Abstract
Methionine aminopeptidase (MetAP) exists in two forms (type I and type II), both of which remove the N-terminal methionine from proteins. It previously has been shown that the type II enzyme is the molecular target of fumagillin and ovalicin, two epoxide-containing natural products that inhibit angiogenesis and suppress tumor growth. By using mass spectrometry, N-terminal sequence analysis, and electronic absorption spectroscopy we show that fumagillin and ovalicin covalently modify a conserved histidine residue in the active site of the MetAP from Escherichia coli, a type I enzyme. Because all of the key active site residues are conserved, it is likely that a similar modification occurs in the type II enzymes. This modification, by occluding the active site, may prevent the action of MetAP on proteins or peptides involved in angiogenesis. In addition, the results suggest that these compounds may be effective pharmacological agents against pathogenic and resistant forms of E. coli and other microorganisms.
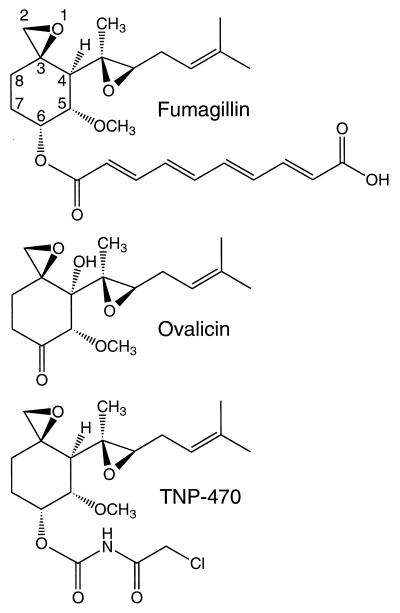
The natural products fumagillin and ovalicin and a related compound, TNP-470, (Fig. 1) have been shown to be potent anti-angiogenic agents (1, 2). These compounds, as well as endostatin and angiostatin, prevent the neovascularization and metastasis of tumors (3–5). Current clinical trials with TNP-470 include patients with cervical cancer, pediatric solid tumors, lymphomas, acute leukemias, and AIDS-related Kaposi’s sarcoma (http://cancertrials.nci.nih.gov and refs. 6–9). Preliminary results suggest that the use of TNP-470, endostatin, and other anti-angiogenesis inhibitors could be a viable approach to avoid drug resistance in cancer therapy (10–12).
Figure 1.
The anti-angiogenesis compounds fumagillin, ovalicin, and TNP-470. The intact epoxide attached to C3 is required for anti-angiogenic activity. Numbering scheme taken from Griffith et al. (14).
The molecular target of fumagillin, ovalicin, and TNP-470 recently was determined to be methionine aminopeptidase type II (MetAP-II) (13, 14). The specific, covalent modification did not block one function of MetAP-II, namely, the prevention of the phosphorylation of the translation initiation factor eIF-2 (14, 15). It did, however, abolish the peptidase activity. This finding strongly implies that the removal of the N-terminal methionine from certain proteins or peptides by MetAP-II is required for angiogenesis. We show here that the MetAP from Escherichia coli, a type I enzyme, also is inhibited by fumagillin and ovalicin and that it is His-79, a conserved residue in the active site, that is covalently modified.
MATERIALS AND METHODS
Expression and Purification of Recombinant E. coli MetAP.
