Abstract
The ankyrin repeat is one of the most frequently observed amino acid motifs in protein databases. This protein–protein interaction module is involved in a diverse set of cellular functions, and consequently, defects in ankyrin repeat proteins have been found in a number of human diseases. Recent biophysical, crystallographic, and NMR studies have been used to measure the stability and define the various topological features of this motif in an effort to understand the structural basis of ankyrin repeat-mediated protein–protein interactions. Characterization of the folding and assembly pathways suggests that ankyrin repeat domains generally undergo a two-state folding transition despite their modular structure. Also, the large number of available sequences has allowed the ankyrin repeat to be used as a template for consensus-based protein design. Such projects have been successful in revealing positions responsible for structure and function in the ankyrin repeat as well as creating a potential universal scaffold for molecular recognition.
Keywords: ankyrin repeat, repeat motif, p16, protei–protein interactions
Proteins containing repeating amino acid sequences have received a great deal of attention recently. Several sequencing projects have shown that relatively short, tandemly repeating motifs are common in many proteins (Heringa 1998; Marcotte et al. 1999). Biochemical and structural characterization reveal that these repeats act as building blocks that stack side by side, forming the underlying architecture of a modular, specific protein-binding interface. Among the most abundant repeat motifs are the ankyrin repeat, leucine rich repeat, armadillo repeat, and tetratrico-peptide repeat. In contrast to other protein–protein interaction domains such as SH2 or SH3, these modular repeats typically do not recognize any specific amino acid sequence or structure. Instead, they form an elongated surface of varying size depending on the number of repeats. Specificity for protein partners is determined by variations in adaptive surface residues. Unlike globular proteins, the structure of repeat domains are dominated by local, short-range interactions, which represent a new paradigm for understanding the principles of protein stability and the mechanism of folding. In addition, a number of proteins with destabilizing mutations in their repeat domains have been implicated in several human diseases.
The ankyrin repeat, a 33-residue sequence motif, was first identified in the yeast cell cycle regulator Swi6/Cdc10 and the Drosophila signaling protein Notch (Breeden and Nasmyth 1987), and was eventually named for the cytoskeletal protein ankyrin, which contains 24 copies of this repeat (Lux et al. 1990). Subsequently, ankyrin repeats have been found in many proteins spanning a wide range of functions. The hand curated database PFAM-A, as of October 2003, contains 9689 ankyrin repeat sequences in 1871 proteins identified from the Swissprot and SP-TrEMBL databases (Bateman et al. 2002), while the SMART database contains 19,276 ankyrin repeat sequences in 3608 proteins identified from the nonredundant protein database (NRDB; Schultz et al. 1998; Letunic et al. 2002). Ankyrin repeat proteins are present in all three superkingdoms including bacteria, archaea, and eukarya, as well as in a number of viral genomes. However, a phylogenetic breakdown of the organisms that contain ankyrin repeats indicates that the majority are found in eukaryotes. Modular protein domains, such as the ankyrin repeat, that act as a scaffold for molecular interactions are likely important for development of the numerous signaling pathways necessary to evolve a more complicated multicellular organism (Marcotte et al. 1999). Some ankyrin repeat proteins have been found in viruses and bacteria, but these are believed to be the result of horizontal gene transfer (Bork 1993).
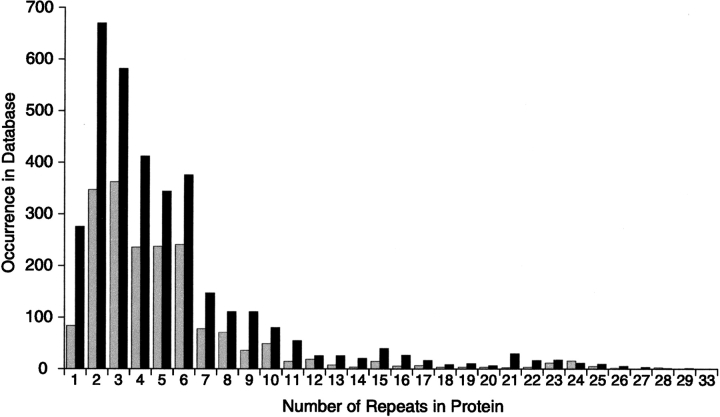
The number of ankyrin repeats found in a single protein varies greatly. Analysis of the SMART and PFAM databases show that the number of repeats per protein reaches from one to 33, with the majority of proteins containing six or fewer repeats (Fig. 1 ▶). The protein with the most ankyrin repeats to date is the ORF EAA39756 from Giardia lamblia, which actually may contain 34 repeats if the PFAM and SMART predictions are combined. Analysis of the SMART database suggests that two repeats are the most commonly occurring number of repeats per protein, while data from the PFAM database suggest three as the most popular number of repeats. Detection of the terminal repeats in an ankyrin repeat domain is difficult due to the divergence of many well-conserved hydrophobic residues. Through evolution, these positions have been replaced with polar residues to facilitate favorable interactions with the solvent, and thus the terminal repeats often deviate from the established consensus sequence. Also, the terminal repeats are frequently truncated, and therefore, the search model may not identify these sequences. Given this limitation, the most popular number of repeats per protein may actually be one or two repeats greater than that shown in Figure 1 ▶. For example, although Figure 1 ▶ shows that a significant number of proteins contain a single ankyrin repeat, we have previously demonstrated that an isolated ankyrin repeat cannot adopt a folded structure unless it shares an interface with another repeat (Zhang and Peng 2000; Mosavi et al. 2002a).
Figure 1.
The distribution of the number of ankyrin repeats per protein (as of October 2003). Analysis was performed on the PFAM database, which contains 9689 ankyrin repeats (gray bars), and the SMART database, which contains 19,276 ankyrin repeats (black bars).
To date, the structures of 13 naturally occurring ankyrin repeat proteins and three designed ones have been solved (Fig. 2 ▶). Ankyrin repeats have been observed to exist by themselves as a single domain protein or in conjunction with other domains in the same protein. Each repeat folds into two antiparallel α-helices followed by a β-hairpin or a long loop. Consecutive repeats stack together to form an L-shaped domain, which resembles a cupped hand with the β-hairpins representing the fingers and the helices as the palm. The overall shape of a typical ankyrin repeat domain is slightly curved, and this feature is particularly apparent in the structure of proteins that contain a large number of repeats (e.g., Fig. 2h ▶).
Figure 2.
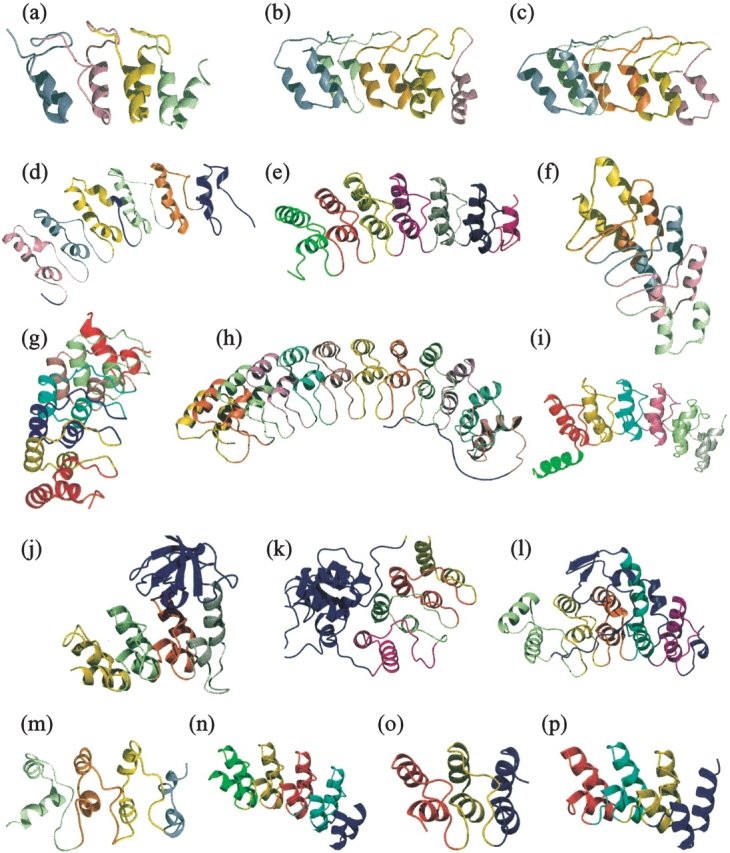
Ankyrin repeat proteins with high-resolution structures in the PDB. The ankyrin repeats have been colored differently to illustrate the packing interactions present in this fold. Cell cycle inhibitors (a) p16 (PDB ID: 1BI7), (b) p18 (1IHB), and (c) p19 (1AP7); (d) Iκ-Bα, inhibitor of Nf-κB (1NFI); (e) Bcl-3, a unique member of the Iκ-B family (1K1A); (f) Transcription factor GABP-β (1AWC); (g) Human oncoprotein Gankyrin (1UOH); (h) D34 region of human Ankyrin-R (1N11); (i) Ankyrin repeat domain of Drosophila signaling protein Notch (1OT8); (j) Tumor suppressor 53BP2 (1YCS); (k) Signaling protein PAP-β (1DCQ); (l) Transcriptional regulator Swi6 (1SW6); (m) Cardiomyogenic hormone myotrophin (2MYO). Designed ankyrin repeat proteins (n) Sank E3_5 (1MJ0), (o) 3ANK (1N0Q), and (p) 4ANK (1N0R).
