cluster of peaks (data not shown). Increasing the laser power (lowering the attenuation) just further distorts the expected isotope pattern for m/z 243 (Fig. 7d). In mixtures of HABA and CHCA, HABA appears to be protecting CHCA from reduction by reacting first.
FIGURE 7.
a: The calculated isotope patterns for [CHCA+Na]+ and [HABA+H]+. b):The observed matrix region for 1:10 HABA:CHCA at two laser attenuations.
The observation of reduction in mass spectrometric desorption processes is not new. For example, several groups12–15 have reported reduction using fast atom bombardment (FAB). With FAB, a mixture of liquid matrix and analyte are splattered (desorbed) into the vacuum by beams of cesium ions or xenon atoms. The process momentarily adds energy to the mix, resulting in possible reactions. In 1987, Williams et al.12 showed that reduction was a common process in FAB. Since FAB does not use 337-nm light, their observations suggest that the close proximity of excited matrix and analyte molecules is all that is needed for reduction to occur. Kyranos and Vouros14 reported that FAB reduction processes depended on the relative redox potentials of the analyte and the matrix. They investigated a series of dyes in several FAB matrices (glycerol, sulfolane, thioglycerol, hydroxyethyl disulfide, m-nitrobenzyl alcohol). Methyl red, which has an azo group, has also been studied using FAB by Florencio and Heerma.15
More recently, Zenobi’s group16 has investigated the reduction of Cu(II) to Cu(I) seen in MALDI spectra when only Cu(II) has been placed on the target. They state that two processes are generally available to accomplish the task. The first is gas-phase charge exchange with matrix molecules such as nicotinic acid, dithranol, and 2,5-dihydroxybenzoic acid. The second is reduction with free electrons generated when the laser hits thin layers of matrix on a metal target. Knochenmuss17 has built a quantitative model of the MALDI process incorporating thermodynamically favored reactions in the plume just after the laser shot hits the matrix–analyte spot and incorporating free electron capture. His model supports the idea that chemicals added to the analyte–matrix mix can dictate what the detector will register. Thus it makes sense that adding a protecting agent (HABA in this report) to the analyte–matrix mixture leads to observation of nonreduced disulfides.
CONCLUSIONS
The mixed matrix HABA:CHCA (1:10) is an excellent choice for MALDI analysis of disulfide-bonded peptides. Not only is the ISD reduction of disulfides to thiols suppressed but the same low-threshold laser power generally used with CHCA is still feasible. Energy transfer from CHCA to HABA is implied by the observation that HABA ions dominate over CHCA in mixed matrices, even when CHCA is present in excess and when the lower laser powers applicable for CHCA are used. CHCA is the matrix of choice for cysteine-containing peptides, as its use leads to clean spectra at low-threshold laser powers. Cysteine-containing peptides are easily generated from solutions of the corresponding disulfides on-target. HABA appears to be protecting disulfides from reduction by being reduced preferentially. The idea of using a mixed matrix and/or adding a protective reagent to a matrix for MALDI-TOF-MS may be of general utility for other redox active compounds.
Acknowledgments
Kristin G. Huwiler was supported by NIH training grant 5T32HL07899 and NIH grant HL54462 to DFM.
Footnotes
REFERENCES
- 1.Patterson SD, Katta V. Prompt fragmentation of disulfide-linked peptides during matrix-assisted laser desorption ionization mass spectrometry. Anal Chem 1994; 66:3727–3732. [DOI] [PubMed] [Google Scholar]
- 2.Crimmins DL, Saylor M, Rush J, Thoma RS. Facile, in situ matrix-assisted laser desorption ionization mass spectrometry analysis and assignment of disulfide pairings in heteropeptide molecules. Anal Biochem 1995; 226:355–361. [DOI] [PubMed] [Google Scholar]
- 3.Huwiler KG, Vestling MM, Annis DS, Mosher DF. Biophysical characterization, including disulfide bond assignments, of the anti-angiogenic type 1 domains of human thrombospondin-1. Biochemistry 2002;41:14329–14339. [DOI] [PubMed] [Google Scholar]
- 4.Jones MD, Patterson SD, Lu HS. Determination of disulfide bonds in highly bridged disulfide-linked peptides by matrix-assisted laser desorption/ionization mass spectrometry with postsource decay. Anal Chem 1998; 70:136–143. [DOI] [PubMed] [Google Scholar]
- 5.Katta V, Chow DT, Rhode MF. Applications of in-source fragmentation of protein ions for direct sequence analysis by delayed extraction MALDI-TOF mass spectrometry. Anal Chem 1998;70:4410–4416. [DOI] [PubMed] [Google Scholar]
- 6.Mhatre R, Woodward J, Zeng C. Strategies for locating disulfide bonds in a monoclonal antibody via mass spectrometry. Rapid Commun Mass Spectrom 1999;13: 2503–2510. [DOI] [PubMed] [Google Scholar]
- 7.Schnaible V, Wefing S, Resemann A, et al. Screening for disulfide bonds in proteins by MALDI in-source decay and LIFT-TOF/TOF-MS. Anal Chem 2002;74:4980–4988. [DOI] [PubMed] [Google Scholar]
- 8.Chait BT, Field FH. A rapid, sensitive mass spectrometric method for investigating microscale chemical reactions of surface adsorbed peptides and proteins. Biochem Biophys Res Commun 1986;134:420–426. [DOI] [PubMed] [Google Scholar]
- 9.Caprioli RM, Fan T. Peptide sequence analysis using exopeptidases with molecular analysis of the truncated polypeptides by mass spectrometry. Anal Biochem 1986;154:596–603. [DOI] [PubMed] [Google Scholar]
- 10.IsoPro 3.0: http://members.aol.com/msmssoft
- 11.ChemPuter: http://www.shef.ac.uk/chemistry/chemputer/isotopes.html
- 12.Williams DH, Findeis AF, Naylor S, Gibson BW. Aspects of the production of FAB and SIMS mass spectra. J Am Chem Soc 1987;109:1980–1986. [Google Scholar]
- 13.Dass C, Desiderio DM. Particle beam induced reactions between peptides and liquid matrices. Anal Chem 1988;60:2723–2729. [DOI] [PubMed] [Google Scholar]
- 14.Kyranos JN, Vouros P. Reduction processes in fast atom bombardment mass spectrometry: Interdependence of analyte and matrix redox potentials. Biomed Environ Mass Spectrom 1990; 19:628–634. [DOI] [PubMed] [Google Scholar]
- 15.Florencio MH, Heerma W. Behaviour of methyl red under fast atom bombardment conditions. Org Mass Spectrom 1993;28:657–664. [Google Scholar]
- 16.Zhang J, Frankevich V, Knochenmuss R, Friess SD. Zenobi R. Reduction of Cu(II) in matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom 2003;14:42–50. [DOI] [PubMed] [Google Scholar]
- 17.Knochenmuss R. A quantitative model of ultraviolet matrix-assisted laser desorption/ionization. J Mass Spectrom 2002;37:867–877. [DOI] [PubMed] [Google Scholar]