A C-terminal poly-His-tagged form of E. coli MetAP was obtained by overexpression in E. coli. The expression vector was constructed by amplifying the E. coli MetAP gene via the overlap extension method of PCR with Vent DNA polymerase (New England Biolabs) (16, 17). The flanking restriction sites, NcoI and XhoI, the mutation Arg-175–Gln and a C-terminal thrombin cleavage site were added with the following primers: 5′ end primer, 5′-GAA TCC ATG GCT ATC TCA ATC-3′; 3′ end primer, 5′-ATC TTA CTC GAG GCT ACC ACG CGG CAC CAG CTC GTC GTG CGA GAT-3′; Arg-175–Gln mutant, 5′-CTT CAT GGA AGC CCT GGC CGA TAC CG-3′ and 5′-GGC TTC CAT GAA GAA CCG CAG G-3′. The resulting product was cloned into the Novagen pet28b expression vector and verified by sequencing. The Arg-175–Gln mutation eliminates a secondary thrombin cleavage site on a surface loop and does not affect activity. Four liter fermentation cultures of BL21(DE3) E. coli cells containing the expression plasmid were grown in Luria–Bertani broth with kanamycin (100 mg/liter) at 37°C. Expression was induced by the addition of isopropyl β-d-thiogalactoside to 1 mM at 1.0 OD600 for 3 hr at 25°C. The cells were lysed by French Press in 100 ml of +T/G buffer [50 mM Hepes, pH 7.9/10% glycerol/0.1% Triton X-100/0.5 M KCl/40 μg/ml DNase/1 mM MgCl2/15 mM methionine/5 mM imidazole/2 Complete/EDTA-free (Boehringer Mannheim) inhibitor tablets] and centrifuged at 40,000 × g for 45 min. The supernatant was loaded onto a 10-ml nitrilotriacetic acid-agarose column (Qiagen) equilibrated with +T/G buffer. After washing with +T/G and −T/G buffer (+T/G buffer without glycerol, Triton X-100, and inhibitor cocktail), MetAP was eluted with −T/G buffer containing 60 mM imidazole directly into 1 ml of 500 mM EDTA, pH 8.0. Additional EDTA was added, if necessary, to give a final concentration of 5 mM. After dialysis at 4°C against 25 mM Hepes buffer, pH 7.9, 150 mM KCl, 15 mM methionine, the poly-His tail was removed by incubation of 100–200 mg of MetAP with no more than 0.25 units/mg of biotinylated thrombin (Novagen) at 15°C for 18–20 hr. The biotinylated thrombin was eliminated by treatment with excess streptavidin agarose (Novagen) prewashed with −T/G buffer. Passage of the protein through another nitrilotriacetic acid-agarose column equilibrated with −T/G resulted in His-tag free protein that was subsequently loaded onto a Superdex 75 Hi-load, prep-grade 16/60 gel filtration column (Pharmacia) equilibrated with 25 mM Hepes, pH 6.8, 25 mM K2SO4, 100 mM NaCl, 1 mM CoCl2, 15 mM methionine. Protein concentrations were determined by absorption at 280 nm with the extinction coefficient of 16,350 M−1⋅cm−1 calculated by using the Genetics Computer Group program peptidesort. Typical yields were 125–200 mg/liter of culture.
The His-79–Ala mutant of the E. coli MetAP was obtained by using the same molecular biology and protein purification procedures. The following primers were used to generate the mutation: 5′-CCG GGA TCC CTG CGC ACA CCA CTT C-3′ and 5′-GGG ATC CCG GAC GAT GCT AAG C-3′. The protein incorporated Co(II) in the same manner as the wild-type enzyme.
Preparation and Purification of MetAP-Fumagillin (MetAP-Fum) and MetAP-Ovalicin Complexes.
E. coli MetAP (120 μM) was treated with a 20-fold molar excess (2.4 mM) of fumagillin (Sigma) or ovalicin (gift from P. Bollinger, Novartis Pharma AG) (dissolved in dimethyl sulfoxide) in 50 mM Hepes, pH 7.5/50 mM KCl/1 mM CoCl2 at 30°C for 30 min. Unreacted fumagillin or ovalicin was removed, and the buffer was changed to 20 mM Hepes, pH 7.4/1 mM CoCl2 by passing the protein through a Pharmacia PD10 column. Electronic absorption spectra were recorded with a Shimadzu (model UV-2101PC) spectrophotometer at room temperature.
Yeast MetAP type I (yMetAP-I) (5.4 μM) (gift from K. Walker and R. Bradshaw, University of California, Irvine) was treated with up to 500-fold excess (2.7 mM) fumagillin or ovalicin under the same reaction conditions as the E. coli MetAP.