Ankyrin repeats have been found in numerous proteins with functions that include cell–cell signaling, cytoskeleton integrity, transcription and cell–cycle regulation, inflammatory response, development, and various transport phenomena. Of the ankyrin repeat proteins that have been characterized, a unifying trait is that they typically function in mediating specific protein–protein interactions. Indeed, no enzymatic function has been detected for any ankyrin repeat domain. The ankyrin repeat is found in a number of biologically important proteins. The family of INK4 tumor suppressors, p15, p16, p18, and p19, as well as 53BP2, a regulator of the tumor suppressor p53, all contain ankyrin repeats. The signaling protein Notch, which is involved in many cell-fate decisions during development, has seven ankyrin repeats. Nf-κB, a transcription factor that regulates inflammatory response is inhibited by IκB, which contains seven ankyrin repeats. Various numbers of ankyrin repeats are found in members of the TRP cation channel family, which include heat and cold sensitive receptors as well as mechanosensory proteins.
Structural features of the ankyrin repeat
The ankyrin repeat has a well-defined structure. With the exception of insertions found mainly in the loop regions, this motif rarely deviates from the canonical helix-loop-helix-β-hairpin/loop fold. The helices are arranged in anti-parallel fashion followed by a loop region that points outward at an approximately 90° angle and forms a β-hairpin in some known ankyrin repeat structures. The helices of one repeat pack against the helices of the adjacent repeat while the β-hairpin/loop region, in some cases, forms a continuous β-sheet. The interrepeat interface mainly consists of hydrophobic interactions stabilizing the helices in addition to a hydrogen bonding network connecting the β-hairpin/ loop region. The outer helix is nine residues long, spanning from positions 15–24 of the canonical sequence, while the inner helix is seven residues long and extends from positions 5–12 (Fig. 3A ▶). The overall stack exhibits a slight curvature resulting from the difference in helix length as well as the interrepeat packing interactions between the two helices. Near the short loop of the helix-loop-helix, the presence of a small side-chain residue at position 10 allows the shorter inner helices to pack more closely against each other while the outer helices are spaced further apart due to midsize residues at positions 17 and 18. Closer to the β-hairpin/loop junction, positions 6, 21, and 22 typically contain residues with large side chains, such as leucine, which lends the ankyrin repeat stack a left-handed superhelical twist. Each repeat is rotated 2–3° counterclockwise relative to the preceding repeat and contributes approximately 13° of pitch to the superhelical turn of the molecule (Michaely et al. 2002). A complete turn of the superhelix would require 32 repeats and have a 35 Å radius (Michel et al. 2001).
Figure 3.
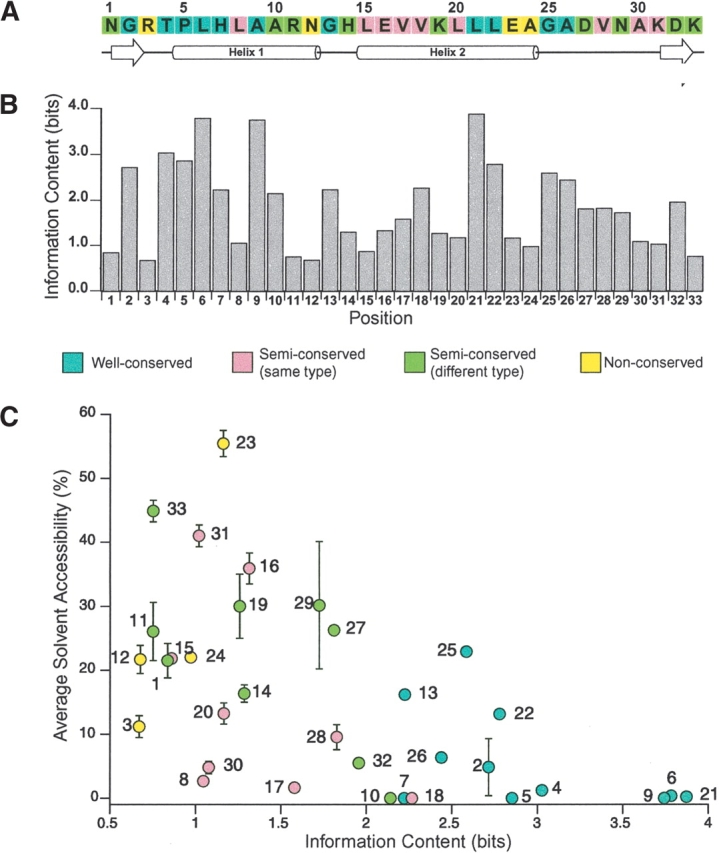
Conservation level and information content in the ankyrin repeat. (A) The ankyrin repeat consensus sequence derived from statistical analysis of ~4400 ankyrin repeats used by Mosavi et al. 2002a, displaying the conservation level of each position as different colors. Corresponding secondary structure elements are displayed below. (B) The information content (bits) for each of the 33 positions in the ankyrin repeat calculated from the current version of PFAM database (containing ~9000 ankyrin repeat sequences) using the program ALPRO (Schneider and Stephens 1990). Higher information content implies that the residue is more conserved within the motif. (C) Comparison of the solvent accessibility of each position to the information content calculated by ALPRO. Average solvent accessibility was calculated for all 33 positions of the two internal repeats (repeat 2 and 3) in the structure of designed protein 4ANK (1N0R) with four identical repeats. Each marker is color coded according to the conservation-level classification of (Mosavi et al. 2002a) and labeled with the position number. Error bars represent the deviation from average for solvent accessibility.
The consensus sequence of the ankyrin repeat contains a number of “signature” residues that define the shape of the repeat. The most prevalent is the TPLH motif at positions 4 through 7. The proline, at position 5, initiates a tight turn responsible for the L-shaped structure of the ankyrin repeat, which is stabilized by side-chain hydrogen bonding interactions of histidine at position 7 in the helix and threonine at position 4 in the β-hairpin/loop. In addition, well-conserved glycines at positions 13 and 25 contribute to helix termination thereby fostering the transition into a tight loop between the helices and the beginning of the long β-hairpin/loop region, respectively. The helical packing interactions of neighboring ankyrin repeats are essentially identical to that of a four-helix bundle. The nonpolar residues at positions 9 and 18 (intra-repeat), positions 8, 10, 17, 20, and 22 (interrepeat) and positions 6 and 21 (both) form the hydrophobic core. For the most part, these positions have relatively low solvent accessibility (Fig. 3C ▶). However, in terminal repeats that are exposed to solvent, these positions are often replaced by polar residues.
For motifs that have a well-conserved structure but many different functions, one might expect the surface residues to exhibit a high degree of variability to accommodate assorted binding interactions. In the ankyrin repeat, the information content or conservation level of each position does correlate somewhat with solvent accessibility. Previously, we classified each of the 33 positions in the ankyrin motif as either well-conserved, semiconserved of the same type, semiconserved of different type, and nonconserved (Fig. 3A ▶; Mosavi et al. 2002a). As a comparison, we have determined the information content (in bits) of each position in this motif using the currently available repeats downloaded from the PFAM database (http://pfam.wustl.edu/) with the program ALPRO (Schneider and Stephens 1990; Fig. 3B ▶). Generally, the degree of information content calculated by ALPRO agrees well with our original classification scheme, particularly for the well-conserved residues, which display some of the highest values. Minor differences between our classification and the ALPRO calculation exist probably because these analyses treat the amino acid distribution in a different manner. Comparison of the information content to the average solvent accessibility for each of the 33 positions (for internal repeats of identical repeat protein, 4ANK) suggests that most positions with high information content have low solvent accessibility (less than 10%), while the positions previously classified as nonconserved (yellow markers) show high solvent accessibility and low information content (Fig. 3C ▶). Interestingly, a number of the semiconserved residues have both low solvent accessibility and information content.
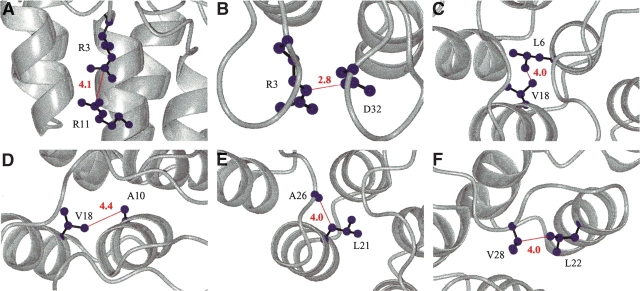
Covariation or correlation analysis has been be used to expose the distance information and energetic coupling between residues in a large protein family (Gobel et al. 1994; Shindyalov et al. 1994; Lockless and Ranganathan 1999; Larson et al. 2000). Using the ankyrin repeat sequences in the PFAM database, we have performed covariation analysis of all 33 positions in this motif in an effort to reveal positions that may be structurally “coupled” to one another. Several pairs of positions had covariation scores significantly above the mean. The majority of these pairs are located in close proximity to one another (<5 Å) and form interactions in the inter-helical interfaces as well as in the β-hairpin/loop region (Fig. 4 ▶). Such geometrical complementarities between residues may be important for the stability of the ankyrin repeat.
Figure 4.
(A–F) Pairs of positions in the ankyrin repeat that have high covariation scores displayed on the structure of 4ANK (1N0R). Covariation was calculated as previously described using ~4400 ankyrin repeat sequences (Mosavi et al. 2002a). The backbone is displayed in gray, whereas the atoms of the residues shown are represented as blue spheres. All distances (red lines) are labeled.
