Abstract
Laccases and other four-copper oxidases are usually constructed of three domains: Domains one and three house the copper sites, and the second domain often helps form a substrate-binding cleft. In contrast to this arrangement, the genome of Streptomyces coelicolor was found to encode a small, four-copper oxidase that lacks the second domain. This protein is representative of a new family of enzymes—the two-domain laccases. Disruption of the corresponding gene abrogates laccase activity in the growth media. We have recombinantly expressed this enzyme, called SLAC, in Escherichia coli and characterized it. The enzyme binds four copper ions/monomer, and UV-visible absorption and EPR measurements confirm that the conserved type 1 copper site and trinuclear cluster are intact. We also report the first known paramagnetic NMR spectrum for the trinuclear copper cluster of a protein from the laccase family. The enzyme is highly stable, retaining activity as a dimer in denaturing gels after boiling and SDS treatment. The activity of the enzyme against 2,6-dimethoxyphenol (DMP) peaks at an unprecedentedly high pH (9.4), whereas the activity against ferrocyanide decreases with pH. SLAC binds negatively charged substrates more tightly than positively charged or uncharged molecules.
Keywords: laccase, NMR, activity, multicopper, oxidase, copper, PPO
The laccase class of enzymes is widely distributed throughout nature and exhibits a wide range of roles and substrate specificity. They are sometimes also referred to as polyphenol oxidases (PPOs) and are a part of the multi-copper oxidase family (Messerschmidt 1997). The first laccase was discovered in the tree Rhus vernicifera more than a hundred years ago (Yoshida 1883). The plant and fungal laccases are similar in that they have low substrate specificity, reacting with a host of derivatized phenols with Km of a few millimolar. The lack of substrate specificity often leaves the natural role of laccases in question, but this versatility has allowed new roles to be found for many in several commercial processes, including delignification, decolorization of industrial waste, and wine clarification (Mayer and Staples 2002).
In contrast, ceruloplasmin (from the plasma of vertebrates) and Fet3p (from Saccharomyces cerevisiae) bind and oxidize Fe(II) specifically. CueO and PcoA are involved with metal homeostasis in Escherichia coli (Rensing and Grass 2003). Ascorbate oxidase, found in many plants, uses ascorbate as a substrate, but for unknown purposes. In this article, all of these proteins will be denoted laccases.
With the recent advent of available genomic data, these enzymes are being isolated from a wide range of organisms, with a large increase in the number of enzymes originating from prokaryotic sources (Claus 2003). Highly conserved features of these proteins include a copper ion bound in a type 1, or blue, site and three more copper ions bound in a trinuclear cluster that is sometimes described as a sum of type 2 and type 3 sites. The polypeptide architecture typically consists of three cupredoxin domains: Disulfide bonds link domain one with domains two and three, and the trinuclear cluster bridges the first and third domains. The reducing substrate is bound in a cleft at the surface and is oxidized by the nearby type 1 site in domain three. Electrons donated by the reducing substrate are transferred to the trinuclear cluster, where dioxygen, the cosubstrate, is reduced to two water molecules (Solomon et al. 1996). Two histidine ligands for the cluster and the cysteine ligand for the type 1 site are consecutive amino acids in sequence (HCH), and thus the distance between the type 1 site and the trinuclear cluster as measured by the number of intervening covalent bonds is quite small.
The substrate-binding clefts of laccases are normally constructed from one loop at the end of domain one, two β-turns from domain three, and several residues from the second domain. A recent crystal structure of the Melanocarpus albomyces laccase (MaL; Hakulinen et al. 2002) implicates several extended loops in domain two that help form a more confining binding pocket, whereas a structure of Trametes versicolor laccase (TvL; Piontek et al. 2002) shows a phenylalanine from domain two in proximity to the substrate. Messerschmidt and coworkers (Messerschmidt et al. 1992) proposed that two tryptophans and a histidine from the second domain in zucchini ascorbate oxidase stabilize the lactone ring of the substrate, ascorbate. Defining the structural constraints for substrate binding is important for applications of these enzymes.
Analysis has suggested that the laccases are the result of replicating a two-domain motif (Messerschmidt and Huber 1990), both domains having cupredoxin folds. Murphy et al. (1997) have predicted that such a precursor could not function as a monomer of two domains. EpoA, a two-domain laccase, has been recently found in Streptomyces griseus but operates as a trimer (Endo et al. 2002). This trimer is stable and active on SDS-PAGE gels, similar to a tetrameric laccase from Gaeumannomyces graminis var. tritici found earlier (Edens et al. 1999).