MetAP-fum and MetAP-ovalicin complexes were purified by RP-HPLC using a Vydac C18 column (218TP5415, 4.6 × 150 mm, The Separations Group) with mobile phases: A, 0.1% trifluoroacetic acid (TFA); B, 70% acetonitrile, 0.1% TFA. Chromatography was performed on a Beckman 126 Gold system with simultaneous detection at 220 and 335 nm and a 30-min gradient from either 65% B to 85% B (E. coli MetAP) or 50% to 80% B (yMetAP) at 0.7 ml/min. Mass analysis of the E. coli MetAP-fum complex was obtained by ionspray mass spectrometry on a Perkin–Elmer Sciex API III+ triple quadrupole mass spectrometer. The potential of the ionspray needle was placed at 5,000 V. Mass analysis of the E. coli and the yeast MetAP-ovalicin complexes was performed by matrix-assisted laser desorption ionization (MALDI) on a custom-built time-of-flight mass spectrometer equipped with a two-stage delayed extraction source. Approximately 1 μl of the sample was mixed with 3 μl of n-octylglucoside/2,5-dihydroxybenzoic acid (E. coli MetAP) or α-cyano-4-hydroxycinnamic acid (yMetAP-I); a 0.5 μl droplet of this mix was deposited onto the sample probe and allowed to air dry. Mass spectra were produced by irradiating the sample with a (337 nm) N2-laser (LSI) with operation of the ion source at 21 kV with a 150 ns/1.8 kV delay. The mass spectrum was recorded as the sum of 30 consecutive spectra, each produced by a single pulse of photons. Ions from an added standard were used for mass calibration.
In a test of the specificity of fumagillin and ovalicin for the active site, E. coli MetAP (34 μM) and yMetAP-I (5.4 μM) were preincubated for 5 min at 30°C with increasing concentrations (5.4 μM to 5.4 mM) of the competitive, peptidomimetic, bestatin-based inhibitor (2S,3R)-3-amino-2-hydroxyheptanoic acid (E. coli MetAP, Ki 24 nM) (18). The extent of fumagillin or ovalicin modification (2.4 or 2.7 mM as previously described) was determined by RP-HPLC after further incubation for 15 min. Formation of both complexes resulted in a clear shift in retention time of the protein and, for fumagillin, the peak had absorbance at 335 nm.
Peptidase Activity Assay.
The enzymatic activity of MetAP was assayed with the substrate norleucine-Ala-Ala-Glu-Glu after a 2-min preincubation with different concentrations of fumagillin at 30°C. The 15-μl reaction mixtures contained 0.05 μg enzyme, 4 mM substrate, 50 mM Hepes, pH 7.5, 50 mM KCl, and 1 mM CoCl2. After 10-min incubation at 30°C, enzymatic activity was quenched by the addition of EDTA, and the products were derivatized with the AccQ-Fluor Reagent Kit (Waters) (19). The extent of the removal of the N-terminal norleucine (a surrogate for methionine) was determined by quantitation of the remaining peptide by using micellar electrokinetic chromatography on a Hewlett Packard HP3D capillary electrophoresis system. The electropherogram-run parameters were: 50 μm × 56 cm extended light path fused-silica capillary; buffer, 100 mM borate, pH 9.2/100 mM SDS/15% methanol; run conditions, 30 kV, 25°C, 50 mbar × 3-sec injection; peak detection at 254 nm.
Proteolytic Digestion of the E. coli MetAP-Fum Complex.
The most efficient yield of proteolytic products was obtained by digestion of the non-RP-HPLC purified MetAP-fum complex. Four nanomoles of E. coli MetAP-fum were digested with endoproteinase Glu-C (Endo Glu-C, Boehringer Mannheim) overnight at room temperature (12.8 μl of MetAP, 200 μl of 100 mM K2PO4, pH 8.0, 10% acetonitrile, 6 μg of Endo Glu-C). The sample was further digested with endoproteinase Lys-C (Endo Lys-C, Achromobacter Protease I, Wako Pure Chemical, Osaka) overnight at 37°C (5 μg of Endo Lys-C in 200 μl of 100 mM K2PO4, 10% acetonitrile, and the addition of DTT to 5 mM). The resulting sample was spun to remove a dark, flocculant precipitate (DTT plus CoCl2). The pellet was washed with 50 μl of 100 mM K2PO4, 10% acetonitrile, and 5 mM DTT, and the supernatants were pooled. The peptide fragments were alkylated by the addition of 20 μl of 100 mM iodoacetic acid at room temperature for 30 min in the dark. The sample was solubilized with 5% acetonitrile, 0.1% TFA.