Despite the high degree of sequence conservation, insertions in the ankyrin repeat motif are not uncommon and are usually found in the loop regions, particularly the long loop preceding the β-hairpin. For example, both Iκβ and Bcl-3 have short insertions between repeats 1 and 2 as well as repeats 3 and 4 in the long loop (Fig. 2d–e ▶). The ankyrin repeat domain of Swi6 contains an insertion between repeats 2 and 3, and another unusually long 40-residue insertion between repeats 3 and 4, both in the long loop (Fig. 2l ▶). Shortening of the helices by deletion of one or two residues has been observed as well. The crystal structures of the INK4 family members p16, p18, and p19, all have a slightly distorted inner helix due to the shortening in the second repeat (Fig. 2a–c ▶).
Folding and stability of ankyrin repeat proteins
Until recently, the field of protein folding has focused mainly on globular proteins. Ankyrin repeats represent a class of protein with an elongated structure and little or no long-range contacts. The modular nature of ankyrin repeat domains presents a number of interesting questions from a folding perspective. The ankyrin repeats stack against one another to form a single domain, yet each of the repeating units must possess some degree of intrinsic stability. Does this mean that ankyrin repeat domains follow a multistate folding pathway populated by intermediates consisting of some repeats folded and others unfolded? Interestingly, the majority of data does not support such a model. Folding studies on a number of naturally occurring ankyrin repeat proteins show that, for the most part, the ankyrin repeat domain follows a two-state folding pathway. Equilibrium folding experiments with human p16 (Tang et al. 1999), rat myotrophin (Mosavi et al. 2002b), and Drosophila Notch (Zweifel and Barrick 2001), all indicate that ankyrin repeat domains exist exclusively in either a folded or unfolded state. The folding transition is highly cooperative and partially folded intermediates are not detectable in the equilibrium folding/unfolding reaction indicating that some type of coupling mechanism must exist among different repeats.
The ankyrin repeat domain of Drosophila Notch has been used as a model system to investigate the long-range coupling among repeats (Zweifel and Barrick 2001; Bradley and Barrick 2002). Notch is a signaling molecule that contains seven ankyrin repeats in the intracellular domain. Experiments on the full ankyrin domain (Nank1–7*) as well as two variants with deletion of one or two C-terminal repeats, Nank1–6* and Nank1–5*, respectively showed all proteins to fold in a two-state manner. However, Nank1–7* had significantly higher stability (ΔGH2O = 8.03 kcal•mole−1) than Nank1–6*(ΔGH2O = 4.14 kcal•mole−1) and Nank1–5*(ΔGH2O = 3.55 kcal•mole−1), indicating that the seventh repeat contributed more to global stability than the sixth repeat (Zweifel and Barrick 2001). These results suggest that although each repeat has some level of intrinsic stability, the effects are felt “globally” over the entire protein. In an effort to uncouple the Notch ankyrin repeats, Nank1–7* constructs were made with a disruptive alanine to glycine substitution in helix 1 of each repeat. Repeat 6 was the only repeat in the Notch ankyrin domain that was severely affected by the substitution and caused the folding of Nank1–7* to go from two-state to multistate. The fact that the long-range cooperativity was interrupted suggests that the interrepeat coupling was weakest through the region of repeat 6 (Bradley and Barrick 2002).
Although the bulk of the equilibrium folding studies on ankyrin repeats strongly suggest that this domain follows a two-state folding pathway, work on human p19 has provided evidence of a partially folded intermediate by NMR studies. Chemical denaturation experiments showed completely superimposable fluorescence and CD chemical denaturation curves. However, a series of 1H-15N HSQC spectra at various urea concentrations revealed the presence of unidentified cross-peaks that were not present in the folded or unfolded states. In addition, urea-induced folding and unfolding transitions monitored by NMR did not intersect at half fraction unfolded, leading to the conclusion that an intermediate with 30% maximum population was present in the equilibrium folding pathway (Zeeb et al. 2002). This intermediate may be an oligomer because of the high concentrations required for NMR studies.
The kinetic folding of ankyrin repeat proteins has been studied for human p16 and p19, both members of the INK4 tumor suppressor family (Tang et al. 1999; Zeeb et al. 2002). Although p16 has four repeats and p19 has five repeats, they share 40% sequence identity and a high degree of structural similarity. Both proteins show three refolding phases at low urea concentrations and one unfolding phase. The slowest phase for both proteins is urea independent and can be accelerated with the addition of a prolyl isomerase such as cyclophilin indicating that proline isomerization is responsible for the rate limiting step. The fast phases result in partial formation of α-helical secondary structure, although the details are not well resolved. The kinetic folding pathway of p16 has also been studied using φ-value analysis (Tang et al. 2003). P16 constructs with core mutations designed to subtly decrease the hydrophobic contacts in each repeat were created. Equilibrium folding studies showed a large number of φ-values close to 1 in the C-terminal two repeats while the φ-values on the N-terminal two repeats were generally smaller. These results indicate that the C-terminal repeats fold first and may form a “structural scaffold” on which to fold the less stable N-terminal repeats. Indeed, truncation studies of p16 showed that the C-terminal two repeats can form an autonomously folded structure while the N-terminal two repeats are disordered in isolation, suggesting that the two subdomains have very different intrinsic stabilities (Zhang and Peng 2000).
Various truncation studies of ankyrin repeat proteins have been used to investigate the minimum folding unit of this motif. Limited proteolysis combined with deletion studies of ankyrin, which contains 24 repeats, suggested that four to six repeats were necessary to form a stably folded structure, and a single repeat was not capable of folding (Michaely and Bennett 1993). The above-mentioned truncation studies of p16 demonstrated that the C-terminal two repeats could form an autonomously folded unit and suggested that two repeats are actually the minimum folding unit of the ankyrin repeat (Zhang and Peng 2000). Work on our consensus designed ankyrin repeat protein supports this conclusion. 2ANK, with two identical ankyrin repeats, adopted a fully folded conformation after substitution of six surface leucine residues with arginine (Mosavi and Peng 2003).
Ankyrin repeat-mediated protein–protein interactions
Ankyrin repeats are present in many biologically important proteins. A search of the PUBMED database with the term “ankyrin repeat” yielded 305 publications (October 2003). We examined each of these studies for detailed evidence of protein–protein interactions mediated by ankyrin repeat domains as detected by yeast two-hybrid, coimmunoprecipitation, in vitro binding assays and GST pull-down experiments. More than 60 such interactions are summarized in Table 1. These proteins and their corresponding complexes are essential for many cellular processes such as cell fate determination, endocytosis, transcription regulation, and cell cycle control.
Table 1.