An important parameter for laccases is the pH-dependence of their activity. For phenolic substrates, the pH-dependence is bell-shaped: An increase with pH is attributed to the pH-dependent decrease in reduction potential of the phenols that formally can be connected to the phenol–phenolate interconversion, whereas the competing atrophy of activity with pH has been ascribed to hydroxide inhibition at the trinuclear cluster (Xu 1997). The maximum of this bell-shaped curve occurs at acidic pHs but has been found as high as pH 9.0 for one laccase. Activity at high pHs is a desired trait for industrial applications.
The above characterization applies to typical laccases with three or more domains. We report the characterization of an enzyme that represents a new family of laccases that possesses only two domains. This laccase lacks the second domain yet exhibits significant activity. We designate the enzyme SLAC, for Small LACcase. SLAC is easily prepared following heterologous expression in E. coli, providing an enzyme that is simple to modify and thus a useful tool to understand activity in all laccases. Experiments show that all three copper sites are intact, and the first NMR spectrum of a laccase trinuclear cluster is presented. SLAC demonstrates resistance to detergents, high thermal stability, and is active as a dimer in both gels and in solution. The activity of SLAC against 2,6-dimethoxyphenol (DMP) reaches a maximum at the unusually high pH of 9.4, suggesting a suitability of the enzyme for industrial processes. The pH-dependencies of DMP activity suggest that SLAC binds negatively charged substrates more tightly, and that the rate of the catalytic step is maximized at pH 8.2.
Results
Alignment
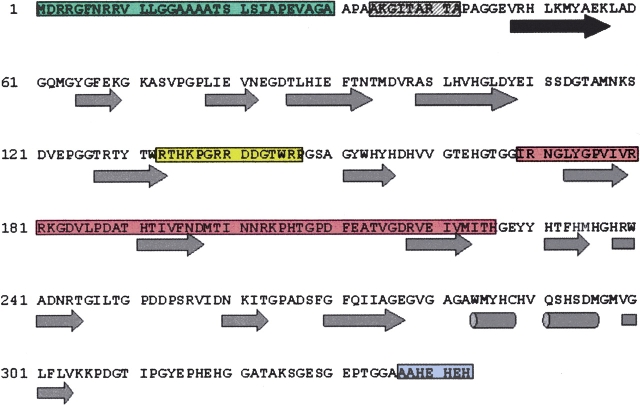
Attempts to align the entire SLAC protein (Fig. 1 ▶) against protein databases resulted in failure because of the large gap penalty of the absent middle domain. Instead, the first and last domains were each aligned, with a portion of the inter-domain region (I169–H206) not included in either alignment. In addition, an insertion (R133–P147, gold-shaded region) not present in the other enzymes was also removed. Interdomain residues T207–H226 are included at the beginning of the second domain alignment to highlight the lack of homology in this region. Alignments of each domain with other laccases appear in Figure 2 ▶. Domain one of SLAC (A31–G168) has an average of 29% identity and 47% conserved homology with the laccases tested, whereas domain two (G227–H343) averages 27% identity and 45% conserved homology with the third domain of the laccases compared (EpoA not included). GenTHREADER (Jones 1999a) predicts that the protein fold of SLAC resembles the folds of both MaL and the Coprinus cinereus laccase (CcL) with >99% confidence. PSIPRED (Jones 1999b) forecasts the same secondary structure as measured in these laccases. It also predicts that the interdomain region consists of three strands and three large random coils (Fig. 1 ▶) without similarity to other laccase domains. SLAC has no cysteines other than the type 1 copper ligand and thus lacks the common disulfide bonds. The only other members of the laccase family that share this feature are EpoA and the six-domain protein ceruloplasmin.
Figure 1.
Analysis of the SLAC sequence. The green box contains the TAT-leader sequence (M1–A30). The hatched gray box contains the starting amino acids of the different N termini that are found in SLAC purified from E. coli (A34–A42). The gold box contains a loop (R133–P147) that is not normally found in laccases and occurs at a position to possibly form a flap over one of the channels leading to the trinuclear cluster. The red box is the approximate region that bridges domains one and two (I169–H226). The mauve box contains the amino acids that are cleaved from the C terminus of SLAC (A337–H343). The secondary structural predictions from PSIPRED, beginning after the TAT-leader sequence, are shown below the amino acid sequence. Arrows represent β-strands, and cylinders represent helices.
Figure 2.