Tandem Mass Spectral Analysis and N-Terminal Sequencing.
The alkylated proteolytic digest was chromatographed on a Perkin–Elmer ABI 140B syringe pump with a 0.3 × 300 mm column packed with Vydac 218TP5 C18 material. Peaks were eluted with a gradient of 20% B to 70% B in 50 min at a flow rate of 5 μl/min (mobile phase A, 0.1% TFA; mobile phase B, 0.1% TFA, 70% acetonitrile). Tandem mass spectrometry was performed by using argon with 10% nitrogen as the collision gas in the second quadrupole. The collision gas thickness was set to 270 × 1012 molecules/cm2, and the collision energy was adjusted to 25 V. Daughter scans were accomplished by selecting the ion of interest (m/z 836) with quadrupole 1 (Q1), colliding the ion with argon in Q2, and scanning Q3 to identify the masses of the fragments produced. Peptide sequence was determined by Edman degradation chemistry by using an Applied Biosystems model 470A automated gas-phase protein sequencer equipped with an Applied Biosystems model 120A phenylthiohydantoin analyzer.
RESULTS
His-79 Is Covalently Modified.
Although fumagillin, ovalicin, and TNP-470 (Fig. 1) contain two epoxides, only the epoxide attached to C3 confers anti-angiogenic activity (14). Changes in the substituent at the C6 position influence potency and bioavailability and also may cause different side effects (1, 14). In the case of fumagillin the extended conjugated double-bond system results in a yellow color. This attribute made it possible to use the absorption at 335 nm to locate the residue in E. coli MetAP modified by this inhibitor.
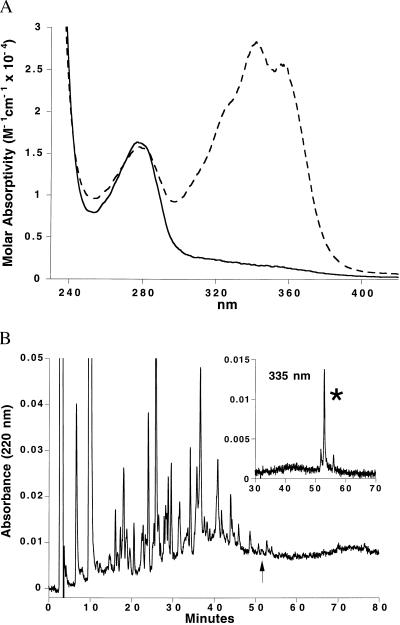
Recombinant E. coli MetAP was irreversibly inactivated by treatment with fumagillin (data not shown). The electronic absorption spectrum of the MetAP-fum complex is shown in Fig. 2A. The mass spectrum of the complex purified by RP-HPLC was consistent with the addition of 1 mol of fumagillin (molecular weight 468.5) per mol of MetAP (MetAP m/z 29630, MetAP-fum m/z 30098). The same stoichiometry was observed for the covalent modification by ovalicin (m/z 29926, ovalicin molecular weight 296.4). Yeast MetAP-I also was modified by only a single ovalicin molecule (yMetAP-I m/z, 43779; yMetAP-I-ovalicin m/z 44075). Consistent with previous reports (13, 14), fumagillin under these reaction conditions did not modify yMetAP-I.
Figure 2.
Analysis of the MetAP-fum complex. (A) Electronic absorption spectrum of E. coli MetAP before (solid line) and after (dashed line) fumagillin treatment and removal of excess fumagillin. The peak at 280 nm derives from the protein aromatic residues whereas the peak at 335 nm matches the absorbance of the conjugated fumagillin double-bond system. The absorption spectrum of the MetAP-ovalicin complex completely lacked the latter peak (not shown). (B) RP-HPLC separation monitored at 220 nm of the MetAP-fum covalent complex after digestion with Endo Glu-C and Endo Lys-C, and treatment with iodoacetic acid. Simultaneous monitoring of the region at 53 min (see arrow) at 335 nm (Inset), uniquely identified the fumagillin-labeled peak (∗) in the complex peptide mixture.