Confirmed protein–protein interactions mediated by ankyrin repeats
| Ankyrin repeat protein | Number of repeats | Function | Partner(s) | Reference(s) |
| Ankyrin | 24 | Intracellular adaptor | α-Na,K-ATPase | (Davis and Bennett 1990; Thevananther et al. 1998) |
| L1CAMs and CD44 | (Davis et al. 1993; Davis and Bennett 1994; Michaely and Bennett 1995; Zhu and Bourguignon 2000) | |||
| Cl−/HCO3-anion exchanger (AE1–3) | (Davis and Bennett 1984; Davis et al. 1991; Morgans and Kopito 1993) | |||
| NaCh | (Srinivasan et al. 1992) | |||
| Clathrin heavy chain | (Michaely et al. 1999) | |||
| Rh and RhAG | (Nicolas et al. 2003) | |||
| ANKRA | 3 | Endocytosis | Megalin | (Rader et al. 2000) |
| Ankrd2 | 4 | Regulation of muscle stress response | Titin | (Miller et al. 2003) |
| BARD1 | 3 | Inhibition of polyadenylation | CstF50 | (Kleiman and Manley 1999) |
| BCL-3 | 7 | Oncoprotein and transcriptional regulator | NFκB | (Wulczyn et al. 1992) |
| JAB1, Pirin, Tip60, Bard1 | (Dechend et al. 1999) | |||
| B3BP | (Watanabe et al. 2003) | |||
| CARP | 4 | Regulation of muscle stress response | YB1 | (Zou et al. 1997) |
| Titin | (Miller et al. 2003) | |||
| cpSRP43 | 4 | Chloroplast signal recognition particle | LHCP | (Jonas-Straube et al. 2001) |
| DARP | 4 | Regulation of muscle stress response | Titan | (Miller et al. 2003) |
| GABP3 | 5 | Transcriptional regulator | GABPα | (Batchelor et al. 1998) |
| Gankyrin | 6 | Oncoprotein and regulator of retinoblastoma protein (Rb) | CDK4/Cyclin-D complex, Rb, and S6-ATPase | (Li and Tsai 2002; Dawson et al. 2002) |
| MAGE-A4 | (Nagao et al. 2003) | |||
| IκB | 6 or 7 | Transcriptional regulators | NFκB | (Hatada et al. 1992; Inoue et al. 1992; Kidd 1992; Rice et al. 1992; Mercurio et al. 1993; Naumann et al. 1993; Dobrzanski et al. 1995; Thompson et al. 1995; Simeonidis et al. 1997; Whiteside et al. 1997; Huxford et al. 1998; Jacobs and Harrison 1998; Malek et al. 2003) |
| PTP-BAS (IκBα only) | (Maekawa et al. 1999) | |||
| Integrin linked kinase | 4 | Cell–cell adhesion | PINCH | (Li et al. 1999; Tu et al. 1999; Velyvis et al. 2001) |
| INK4 | 3, 4, or 5 | Tumor suppressors and cell-cycle regulators | Cdk4/6 | (Serrano et al. 1993; Hannon and Beach 1994; Hirai et al. 1995; Russo et al. 1998) |
| NFκB (p16 only) | (Wolff and Naumann 1999) | |||
| Tax (p16 and p15 only) | (Suzuki et al. 1996, 1999) | |||
| Mbp1 | 4 | Late G1 transcription factor | Clb2/Cdc28 kinase | (Siegmund and Nasmyth 1996) |
| Myotrophin/V1 | 3 | Development | CP | (Taoka et al. 2003) |
| NFκB | (Sivasubramanian et al. 1996) | |||
| MYPT1 | 8 | Regulation of myosin phosphorylation | Protein phosphatase-1c | (Tanaka et al. 1998) |
| NPR1 | 4 | Transcriptional regulator | AHBP-1b and TGA6 | (Zhang et al. 1999) |
| Notch | 7 | Cell-fate determination | Deltex | (Diederich et al. 1994; Matsuno et al. 1995) |
| CSL | (Kato et al. 1997; Petcherski and Kimble 2000; Tani et al. 2001) | |||
| PCAF and GCN5 | (Kurooka and Honjo 2000) | |||
| SKIP | (Zhou et al. 2000) | |||
| YY1 | (Yeh et al. 2003) | |||
| MAML1 | (Wu et al. 2000; Nam et al. 2003) | |||
| P85 | 6 | Regulation of actin cytoskeleton | Protein Phosphatase-1Δ | (Tan et al. 2001) |
| RNase L | 9 | Ribonuclease | 2′–5′ oligoadenylates | (Zhou et al. 1993; Dong and Silverman 1997) |
| RFXANK | 4 | Transcriptional regulator | RFXAP and CIITA | (Nekrep et al. 2001) |
| Shank | 7 | Synapse density scaffolding | Sharpin | (Lim et al. 2001) |
| α-fodrin | (Bockers et al. 2001) | |||
| Swi4 | 4 | Late G1 transcription factor | Clb2/Cdc28 kinase | (Siegmund and Nasmyth 1996) |
| Swi6 | 4 | Late G1 transcription factor | Stb1 | (Ho et al. 1999) |
| Tankyrase | 24 | Poly(ADP-ribose) polymerase | TRF1 | (Smith et al. 1998) |
| IRAP | (Chi and Lodish 2000) | |||
| Grb14 | (Lyons et al. 2001) | |||
| TAB182 | (Seimiya and Smith 2002) | |||
| NuMA | (Sbodio and Chi 2002) | |||
| Mcl-1 | (Bae et al. 2003) | |||
| Tvl-1 | 4 | Signal transduction adaptor | Raf-1 | (Lin et al. 1999) |
| 53BP2 | 4 | Tumor suppression | p53 | (Iwabuchi et al. 1994; Gorina and Pavletich 1996) |
| Protein phosphatase-1 | (Helps et al. 1995) | |||
| Bcl-2 | (Naumovski and Cleary 1996) | |||
| NFκB | (Yang et al. 1999) | |||
To date, eight co-crystal structures consisting of six ankyrin repeat proteins have been solved at atomic resolution and point to a common mode of binding for this motif. In each case, the ankyrin repeat binding interface is comprised almost exclusively of the concave inner surface made up of the β-hairpin/loop region and inner short helices. Figure 5 ▶ illustrates the contact surfaces of four different ankyrin repeat proteins. Four structures of the INK4 family of tumor suppressors in complex with Cdk6 have been solved: two of p19 and one each of p16 and p18. All three proteins have very similar contacts in the interface with Cdk6 mostly involving H-bonds and salt bridges (Fig. 5C ▶). The electrostatic complementarity of the interface is likely responsible for the specificity of p16 and p19 for Cdk4 and Cdk6. Similar to p16 or p19/Cdk6 binding, the interface of the inhibitor IκB-α with transcription factor NF-κB is heavily influenced by contributions from the inner helices (Fig. 5B ▶). In contrast, the structures of the transcription factor complex GABP-α/GABP-β (Fig. 5A ▶) and the tumor suppressor p53 with regulator 53BP2 (Fig. 5D ▶) show the β-hairpin/loops to be the key participant in forming the binding interface. The 53BP2/p53 interface, which also includes a SH3 domain (upper red patch in Fig. 5D ▶), uses only the β-hairpin/loop of the fourth ankyrin repeat. GABP-β, on the other hand, includes portions of the inner helices in the interface with GABP-α.
Figure 5.
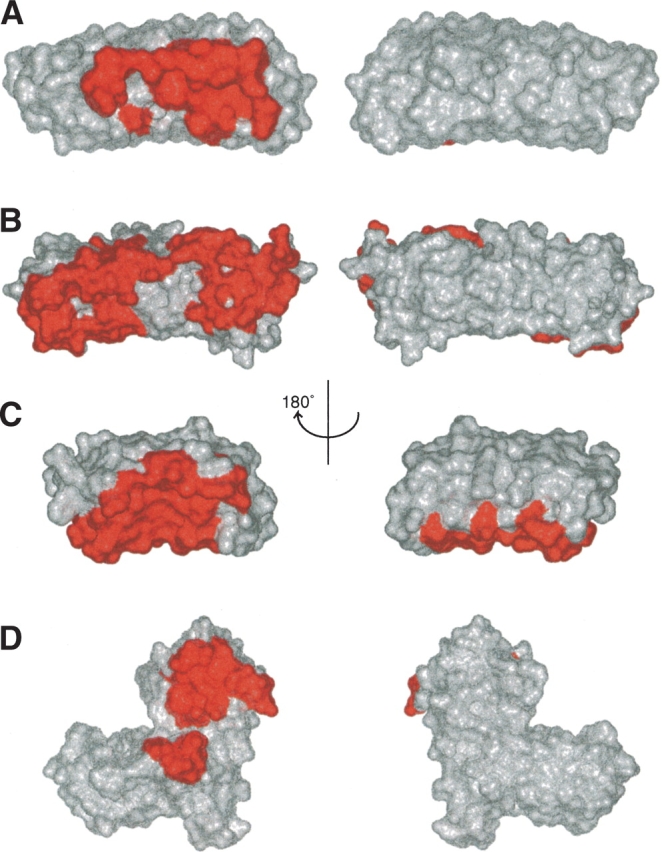
Locations of the interaction interface in four different ankyrin repeat co-crystal structures. The residues involved in the interaction with protein partners are colored red in the surface representations. The β-hairpin/loop region is pointing directly towards reader (left) and away from the reader (right). (A) The interface between GABP-β and GABP-α from the co-crystal structure 1AWC, (B) the interface between Iκ-Bα and NFκ-B from the co-crystal structure 1NFI, (C) the interface between p16 and Cdk6 from the co-crystal structure 1BI7, (D) the interface between 53BP2 and p53 from the co-crystal structure 1YCS. Note that the lower red patch is the ankyrin repeat interface, while the upper patch is the SH3 domain interface.
The fact that ankyrin repeat proteins are found in such a diverse set of cellular functions can be attributed at least partially to the presence of other signaling, protein–protein interaction, and catalytic domains in various ankyrin repeat-containing proteins. For example, Shank contains SH3, SAM, and PDZ domains in addition to seven ankyrin repeats. Tankyrase, has 19–24 ankyrin repeats, a SAM domain, and a PARP domain that can catalyze the addition of poly ADP-ribose chains onto an acceptor molecule. In signal transduction, different domains often work in a synergetic or cooperative manner. For example, an ankyrin repeat domain may help recruit a substrate to a catalytic domain, or stable complex formation may require the interaction of more than one domain bound to the same molecule. These types of mechanisms are potentially important for signal diversity and integration (Prehoda et al. 2000; Dueber et al. 2003).
Disease-related defects in ankyrin repeat proteins
A number of ankyrin repeat proteins have been linked to human diseases; however, only a small fraction have been characterized using genetic, biochemical, or biophysical methods. The INK4a gene, which encodes the mammalian cell cycle inhibitor p16, is a frequent site of mutations that are strongly associated with cancer (Ruas and Peters 1998; Sherr and Roberts 1999). Recently, Marc Greenblatt’s group at the University of Vermont developed an interactive database detailing each human p16 mutation (https://biodesktop.uvm.edu/perl/p16#1). Mapping of the more than 150 cancer-related missense mutations on the three dimensional structure correlates with a myriad of defects including (1) loss of a ligand-binding residue, (2) perturbed secondary structure and thermodynamic stability, and (3) large conformational changes in tertiary structure associated with severe aggregation. Furthermore, native p16 lacks many of the stabilizing hydrogen bonds typically seen in the β-hairpin/loop region, which may account for the unusually low thermodynamic stability (~3.3 kcal/mole) and high backbone flexibility that leave the protein particularly vulnerable to even subtle mutations (Tevelev et al. 1996).