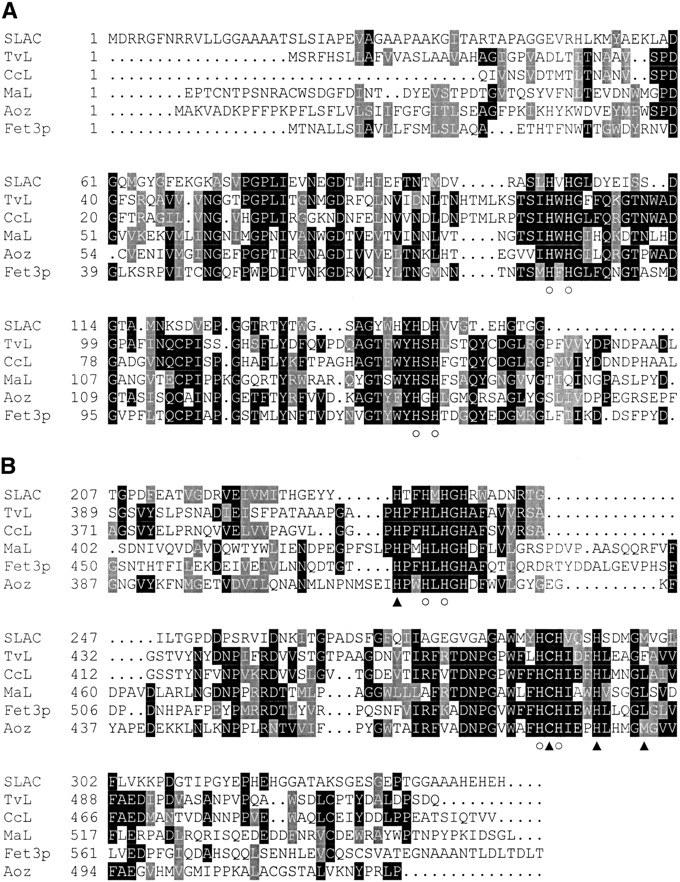
(A) Multiple sequence alignment of SLAC domain one with the first domain of other laccases. The proteins and the accession numbers are SLAC (CAB45586), Trametes versicolor laccase (TvL, A35883), Coprinus cinereus laccase (CcL, 1A65A), Melanocarpus albomyces laccase (MaL, 1GW0A), Saccharomyces cerevisiae Fet3p (Fet3p, CAA89768), and cucumber ascorbate oxidase (Aoz, KSKVAO). Ligands to the type 1 copper site are indicated by filled triangles at the bottom of their columns, and ligands to the trinuclear cluster are indicated by open circles. Black-shaded regions highlight identical amino acids, and gray-shaded regions highlight regions of conserved homology. (B) Multiple sequence alignment of the second domain of SLAC with the third domains of three-domain laccases.
One of the significant results of the alignment is that all the copper ligands are conserved, including a methionine in the predicted axial position of the type 1 site (M298). Other residues that usually form important hydrogen-bonds in the active sites are also present (D113 and D259 in SLAC correspond to D77 and D424 in TvL). It is difficult to comment on the loops that traditionally make up the substrate-binding pocket, because these motifs appear at the end of the first domain and the beginning of the third domain in other laccases. These regions have little homology within the family of proteins. The 15 amino acids (R133–P147) that were left out of alignments are located just prior to a turn normally found in the laccases at the mouth of a channel that leads to the trinuclear cluster. Because there are normally two channels to the trinuclear cluster, it is possible that the enzyme may function even if these residues block one of the openings.