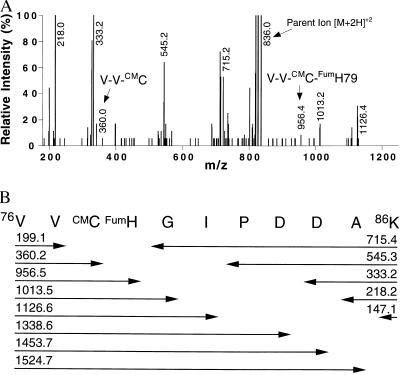
Sequential proteolytic digestion of E. coli MetAP-fum with the endoproteinases Endo Glu-C and Endo Lys-C and alkylation with iodoacetic acid revealed one main product (Fig. 2B) with absorbance at 335 nm. The combined results from N-terminal sequencing and mass spectral analysis yielded the sequence 76VV*C*HGIPDDAK86 (m/z 1670) where *C (Cys-78) and *H (His-79) were derivatized as discussed below.
Tandem mass spectrometry was used to determine which residue was modified by fumagillin. The observed m/z peaks in the fragmentation mass spectrum (Fig. 3A) of the doubly charged precursor ion ([M+2H]+2, m/z 836) correspond directly to the theoretical values (Fig. 3B) for the addition of the fumagillin to His-79 and S-carboxymethylation of Cys-78. Fragments that would coincide with the opposite alkylation pattern were not observed. Remarkably, within the whole protein only His-79 is modified even though other potential nucleophiles (Cys-78, Asp-83, and Asp-84) are nearby in the amino acid sequence. Moreover, the His-79–Ala mutant of E. coli MetAP did not show absorbance at 335 nm or other indications of covalent modification (i.e., change in retention time by RP-HPLC) by fumagillin or ovalicin treatment. RP-HPLC analysis (data not shown) showed that the formation of the E. coli MetAP-fum, E. coli MetAP-ovalicin, and the yMetAP-I-ovalicin complexes could be blocked by the addition of a substrate-like, competitive inhibitor. For example, less than 40% of E. coli MetAP was complexed with fumagillin or ovalicin in the presence of 120 μM competitor. Sixty percent of yMetAP-I was complexed with ovalicin at 54 μM competitor. Higher concentrations of competitor gave almost complete protection against covalent modification.
Figure 3.
Mass spectral analysis of the isolated, fumagillin-linked peptide. (A) A subset of the observed mass fragmentation spectra of the MetAP 76–86 polypeptide containing the fumagillin covalent adduct. The m/z ions are the result of cleavage at the peptide bonds. Fragments with m/z 147.2, 199.2, 1338.8, 1453.6, and 1524.8 values (not shown) also were observed and assigned to the theoretical fragmentation pattern shown in B. Peak heights are normalized to m/z 218. (B) Summary of the theoretical peptide fragmentation pattern with corresponding molecular weights. The spectrum in A is consistent with the addition of fumagillin to His-79 (FumH) and the alkylation of Cys-78 (CMC).
Spectroscopic Evidence for Binding in the Active Site.
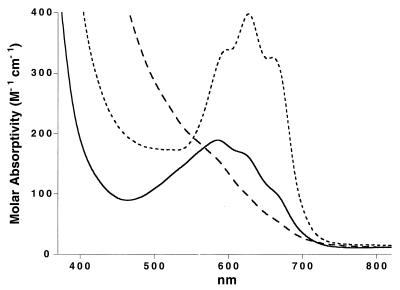
As shown in Fig. 4, the nonmodified enzyme has several weak absorption bands between 550 and 700 nm, which arise from d→d transitions of the d7 Co(II) ions (26, 27). The electronic spectrum of the MetAP-fum covalent complex has significantly decreased absorption in this region. This observation suggests that one or both of the Co(II) ions may be displaced upon complex formation. In marked contrast, the MetAP-ovalicin covalent complex has dramatically increased absorption centered at 627 nm. The red shift in the λmax and the increase in the molar absorptivity are diagnostic of a significant perturbation of the Co(II) coordination environment and are consistent with a decrease in the coordination number and an alteration of the coordination geometry (26, 27). Such changes could include the displacement of the metal-bridging μ-hydroxo ligand by the C6 keto oxygen atom of ovalicin, as illustrated in Fig. 5A. It is possible that an additional solvent ligand (labeled Wat in Fig. 5A) also might be displaced.
Figure 4.