Naturally occurring mutations in p16 associated with familial melanoma have been well characterized, including R24P, D74N, R87P, P81L, P114L, G101W, and V126D. These seven mutants represent a cross-section of the various defects that interfere with the biological function of this important tumor suppressor molecule. Residues D74 and R87 are located at positions that directly interact with the cyclin-dependent kinases (Russo et al. 1998). These positions are completely conserved among INK4 family members and p16 orthologs. In p16, D74 is located in the loop of repeat 2, while R87 is in the inner helix of repeat 3. These mutations disrupt the electrostatic interactions and hydrogen-bonding network at the binding interface, which in turn, reduce the ligand binding affinity and cell-cycle inhibitory function (Koh et al. 1995; Reymond and Brent 1995; Parry and Peters 1996; Ruas et al. 1999). Indeed, biophysical studies show that D74N retains a native-like structure (Tevelev et al. 1996; Tang et al. 1999). The aspartate to proline substitution at position 87 also effectively “breaks” the α-helix, thereby reducing the secondary structure of p16 as indicated by CD studies (Zhang and Peng 1996). The mutation R24P removes electrostatic and spatial constraints located between the α-helices of repeat 1, which are important in coordinating the neighboring contact residues R22 and E26. In addition to structural perturbations such as reduced helical content and stability, R24P also forms higher order oligomers, which may reduce kinase binding and cell cycle inhibition (Tang et al. 1999; Walker et al. 1999; Cammett et al. 2003).
In contrast to the above point mutations that introduce proline, P81L and P114L remove the original torsional constraints imposed at these positions. These mutations are located in the highly conserved TPLH motif in the third and fourth repeat, respectively. Both mutants are classified as molten globules based on retention of wild-type–like secondary structure but loss of a cooperative thermal unfolding transition. Additional defects include increased hydrophobic surface exposure and greater susceptibility to proteolysis (Tevelev et al. 1996; Zhang and Peng 1996; Tang et al. 1999; Cammett et al. 2003). Interestingly, P81L remains monomeric while P114L forms higher order oligomers and aggregates. These mutants have greatly reduced affinity for Cdk4 and Cdk6 and are deficient in cell cycle inhibitory function (Lukas et al. 1995; Reymond and Brent 1995; Walker et al. 1999).
Mutations G101W and V126D represent a unique class of temperature sensitive mutations that display diminished biological function as the temperature increases from 30°C to 42°C (Parry and Peters 1996; Ruas et al. 1999). The glycine at position 101 is important for termination of helix-2 in the third repeat. Introduction of the bulky hydrophobic tryptophan side chain disrupts this event at a well-conserved position in the ankyrin consensus (Fig. 3A ▶, position 25). G101W is a molten globule, rapidly proteolyzed and forms large aggregates during sedimentation equilibrium analysis (Zhang and Peng 1996). The side chain of valine, at position 126 in helix-2 of the last repeat, protrudes into the hydrophobic core of repeats 3 and 4. V126D displays reduced secondary structure, increased hydrophobic surface area, and higher order oligomer formation, indicating that substitution with the charged aspartate disrupts the hydrophobic contacts in the C-terminal half of the protein (Tevelev et al. 1996; Zhang and Peng 1996; Tang et al. 1999; Cammett et al. 2003). Aggregation is a process that is accelerated at higher temperature, leading to reduction in the concentration of available p16, which may explain the temperature sensitive phenotype of these mutant proteins (Walker et al. 1999; Becker et al. 2001).
Taken together, these studies illustrate how each mutation contributes to the global destabilization and functional inactivation of p16. Indeed, amino acid substitutions that enhance the stability of p16 without altering its structure or function show increased resistance to the destabilizing effects of different oncogenic mutations, independent of where they occur in the molecule. Recently, a hyperstable variant of wild-type p16 was shown to retain wild type-like Cdk4 binding activity in the presence of the melanoma-associated mutations R24P, P81L, or V126D (Cammett et al. 2003). This apparent global suppressor effect indicates that increasing the thermodynamic stability of p16 could be a general strategy to restore function to defective mutant proteins.
The Notch family of single-pass transmembrane receptors are involved in cell fate determination. The intracellular domain of Notch contains seven ankyrin repeats that may be necessary for recruitment of transcriptional coactivators when the intracellular domain is translocated into the nucleus (Table 1). An alanine to threonine substitution in the second repeat of human Notch3 is associated with CADASIL, a hereditary adult-onset condition causing stroke and dementia (Joutel et al. 1996). This mutation has been mapped onto the structure of highly homologous Drosophila Notch and biophysical characterizations show that the secondary structure and unfolding transition of the mutant protein did not deviate significantly from wild type (Zweifel et al. 2003). These studies strongly suggest that the CADASIL mutation, which is located on the surface of the protein, disrupts specific contacts at the protein–protein interaction interface.
Bare lymphocyte syndrome (BLS) is a disease in which CD4(+) lymphocytes fail to present antigenic peptides to the cellular surface, causing a combined immunodeficiency. Individuals with this disease have defects in at least one of four transcriptional regulatory elements, including the RFXANK gene. RFXANK forms a heterotrimeric complex with RFXAP and RFX5, which can then bind DNA and recruit CIITA to trigger transcription of MHCII genes (Masternak et al. 1998; DeSandro et al. 2000). In the absence of a high-resolution structure for RFXANK, modeling the C-terminal four ankyrin repeats onto the existing crystal structure of GABP-β and Swi6, followed by subsequent mutagenesis studies allowed Nekrep et al. (2001) to identify residues participating in the protein–protein interaction interface. Interestingly, mutagenesis studies indicated that RFXAP and CIITA bind to opposite surfaces of RFXANK, and that RFXAP interacts along two different surfaces of the ankyrin repeats. The BLS-associated mutation D121V is located in the β-hairpin loop of the first repeat and completely inactivates RFXANK (Wiszniewski et al. 2003). This mutation may replace a contact residue and reduce the affinity for one of the protein–protein interaction partners (Table 1). Mutation L195P is located in the inner helix of the third repeat of RFXANK and presumably disrupts the helix, abolishing the interaction with binding partners RFX5 and RFXAP (Nekrep et al. 2001). Taken together, these three proteins represent testable models of disease-related ankyrin repeat proteins that are amenable to further biophysical and structural characterization.
Consensus design studies
The large and diverse collection of ankyrin repeat proteins illustrates the versatility of this particular motif. Throughout evolution, the ankyrin repeat has retained its stacked L-shaped structure as a scaffold for various protein–protein interactions. Consequently, this motif can naturally serve as a template or building block for protein engineering and design studies. In particular, it is an especially attractive model for consensus-based protein design because of the large number of currently available sequences. Recent efforts at designing ankyrin repeat proteins using sequence-based strategies have been quite successful (Mosavi et al. 2002a; Binz et al. 2003; Kohl et al. 2003; for reviews, see Main et al. 2003; Tripp and Barrick 2003).
Consensus-based design strategies have been previously applied to a number of proteins with excellent results. The zinc finger was one of the first motifs to be successfully designed using the existing sequences to engineer DNA binding specificity (Desjarlais and Berg 1993). Early design studies on the coiled-coil to understand the mechanisms of oligomerization, and the contribution of various positions to stability incorporated knowledge of available sequences (Harbury et al. 1993; Zhou et al. 1994). Sequence information has also been used to engineer greater thermostability in proteins. Alignment of 13 fungal phytase sequences was used to create a consensus phytase with normal enzymatic activity but significantly higher thermostability (Lehmann et al. 2000). Finally, a 33-residue WW domain consensus sequence was determined from approximately 60 known WW domain sequences and high resolution structural studies showed this “prototype” WW domain to be well-folded (Macias et al. 2000).
Unlike the protein domains previously chosen for consensus design, the ankyrin repeat has a significantly larger number of identified sequences available. In addition, the modular nature provides an excellent “generic” surface that can theoretically interact with any protein. We performed statistical analysis on ~4400 ankyrin repeat sequences downloaded from the PFAM database to create a consensus or idealized ankyrin repeat sequence (Mosavi et al. 2002a). Our analyses consisted of classifying the conservation-level of each position based on the amino acid occurrence at that position, the amino acid distribution over the entire sequence, and the secondary structure elements corresponding to each position (Fig. 3A ▶). We used this sequence to construct proteins consisting of one, two, three, or four identical repeats (1ANK, 2ANK, 3ANK, and 4ANK respectively). Biophysical characterization showed 3ANK and 4ANK to be fully folded and monomeric, while 2ANK was partially folded and 1ANK was completely unfolded. Thermal denaturation studies on 3ANK and 4ANK revealed that these designed proteins had higher thermostability than any previously characterized, naturally occurring ankyrin repeat proteins of equivalent size. High-resolution X-ray crystal structures of 3ANK and 4ANK showed well-packed highly regular ankyrin repeat structures with a periodic network of hydrogen bonding (Fig. 2o–p ▶). These results demonstrate that a statistically derived consensus sequence contains all the necessary structural information for the ankyrin repeat fold.
Subsequently, similar work utilizing consensus design on ankyrin repeats with some significant differences was published by Binz et al. They started with statistical analysis of 229 repeats from the SMART database, and included information from 10 existing high-resolution structures of ankyrin repeat proteins to create a consensus ankyrin sequence (Binz et al. 2003). Their strategy incorporated “capping” repeats derived from the structure of GABP-β to flank the N and C termini of the protein. In addition, for the internal repeats, they allowed positions in the shorter helix and β-hairpin region (2, 3, 5, 13, 14, and 33) to contain any amino acid except glycine, proline, or cysteine. Six randomly selected library members with four, five, or six repeats were characterized biophysically showing well-folded structures with high stability. X-ray crystallography of E3_5 (a five-repeat library member) showed well-packed side chains adopting a regular ankyrin repeat fold (Fig. 2n ▶; Kohl et al. 2003).