Recombinant expression and purification
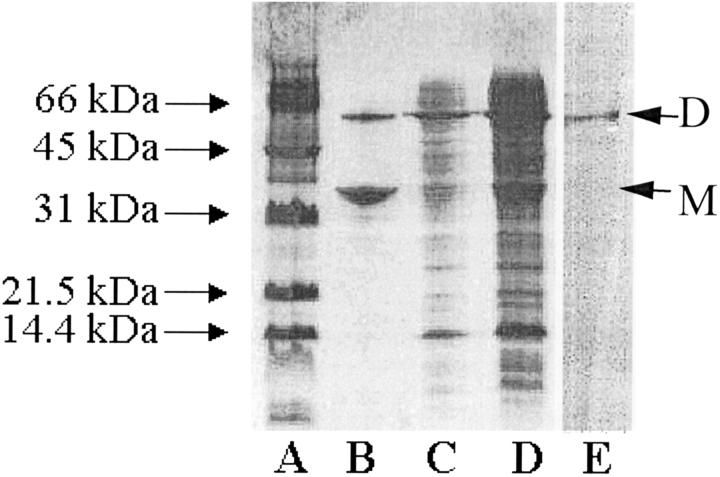
To produce SLAC easily, we cloned the gene into pET20b to make pSLAC1 and transformed the final construct into BL21(DE3). Upon induction with IPTG, a band of ~32 kDa mass appears on SDS-PAGE. The presence of a Streptomyces TAT-leader sequence in the gene led to the expectation of SLAC in the periplasmic fraction (Berks 1996). However, all protein is found in the cytoplasm. Dialysis and anion exchange chromatography purified the protein to >97%. A dimeric band that is thermostable appears on the gels. The band representing the dimer shows activity, but the monomer is inactive (Fig. 3 ▶). Gel filtration experiments (data not shown) confirm a mass representative of a dimer of 69 ± 8 kDa. The predicted pI of the apoenzyme is 6.2, whereas the measured pI of the four-copper oxidized form was 8.2 ± 0.2 and the four-copper reduced form was 7.3 ± 0.2. Mass spectrometry indicates a mass around 32 kDa, much lower than expected (36,875 Da). N-terminal sequencing shows that the polypeptide lacks an N-terminal stretch with varying length corresponding to the first 34–42 amino acids, and C-terminal sequencing shows that a seven-amino-acid fragment is also missing.
Figure 3.
SDS-PAGE gel of SLAC expression, purification, and activity. Lanes A–D are stained with Coomassie blue. (A) Molecular weight standard. (B) Purified SLAC—both monomer and dimer bands are observed. (C) Diluted copper-incubated cytoplasmic fraction of E. coli that has been transformed with pSLAC1, grown, and induced. (D) Cytoplasmic fraction from C directly after cell lysis and before dilution and copper incubation shows both monomeric and dimeric bands. (E) Purified SLAC, identical to lane B, stained by activity against DOPA, showing that activity arises from the dimer. The labels D and M on the right side of the gel designate the positions of the dimer and the monomer bands, respectively.
Streptomyces coelicolor characterization
Activity against DMP was measured for the media of the wild type and gene-disruption mutant. The wild type demonstrated significant activity, whereas the gene-disruption mutant showed no activity (data not shown).
Characterization of recombinant SLAC
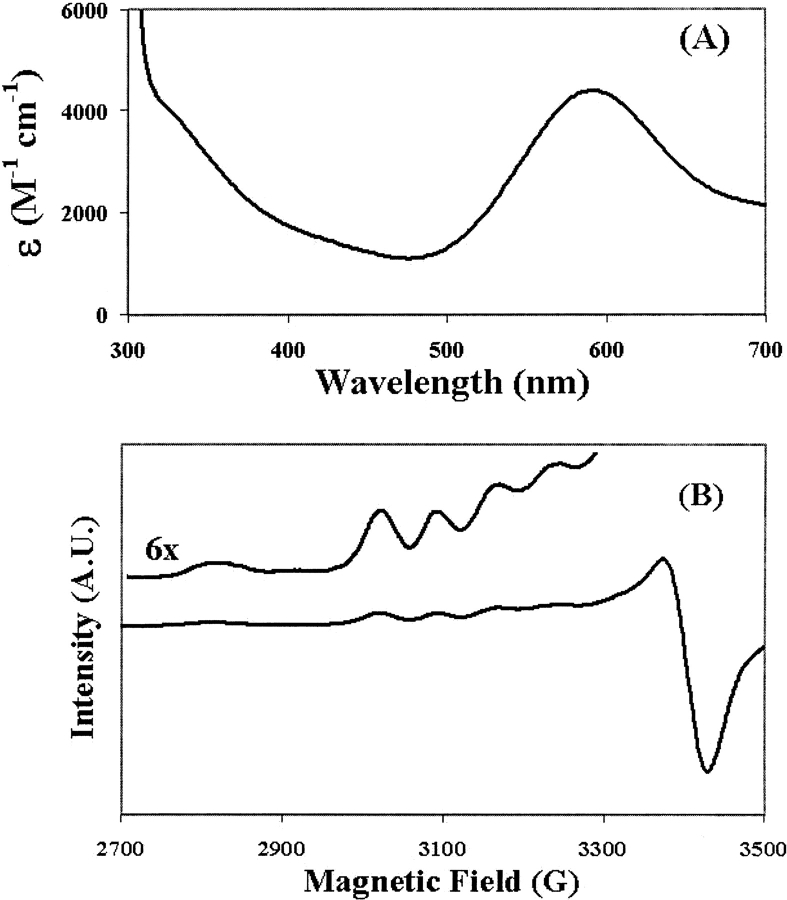
The first property of the enzyme that was noticed was the dramatic blue color of the holoenzyme that is generated by convection of copper-incubated solution with the atmosphere. The type 1 site appears typical in UV-visible (λmax = 590 nm, ɛ = 4400 M−1 cm−1) spectra and demonstrates an axial EPR (g|| = 2.23, A|| = 85 × 10−4 cm−1) spectrum (Fig. 4 ▶). The so-called type 2 copper is EPR-detectable and also axial, but its overlap with the type 1 signal prevents accurate determination of the g-values or the hyperfine splitting. The type 3 copper site dinuclear transition is seen in the UV-visible spectra at 330 nm. BCA quantitation finds 4.0 ± 0.4 copper ions per SLAC monomer, which is confirmed by atomic absorption. The quantification and spectroscopy show that the four copper ions common to laccases and required for activity are present. We have also measured the paramagnetic NMR of the protein. We discern several paramagnetically shifted peaks due to the copper sites, including several with anti-Curie behavior (Fig. 5 ▶).