Electronic absorption spectra from the d→d transitions of the Co(II) ions in the active site of E. coli MetAP (solid line), and the MetAP-fum (long dashed line) and MetAP-ovalicin (short dashed line) covalent complexes. The molar absorptivities were calculated with respect to protein concentration.
Figure 5.
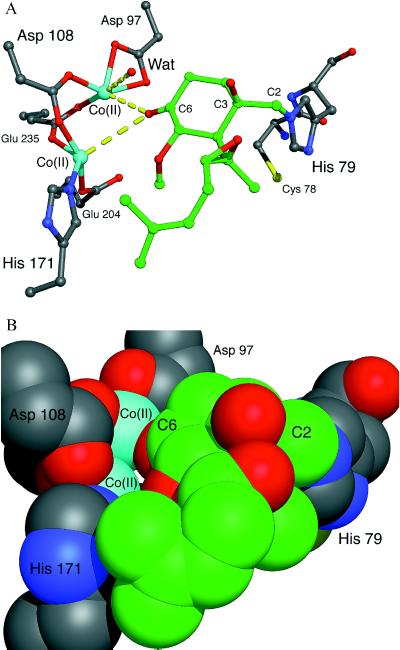
Hypothetical model of ovalicin bound covalently to His-79 in the active site of E. coli MetAP. (A) An energy-minimized model of the complex was made assuming sp3 atoms at C2 and C3 after formation of the covalent bond between C2 and Nɛ2 of His-79. The His-79-ovalicin complex was rotated about the His-79 Cα-Cβ and Cβ-Cγ bonds until the C6 keto oxygen atom of ovalicin was within approximately 2.5 Å of each Co(II) ion. As illustrated via a space filling model (B), access to the metal center is almost completely occluded. Color scheme: protein carbon atoms (gray), ovalicin carbon atoms (green), nitrogen (blue), oxygen (red), cobalt (cyan), and sulfur (yellow). Energy minimizations were performed by using the discover program in the InsightII package (Biosym Technologies, San Diego/Molecular Simulations, Waltham, MA). The figure was made by using midasplus (University of California, San Francisco Computer Graphics Laboratory).
DISCUSSION
X-ray crystallography has demonstrated that the prototypical MetAP from the prokaryote E. coli, a type I enzyme, contains a core catalytic domain (20). Sequence comparisons suggested that the eukaryotic type II enzymes contain a similar catalytic domain and, in addition, have a large insertion of approximately 60 residues within a surface loop (21–23). This suggestion has been confirmed directly by the recently determined structures of the MetAP-II from Pyrococcus furiosis (Brookhaven Protein Data Bank, ID code 1xgs) and the aminopeptidase P from E. coli (24, 25). In addition, some type I and type II MetAPs display large N-terminal extensions that contain either zinc-finger motifs or poly-lysine and aspartic acid blocks (21–23). The variety seen in these domains suggests that these MetAPs may have different physiological roles and subcellular localization. The yMetAP-II and the human MetAP-II were identified as the molecular target of fumagillin and fumagillin-related compounds (13, 14). It was reported previously that these compounds did not interact with the type I enzymes (13, 14). For example, 10 μM of ovalicin did not inhibit yMetAP-I (14). By using higher concentrations and longer incubation times, however, we found that ovalicin does inhibit yMetAP-I and E. coli MetAP, both type I enzymes. It might be noted, however, that although fumagillin modified E. coli MetAP, it did not modify the yMetAP-I. These similarities and differences are consistent with all of the methionine aminopeptidases having similar active sites and mechanisms of action but, at the same time, having differences in individual active site residues, which could modulate the behavior toward different substrates and inhibitors.
One side of the active site cavity is composed of a catalytically essential binuclear metal center created by five strictly conserved ligands: two aspartic acids (Asp-97 and Asp-108, E. coli numbering), two glutamic acids (Glu-204 and Glu-235), and one histidine (His-171) (20). The only other invariant amino acids in the active site are His-79 and His-178, located on opposite sides of the cavity and slightly above the metal center (20, 21).