Design of “generic” ankyrin repeat proteins is possible using consensus sequence design, as demonstrated by these two independent studies. Despite the variations in design strategy, 3ANK, 4ANK, and E3_5 formed well-folded proteins and displayed higher stability than any previously characterized ankyrin repeat proteins. These results demonstrate that statistical analysis of sequence databases is an effective way to design highly stable protein frameworks that serve as a foundation for more specific protein–protein interactions. As more sequence data become available, additional protein folds can be manipulated using this method.
Conclusions
Tandemly repeating amino acid sequences are becoming increasingly prominent as a scaffold for protein–protein interactions. Ankyrin repeats, in particular, are one of the most popular motifs of this class, and have been found in a number of biologically interesting proteins. A thorough understanding of the folding and stability of ankyrin repeats will ultimately expose the underlying mechanism of ankyrin repeat mediated protein–protein interactions, and lay the foundation for future design and engineering studies, which may ultimately help to unravel intricate cell signaling mechanisms.
Acknowledgments
Work in the author’s laboratory was supported by grants from the NIH (R01-GM68830) and American Cancer Society (GMC-103045). Z.-Y.P. is an American Cancer Society Research Scholar.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03554604.
References
- Bae, J., Donigian, J.R., and Hsueh, A.J.W. 2003. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 278 5195–5204. [DOI] [PubMed] [Google Scholar]
- Batchelor, A.H., Piper, D.E., de la Brousse, F.C., McKnight, S.L., and Wolberger, C. 1998. The structure of GABPα/β: An ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279 1037–1041. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T.M., Rizos, H., Kefford, R.F., and Mann, G.J. 2001. Functional impairment of melanoma-associated p16INK4a mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin. Cancer Res. 7 3282–3288. [PubMed] [Google Scholar]
- Binz, H.K., Stumpp, M.T., Forrer, P., Amstutz, P., and Pluckthun, A. 2003. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 332 489–503. [DOI] [PubMed] [Google Scholar]
- Bockers, T.M., Mameza, M.G., Kreutz, M.R., Bockmann, J., Weise, C., Buck, F., Richter, D., Gundelfinger, E.D., and Kreienkamp, H.-J. 2001. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein α-fodrin. J. Biol. Chem. 276 40104–40112. [DOI] [PubMed] [Google Scholar]
- Bork, P. 1993. Hundreds of ankyrin-like repeats in functionally diverse proteins: Mobile modules that cross phyla horizontally? Proteins 17 363–374. [DOI] [PubMed] [Google Scholar]
- Bradley, C.M. and Barrick, D. 2002. Limits of cooperativity in a structurally modular protein: Response of the Notch ankyrin domain to analogous ala-nine substitutions in each repeat. J. Mol. Biol. 324 373–386. [DOI] [PubMed] [Google Scholar]
- Breeden, L. and Nasmyth, K. 1987. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature 329 651–654. [DOI] [PubMed] [Google Scholar]
- Cammett, T.J., Luo, L., and Peng, Z.-y. 2003. Design and characterization of a hyperstable p16INK4a that retsores Cdk4 binding activity when combined with oncogenic mutations. J. Mol. Biol. 327 285–297. [DOI] [PubMed] [Google Scholar]
- Chi, N.-W. and Lodish, H.F. 2000. Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275 38437–38444. [DOI] [PubMed] [Google Scholar]
- Davis, J. and Bennett, V. 1984. Brain ankyrin. A membrane-associated protein with binding sites for spectrin, tubulin, and the cytoplasmic domain of the erythrocyte anion channel. J. Biol. Chem. 259 13550–13559. [PubMed] [Google Scholar]
- ———. 1990. The anion exchanger and Na+K(+)-ATPase interact with distinct sites on ankyrin in in vitro assays. J. Biol. Chem. 265 17252–17256. [PubMed] [Google Scholar]
- ———. 1994. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 269 27163–27166. [PubMed] [Google Scholar]
- Davis, L., Otto, E., and Bennett, V. 1991. Specific 33-residue repeat(s) of erythrocyte ankyrin associate with the anion exchanger. J. Biol. Chem. 266 11163–11169. [PubMed] [Google Scholar]
- Davis, J.Q., McLaughlin, T., and Bennett, V. 1993. Ankyrin-binding proteins related to nervous system cell adhesion molecules: Candidates to provide transmembrane and intercellular connections in adult brain. J. Cell. Biol. 121 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, S., Apcher, S., Mee, M., Higashitsuji, H., Baker, R., Uhle, S., Dubiel, W., Fujita, J., and Mayer, R.J. 2002. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J. Biol. Chem. 277 10893–10902. [DOI] [PubMed] [Google Scholar]
- Dechend, R., Hirano, F., Lehmann, K., Heissmeyer, V., Ansieau, S., Wulczyn, F., Scheidereit, C., and Leutz, A. 1999. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 18 3316–3323. [DOI] [PubMed] [Google Scholar]
- DeSandro, A.M., Nagarajan, U.M., and Boss, J.M. 2000. Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20 6587–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais, J.R. and Berg, J.M. 1993. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl. Acad. Sci. 90 2256–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich, R., Matsuno, K., Hing, H., and Artavanis-Tsakonas, S. 1994. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development 120 473–481. [DOI] [PubMed] [Google Scholar]
- Dobrzanski, P., Ryseck, R.P., and Bravo, R. 1995. Specific inhibition of RelB/ p52 transcriptional activity by the C-terminal domain of p100. Oncogene 10 1003–1007. [PubMed] [Google Scholar]
- Dong, B. and Silverman, R.H. 1997. A bipartite model of 2–5A-dependent RNase L. J. Biol. Chem. 272 22236–22242. [DOI] [PubMed] [Google Scholar]
- Dueber, J.E., Yeh, B.J., Chak, K., and Lim, W.A. 2003. Reprogramming control of an allosteric signaling switch through modular recombination. Science 301 1904–1908. [DOI] [PubMed] [Google Scholar]
- Gobel, U., Sander, C., Schneider, R., and Valencia, A. 1994. Correlated mutations and residue contacts in proteins. Proteins 18 309–317. [DOI] [PubMed] [Google Scholar]
- Gorina, S. and Pavletich, N.P. 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274 1001–1005. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. and Beach, D. 1994. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature 371 257–261. [DOI] [PubMed] [Google Scholar]
- Harbury, P.B., Zhang, T., Kim, P.S., and Alber, T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262 1401–1407. [DOI] [PubMed] [Google Scholar]
- Hatada, E., Nieters, A., Wulczyn, F., Naumann, M., Meyer, R., Nucifora, G., McKeithan, T., and Scheidereit, C. 1992. The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc. Natl. Acad. Sci. 89 2489–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps, N.R., Barker, H.M., Elledge, S.J., and Cohen, P.T.W. 1995. Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett. 377 295–300. [DOI] [PubMed] [Google Scholar]
- Heringa, J. 1998. Detection of internal repeats: How common are they? Curr. Opin. Struct. Biol. 8 338–345. [DOI] [PubMed] [Google Scholar]
- Hirai, H., Roussel, M., Kato, J., Ashmun, R., and Sherr, C. 1995. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol. Cell. Biol. 15 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., Costanzo, M., Moore, L., Kobayashi, R., and Andrews, B.J. 1999. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19 5267–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxford, T., Huang, D., Malek, S., and Ghosh, G. 1998. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 95 759–770. [DOI] [PubMed] [Google Scholar]
- Inoue, J., Kerr, L.D., Kakizuka, A., and Verma, I.M. 1992. IκBγ, a 70 kd protein identical to the C-terminal half of p110 NF-κB: A new member of the IκB family. Cell 68 1109–1120. [DOI] [PubMed] [Google Scholar]
- Iwabuchi, K., Bartel, P., Li, B., Marraccino, R., and Fields, S. 1994. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. 91 6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, M. and Harrison, S. 1998. Structure of an IκBα/NF-κB complex. Cell 95 749–758. [DOI] [PubMed] [Google Scholar]
- Jonas-Straube, E., Hutin, C., Hoffman, N.E., and Schunemann, D. 2001. Functional analysis of the protein-interacting domains of chloroplast SRP43. J. Biol. Chem. 276 24654–24660. [DOI] [PubMed] [Google Scholar]
- Joutel, A., Corpechot, C., Ducros, A., Vahedi, K., Chabriat, H., Mouton, P., Alamowitch, S., Domenga, V., Cecillion, M., Marechal, E., et al. 1996. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383 707–710. [DOI] [PubMed] [Google Scholar]
- Kato, H., Taniguchi, Y., Kurooka, H., Minoguchi, S., Sakai, T., Nomura-Okazaki, S., Tamura, K., and Honjo, T. 1997. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124 4133–4141. [DOI] [PubMed] [Google Scholar]
- Kidd, S. 1992. Characterization of the Drosophila cactus locus and analysis of interactions between cactus and dorsal proteins. Cell 71 623–635. [DOI] [PubMed] [Google Scholar]
- Kleiman, F.E. and Manley, J.L. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285 1576–1579. [DOI] [PubMed] [Google Scholar]
- Koh, J., Enders, G.H., Dynlacht, B.D., and Harlow, E. 1995. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 375 506–510. [DOI] [PubMed] [Google Scholar]
- Kohl, A., Binz, H.K., Forrer, P., Stumpp, M.T., Pluckthun, A., and Grutter, M.G. 2003. Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. 100 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka, H. and Honjo, T. 2000. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275 17211–17220. [DOI] [PubMed] [Google Scholar]
- Larson, S.M., Di Nardo, A.A., and Davidson, A.R. 2000. Analysis of covariation in an SH3 domain sequence alignment: Applications in tertiary contact prediction and the design of compensating hydrophobic core substitutions. J. Mol. Biol. 303 433–446. [DOI] [PubMed] [Google Scholar]
- Lehmann, M., Kostrewa, D., Wyss, M., Brugger, R., D’Arcy, A., Pasamontes, L., and van Loon, A.P. 2000. From DNA sequence to improved functionality: Using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 13 49–57. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. and Tsai, M. 2002. Novel insights into the INK4-CDK4/6-Rb pathway: Counter action of gankyrin against INK4 proteins regulates the CDK4-mediated phosphorylation of Rb. Biochemistry 41 3977–3983. [DOI] [PubMed] [Google Scholar]
- Li, F., Zhang, Y., and Wu, C. 1999. Integrin-linked kinase is localized to cell–matrix focal adhesions but not cell–cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J. Cell. Sci. 112 4589–4599. [DOI] [PubMed] [Google Scholar]
- Lim, S., Sala, C., Yoon, J., Park, S., Kuroda, S., Sheng, M., and Kim, E. 2001. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 17 385–397. [DOI] [PubMed] [Google Scholar]
- Lin, J.-H., Makris, A., McMahon, C., Bear, S.E., Patriotis, C., Prasad, V.R., Brent, R., Golemis, E.A., and Tsichlis, P.N. 1999. The ankyrin repeat-containing adaptor protein Tvl-1 is a novel substrate and regulator of Raf-1. J. Biol. Chem. 274 14706–14715. [DOI] [PubMed] [Google Scholar]
- Lockless, S.W. and Ranganathan, R. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286 295–299. [DOI] [PubMed] [Google Scholar]
- Lukas, J., Parry, D., Aagaard, L., Mann, D.J., Bartkova, J., Strauss, M., Peters, G., and Bartek, J. 1995. Retinoblastoma–protein-dependent cell-cycle inhibition by tumour suppressor p16. Nature 375 503–506. [DOI] [PubMed] [Google Scholar]
- Lux, S.E., John, K.M., and Bennett, V. 1990. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature 344 36–42. [DOI] [PubMed] [Google Scholar]
- Lyons, R.J., Deane, R., Lynch, D.K., Ye, Z.-S.J., Sanderson, G.M., Eyre, H.J., Sutherland, G.R., and Daly, R.J. 2001. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 276 17172–17180. [DOI] [PubMed] [Google Scholar]
- Macias, M.J., Gervais, V., Civera, C., and Oschkinat, H. 2000. Structural analysis of WW domains and design of a WW prototype. Nat. Struct. Biol. 7 375–379. [DOI] [PubMed] [Google Scholar]
- Maekawa, K., Imagawa, N., Naito, A., Harada, S., Yoshie, O., and Takagi, S. 1999. Association of protein-tyrosine phosphatase PTP-BAS with the transcription-factor-inhibitory protein IκBα through interaction between the PDZ1 domain and ankyrin repeats. Biochem. J. 337 179–184. [PMC free article] [PubMed] [Google Scholar]
- Main, E.R., Jackson, S.E., and Regan, L. 2003. The folding and design of repeat proteins: reaching a consensus. Curr. Opin. Struct. Biol. 13 482–489. [DOI] [PubMed] [Google Scholar]
- Malek, S., Huang, D.-B., Huxford, T., Ghosh, S., and Ghosh, G. 2003. X-ray crystal structure of an IκBβ x NF-κB p65 homodimer complex. J. Biol. Chem. 278 23094–23100. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Yeates, T.O., and Eisenberg, D. 1999. A census of protein repeats. J. Mol. Biol. 293 151–160. [DOI] [PubMed] [Google Scholar]
- Masternak, K., Barras, E., Zufferey, M., Conrad, B., Corthals, G., Aebersold, R., Sanchez, J.C., Hochstrasser, D.F., Mach, B., and Reith, W. 1998. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 20 273–277. [DOI] [PubMed] [Google Scholar]
- Matsuno, K., Diederich, R., Go, M., Blaumueller, C., and Artavanis-Tsakonas, S. 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121 2633–2644. [DOI] [PubMed] [Google Scholar]
- Mercurio, F., DiDonato, J.A., Rosette, C., and Karin, M. 1993. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 7 705–718. [DOI] [PubMed] [Google Scholar]
- Michaely, P. and Bennett, V. 1993. The membrane-binding domain of ankyrin contains four independently folded subdomains, each comprised of six ankyrin repeats. J. Biol. Chem. 268 22703–22709. [PubMed] [Google Scholar]
- ———. 1995. Mechanism for binding site diversity on ankyrin. J. Biol. Chem. 270 31298–31302. [DOI] [PubMed] [Google Scholar]
- Michaely, P., Kamal, A., Anderson, R.G.W., and Bennett, V. 1999. A requirement for ankyrin binding to clathrin during coated pit budding. J. Biol. Chem. 274 35908–35913. [DOI] [PubMed] [Google Scholar]
- Michaely, P., Tomchick, D.R., Machius, M., and Anderson, R.G. 2002. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 21 6387–6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Soler-Lopez, M., Petosa, C., Cramer, P., Siebenlist, U., and Muller, C.W. 2001. Crystal structure of the ankyrin repeat domain of Bcl-3: A unique member of the IκB protein family. EMBO J. 20 6180–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.K., Bang, M.-L., Witt, C.C., Labeit, D., Trombitas, C., Watanabe, K., Granzier, H., McElhinny, A.S., Gregorio, C.C., and Labeit, S. 2003. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 333 951–964. [DOI] [PubMed] [Google Scholar]
- Morgans, C. and Kopito, R. 1993. Association of the brain anion exchanger, AE3, with the repeat domain of ankyrin. J. Cell. Sci. 105 1137–1142. [DOI] [PubMed] [Google Scholar]
- Mosavi, L.K. and Peng, Z.Y. 2003. Structure-based substitutions for increased solubility of a designed protein. Protein Eng. 16 739–745. [DOI] [PubMed] [Google Scholar]
- Mosavi, L.K., Minor Jr., D.L., and Peng, Z.Y. 2002a. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. 99 16029–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi, L.K., Williams, S., and Peng, Z.Y. 2002b. Equilibrium folding and stability of myotrophin: A model ankyrin repeat protein. J. Mol. Biol. 320 165–170. [DOI] [PubMed] [Google Scholar]
- Nagao, T., Higashitsuji, H., Nonoguchi, K., Sakurai, T., Dawson, S., Mayer, R.J., Itoh, K., and Fujita, J. 2003. MAGE-A4 interacts with the liver oncoprotein gankyrin and suppresses its tumorigenic activity. J. Biol. Chem. 278 10668–10674. [DOI] [PubMed] [Google Scholar]
- Nam, Y., Weng, A.P., Aster, J.C., and Blacklow, S.C. 2003. Structural requirements for assembly of the CSL•intracellular Notch1•Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278 21232–21239. [DOI] [PubMed] [Google Scholar]
- Naumann, M., Nieters, A., Hatada, E.N., and Scheidereit, C. 1993. NF-κB precursor p100 inhibits nuclear translocation and DNA binding of NF-κB/ rel-factors. Oncogene 8 2275–2281. [PubMed] [Google Scholar]
- Naumovski, L. and Cleary, M. 1996. The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol. Cell. Biol. 16 3884–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrep, N., Geyer, M., Jabrane-Ferrat, N., and Peterlin, B.M. 2001. Analysis of ankyrin repeats reveals how a single point mutation in RFXANK results in bare lymphocyte syndrome. Mol. Cell. Biol. 21 5566–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, V., Le Van Kim, C., Gane, P., Birkenmeier, C., Cartron, J.-P., Colin, Y., and Mouro-Chanteloup, I. 2003. Rh-RhAG/Ankyrin-R, a new interaction site between the membrane bilayer and the red cell skeleton, is impaired by Rh(null)-associated mutation. J. Biol. Chem. 278 25526–25533. [DOI] [PubMed] [Google Scholar]
- Parry, D. and Peters, G. 1996. Temperature-sensitive mutants of p16CDKN2 associated with familial melanoma. Mol. Cell. Biol. 16 3844–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petcherski, A. and Kimble, J. 2000. LAG-3 is a putative transcriptional activator in the C. elegans notch pathway. Nature 405 364–368. [DOI] [PubMed] [Google Scholar]
- Prehoda, K.E., Scott, J.A., Mullins, R.D., and Lim, W.A. 2000. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science 290 801–806. [DOI] [PubMed] [Google Scholar]
- Rader, K., Orlando, R., Lou, X., and Farquhar, M. 2000. Characterization of ANKRA, a novel ankyrin repeat protein that interacts with the cytoplasmic domain of megalin. J. Am. Soc. Nephrol. 11 2167–2178. [DOI] [PubMed] [Google Scholar]
- Reymond, A. and Brent, R. 1995. p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene 11 1173–1178. [PubMed] [Google Scholar]
- Rice, N.R., MacKichan, M.L., and Israel, A. 1992. The precursor of NF-κB p50 has IκB-like functions. Cell 71 243–253. [DOI] [PubMed] [Google Scholar]
- Ruas, M. and Peters, G. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta. 1378 F115–F177. [DOI] [PubMed] [Google Scholar]