There are four independent lines of evidence showing that fumagillin and its congeners bind within the active site of E. coli MetAP. First, the inhibitors specifically and covalently modify His-79, which is within the active site. Second, the His-79–Ala mutant was not modified. Third, the inhibitors perturb the spectrum of the catalytically essential metal center. Fourth, the addition of fumagillin and ovalicin to MetAP was blocked by preincubation with a high-affinity, substrate-like inhibitor.
Model of the MetAP-Ovalicin Complex.
The covalent attachment at His-79 and the perturbation of the Co(II) metal center prompted us to model ovalicin into the active site of E. coli MetAP. The crystal structure of E. coli MetAP (20) recently extended to 1.9 Å resolution in a new crystal form (W.T.L. and B.W.M., unpublished data), shows that the ɛ-nitrogen atom (Nɛ2) of His-79 is approximately 8 Å from the active site binuclear Co(II) center. As illustrated in Fig. 5A, ovalicin spans this space in a very plausible manner.
The model assumes (i) covalent attachment between Nɛ2 of His-79 and the C2 of ovalicin after a base-catalyzed reaction (28), (ii) minimized van der Waals overlap of ovalicin and the protein, and (iii) an interaction between the C6 keto oxygen of ovalicin and the binuclear Co(II) center reminiscent of the peptidyl keto group at the scissile peptide bond. The latter interaction is possible only for ovalicin because both fumagillin and TNP-470 have extended substituents at the C6 position (Fig. 1). This model is consistent with the differences in the spectroscopic properties of the MetAP-fum and ovalicin complexes (Fig. 4). It also is consistent with the observation that the different inhibitors have different binding affinities. [The IC50 for ovalicin against human MetAP-II is 0.4 nM and for TNP-470 is 1.0 nM (14). The IC50 value for fumagillin currently is unknown.] The model, however, does not preclude the possibility of a different orientation of ovalicin if structural changes occur in the active site upon binding. For example, ovalicin and other fumagillin analogs could bind with the conserved hydrophobic C4 substituent in the putative methionine binding pocket. Formation of the covalent complex presumably inactivates the enzyme by steric exclusion of substrate from the active site (Fig. 5B).
Implications of the Inactivation of MetAP by Fumagillin-Related Compounds.
The high degree of conservation of the active site region of MetAPs suggests that the histidine in the type II enzymes that is structurally equivalent to His-79 in the E. coli enzyme (i.e., His-231 in human and His-174 in yMetAP-II) is modified by fumagillin-related compounds. Experimental results imply that the inhibition of MetAP peptidase activity disrupts the intricate balance between positive and negative regulators of angiogenesis (29, 30). Although these compounds have been used primarily in cancer therapy, they also may be effective in noncancerous diseases of aberrant vascularization (29, 30) and the treatment of pathogenic microorganisms, including E. coli (31–34) and microsporidia in AIDS patients (35).
Acknowledgments
We thank David Madden for enthusiastic help and Elisabeth Barofsky and Donald A. Griffin for mass spectrometric analysis (Environmental Health Science Center at Oregon State University, National Institute and Environmental Health Science Grant ES00210). We thank Dr. Daniel Rich (University of Wisconsin-Madison) for the peptidomimetic inhibitor and Dr. Pietro Bollinger (Novartis Pharma AG) for the ovalicin sample. We also thank Dr. Ken Walker and Dr. Ralph Bradshaw for the yeast MetAP-I sample and for insightful comments on the manuscript. This work was supported in part by the National Research Service Award F32-GM17536 (W.T.L.) and Research Grant GM20066 (B.W.M.) from the National Institutes of Health.
ABBREVIATIONS
- MetAP
methionine aminopeptidase
- MetAP-fum
MetAP-fumagillin
- yMetAP-I
yeast MetAP type I
- TFA
trifluoroacetic acid
References
- 1. Ingber D, Fujita T, Kishimoto S, Sudo K, Kanamaru T, Brem H, Folkman J. Nature (London) 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 2.Corey E J, Guzman-Perez A, Noe M C. J Am Chem Soc. 1994;116:12109–12110. [Google Scholar]
- 3.O’Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly M S, Holmgren L, Shing Y, Chen C, Rosenthal R A, Moses M, Lane W S, Cao Y, Sage E H, Folkman J. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 5.Zetter B R. Annu Rev Med. 1998;49:407–424. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 6.Borman, S. (1997) Chem. Eng. News, June 16, 39.
- 7.Hawkins M J. Curr Opin Oncol. 1995;7:90–93. [PubMed] [Google Scholar]
- 8.Twardowski P, Gradishar W J. Curr Opin Oncol. 1997;9:584–589. doi: 10.1097/00001622-199711000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Dezube B J, Von Roenn J H, Holden-Wiltse J, Cheung T W, Remick S C, Cooley T P, Moore J, Sommadossi J P, Shriver S L, Suckow C W, Gill P S. J Clin Oncol. 1998;16:1444–1449. doi: 10.1200/JCO.1998.16.4.1444. [DOI] [PubMed] [Google Scholar]
- 10.Brem H, Goto F, Budson A, Saunders L, Folkman J. Surg Forum. 1994;45:674–677. [Google Scholar]
- 11.Boehm T, Folkman J, Browder T, O’Reilly M S. Nature (London) 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 12.Kerbel R S. Nature (London) 1997;390:335–336. doi: 10.1038/36978. [DOI] [PubMed] [Google Scholar]
- 13.Sin N, Meng L, Wang M Q, Wen J J, Bornmann W G, Crews C M. Proc Natl Acad Sci USA. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith E C, Su Z, Turk B E, Chen S, Chang Y H, Wu Z, Biemann K, Liu J O. Chem Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 15.Ray M K, Datta B, Chakraborty A, Chattopadhyay A, Meza-Keuthen S, Gupta N K. Proc Natl Acad Sci USA. 1992;89:539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Bassat A, Bauer K, Chang S Y, Myambo K, Boosman A, Chang S. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Lim S, Rich D H. In: Peptides: Chemistry, Structure, and Biology. Hodges R S, Smith J A, editors. Leiden, The Netherlands: ESCOM; 1994. pp. 625–627. [Google Scholar]
- 19.Zuo S, Guo Q, Chang Y. Anal Biochem. 1994;222:514–516. doi: 10.1006/abio.1994.1529. [DOI] [PubMed] [Google Scholar]
- 20.Roderick S L, Matthews B W. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 21.Bazan J F, Weaver L H, Roderick S L, Huber R, Matthews B W. Proc Natl Acad Sci USA. 1994;91:2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arfin S M, Kendall R L, Hall L, Weaver L H, Stewart A E, Matthews B W, Bradshaw R A. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradshaw R A, Brickey W W, Walker K W. Trends Biochem Sci. 1998;23:263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 24.Tahirov T H, Oki H, Tsukihara T, Ogasahara K, Izu Y, Tsunasawa S, Kato I, Yutani K. Acta Crystallogr D. 1997;53:798–801. doi: 10.1107/S0907444997006793. [DOI] [PubMed] [Google Scholar]
- 25.Wilce M C, Bond C S, Dixon N E, Freeman H C, Guss J M, Lilley P E, Wilce J A. Proc Natl Acad Sci USA. 1998;95:3472–3477. doi: 10.1073/pnas.95.7.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertini I, Luchinat C. Adv Inorg Biochem. 1984;6:71–111. [PubMed] [Google Scholar]
- 27.Maret W, Vallee B L. Methods Enzymol. 1993;226:52–71. doi: 10.1016/0076-6879(93)26005-t. [DOI] [PubMed] [Google Scholar]
- 28.Morrison R T, Boyd R N. Organic Chemistry. Boston: Allyn and Bacon; 1987. pp. 713–721. [Google Scholar]
- 29.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 30.Folkman J. N Engl J Med. 1995;333:1757–1763. doi: 10.1056/NEJM199512283332608. [DOI] [PubMed] [Google Scholar]
- 31.Chang S Y, McGary E C, Chang S. J Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C G, Kukral A M, Miller J L, Movva N R. J Bacteriol. 1989;171:5215–5217. doi: 10.1128/jb.171.9.5215-5217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo S, Guo Q, Ling C, Chang Y H. Mol Gen Genet. 1995;246:247–253. doi: 10.1007/BF00294688. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Chang Y H. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coyle C, Kent M, Tanowitz H B, Wittner M, Weiss L M. J Infect Dis. 1998;177:515–518. doi: 10.1086/517390. [DOI] [PubMed] [Google Scholar]