- Ruas, M., Brookes, S., McDonald, N.Q., and Peters, G. 1999. Functional evaluation of tumour-specific variants of p16INK4a/CDKN2A: Correlation with protein structure information. Oncogene 18 5423–5434. [DOI] [PubMed] [Google Scholar]
- Russo, A.A., Tong, L., Lee, J.-O., Jeffrey, P.D., and Pavletich, N.P. 1998. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395 237–243. [DOI] [PubMed] [Google Scholar]
- Sbodio, J.I. and Chi, N.-W. 2002. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277 31887–31892. [DOI] [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, R.M. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya, H. and Smith, S. 2002. The telomeric Poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 277 14116–14126. [DOI] [PubMed] [Google Scholar]
- Serrano, M., Hannon, G.J., and Beach, D. 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366 704–707. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J. and Roberts, J.M. 1999. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13 1501–1512. [DOI] [PubMed] [Google Scholar]
- Shindyalov, I.N., Kolchanov, N.A., and Sander, C. 1994. Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations? Protein Eng. 7 349–358. [DOI] [PubMed] [Google Scholar]
- Siegmund, R. and Nasmyth, K. 1996. The Saccharomyces cerevisiae start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol. Cell. Biol. 16 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonidis, S., Liang, S., Chen, G., and Thanos, D. 1997. Cloning and functional characterization of mouse IκBvarɛ. Proc. Natl. Acad. Sci. 94 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian, N., Adhikary, G., Sil, P.C., and Sen, S. 1996. Cardiac myotrophin exhibits rel/NF-κB interacting activity in vitro. J. Biol. Chem. 271 2812–2816. [DOI] [PubMed] [Google Scholar]
- Smith, S., Giriat, I., Schmitt, A., and de Lange, T. 1998. Tankyrase, a poly-(ADP-Ribose) polymerase at human telomeres. Science 282 1484–1487. [DOI] [PubMed] [Google Scholar]
- Srinivasan, Y., Lewallen, M., and Angelides, K. 1992. Mapping the binding site on ankyrin for the voltage-dependent sodium channel from brain. J. Biol. Chem. 267 7483–7489. [PubMed] [Google Scholar]
- Suzuki, T., Kitao, S., Matsushime, H., and Yoshida, M. 1996. HTLV-1 Tax protein interacts with cyclin-dependent kinase inhibitor p16INK4A and counteracts its inhibitory activity towards CDK4. EMBO J. 15 1607–1614. [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Narita, T., Uchida-Toita, M., and Yoshida, M. 1999. Down-regulation of the INK4 family of cyclin-dependent kinase inhibitors by tax protein of HTLV-1 through two distinct mechanisms. Virology 259 384–391. [DOI] [PubMed] [Google Scholar]
- Tan, I., Ng, C.H., Lim, L., and Leung, T. 2001. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 276 21209–21216. [DOI] [PubMed] [Google Scholar]
- Tanaka, J., Ito, M., Feng, J., Ichikawa, K., Hamaguchi, T., Nakamura, M., Hartshorne, D., and Nakano, T. 1998. Interaction of myosin phosphatase target subunit 1 with the catalytic subunit of type 1 protein phosphatase. Biochemistry 37 16697–16703. [DOI] [PubMed] [Google Scholar]
- Tang, K.S., Guralnick, B.J., Wang, W.K., Fersht, A.R., and Itzhaki, L.S. 1999. Stability and folding of the tumour suppressor protein p16. J. Mol. Biol. 285 1869–1886. [DOI] [PubMed] [Google Scholar]
- Tang, K.S., Fersht, A.R., and Itzhaki, L.S. 2003. Sequential unfolding of ankyrin repeats in tumor suppressor p16. Structure (Camb) 11 67–73. [DOI] [PubMed] [Google Scholar]
- Tani, S., Kurooka, H., Aoki, T., Hashimoto, N., and Honjo, T. 2001. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 29 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka, M., Ichimura, T., Wakamiya-Tsuruta, A., Kubota, Y., Araki, T., Obinata, T., and Isobe, T. 2003. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J. Biol. Chem. 278 5864–5870. [DOI] [PubMed] [Google Scholar]
- Tevelev, A., Byeon, I.-J.L., Selby, T., Ericson, K., Kim, H.-J., Kraynov, V., and Tsai, M.-D. 1996. Tumor suppressor p16INK4a: Structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry 35 9475–9487. [DOI] [PubMed] [Google Scholar]
- Thevananther, S., Kolli, A.H., and Devarajan, P. 1998. Identification of a novel ankyrin isoform (AnkG190) in kidney and lung that associates with the plasma membrane and binds α-Na, K-ATPase. J. Biol. Chem. 273 23952–23958. [DOI] [PubMed] [Google Scholar]
- Thompson, J.E., Phillips, R.J., Erdjument-Bromage, H., Tempst, P., and Ghosh, S. 1995. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell 80 573–582. [DOI] [PubMed] [Google Scholar]
- Tripp, K.W. and Barrick, D. 2003. Folding by consensus. Structure (Camb) 11 486–487. [DOI] [PubMed] [Google Scholar]
- Tu, Y., Li, F., Goicoechea, S., and Wu, C. 1999. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 19 2425–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velyvis, A., Yang, Y., Wu, C., and Qin, J. 2001. Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain. J. Biol. Chem. 276 4932–4939. [DOI] [PubMed] [Google Scholar]
- Walker, G.J., Gabrielli, B.G., Castellano, M., and Hayward, N.K. 1999. Functional reassessment of p16 variants using a transfection-based assay. Int. J. Cancer 82 305–312. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Wachi, S., and Fujita, T. 2003. Identification and characterization of BCL-3-binding protein: Implications for transcription and DNA repair or recombination. J. Biol. Chem. 278 26102–26110. [DOI] [PubMed] [Google Scholar]
- Whiteside, S.T., Epinat, J.-C., Rice, N.R., and Israel, A. 1997. IκB ɛ, a novel member of the Iκ B family, controls RelA and cRel NF-κB activity. EMBO J. 16 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewski, W., Fondaneche, M.C., Louise-Plence, P., Prochnicka-Chalufour, A., Selz, F., Picard, C., Le Deist, F., Eliaou, J.F., Fischer, A., and Lisowska-Grospierre, B. 2003. Novel mutations in the RFXANK gene: RFX complex containing in-vitro-generated RFXANK mutant binds the promoter without transactivating MHC II. Immunogenetics 54 747–755. [DOI] [PubMed] [Google Scholar]
- Wolff, B. and Naumann, M. 1999. INK4 cell cycle inhibitors direct transcriptional inactivation of NF-κB. Oncogene 18 2663–2666. [DOI] [PubMed] [Google Scholar]
- Wu, L., Aster, J.C., Blacklow, S.C., Lake, R., Artavanis-Tsakonas, S., and Griffin, J.D. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26 484–489. [DOI] [PubMed] [Google Scholar]
- Wulczyn, F.G., Naumann, M., and Scheidereit, C. 1992. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. Nature 358 597–599. [DOI] [PubMed] [Google Scholar]
- Yang, J.P., Hori, M., Takahashi, N., Kawabe, T., Kato, H., and Okamoto, T. 1999. NF-κB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene 18 5177–5186. [DOI] [PubMed] [Google Scholar]
- Yeh, T.-S., Lin, Y.-M., Hsieh, R.-H., and Tseng, M.-J. 2003. Association of transcription factor YY1 with the high molecular weight notch complex suppresses the transactivation activity of notch. J. Biol. Chem. 278 41963–41969. [DOI] [PubMed] [Google Scholar]
- Zeeb, M., Rosner, H., Zeslawski, W., Canet, D., Holak, T.A., and Balbach, J. 2002. Protein folding and stability of human CDK inhibitor p19(INK4d). J. Mol. Biol. 315 447–457. [DOI] [PubMed] [Google Scholar]
- Zhang, B. and Peng, Z.-y. 1996. Defective folding of mutant p16INK4a proteins encoded by tumor-derived alleles. J. Biol. Chem. 271 28734–28737. [PubMed] [Google Scholar]
- ———. 2000. A minimum folding unit in the ankyrin repeat protein p16(INK4). J. Mol. Biol. 299 1121–1132. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. 1999. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. 96 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, A., Hassel, B., and Silverman, R. 1993. Expression cloning of 2–5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell 72 753–765. [DOI] [PubMed] [Google Scholar]
- Zhou, N.E., Kay, C.M., and Hodges, R.S. 1994. The role of interhelical ionic interactions in controlling protein folding and stability. De novo designed synthetic two-stranded α-helical coiled-coils. J. Mol. Biol. 237 500–512. [DOI] [PubMed] [Google Scholar]
- Zhou, S., Fujimuro, M., Hsieh, J.J.-D., Chen, L., Miyamoto, A., Weinmaster, G., and Hayward, S.D. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D. and Bourguignon, L. 2000. Interaction between CD44 and the repeat domain of ankyrin promotes hyaluronic acid-mediated ovarian tumor cell migration. J. Cell Physiol. 183 182–195. [DOI] [PubMed] [Google Scholar]
- Zou, Y., Evans, S., Chen, J., Kuo, H., Harvey, R., and Chien, K. 1997. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2–5 homeobox gene pathway. Development 124 793–804. [DOI] [PubMed] [Google Scholar]
- Zweifel, M.E. and Barrick, D. 2001. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding. Biochemistry 40 14357–14367. [DOI] [PubMed] [Google Scholar]
- Zweifel, M.E., Leahy, D.J., Hughson, F.M., and Barrick, D. 2003. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 12 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]