Figure 4.
(A) UV–visible absorption spectrum of SLAC. The extinction coefficient, ɛ, is plotted on the ordinate axis. (B) Plot of the derivative of the EPR spectrum of SLAC, with an expansion of the g|| region inset.
Figure 5.
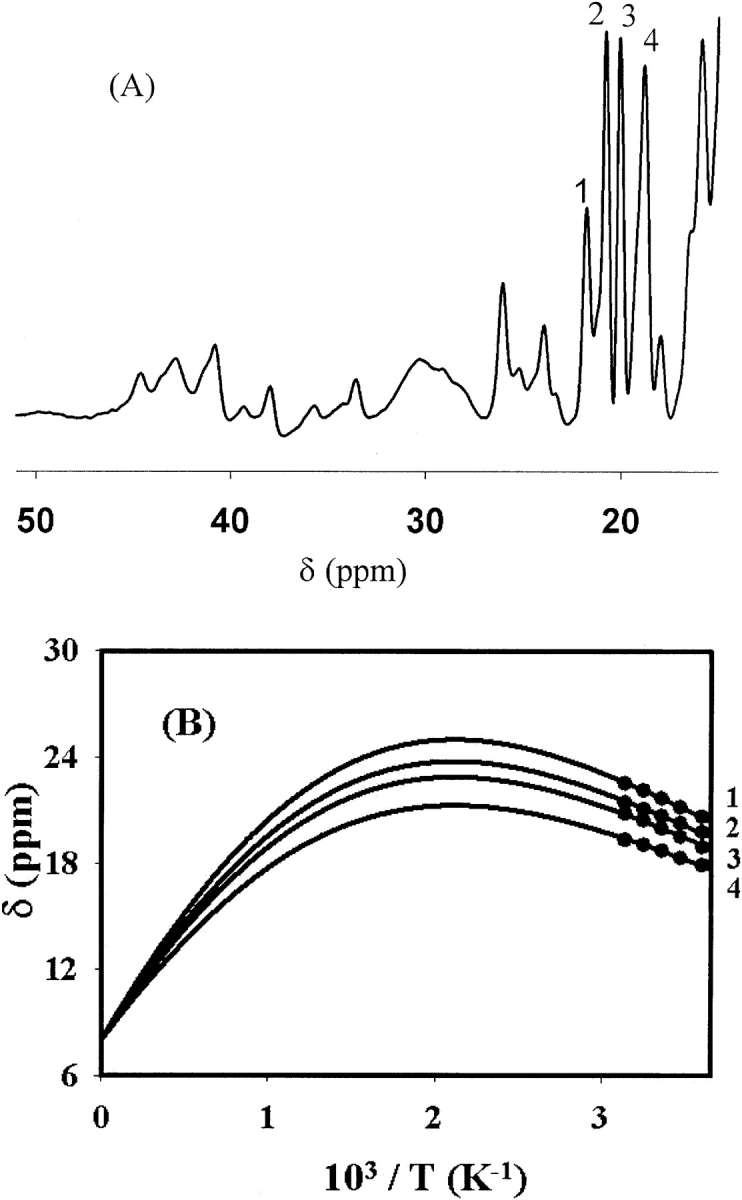
(A) Downfield region of the super-WEFT NMR spectrum of SLAC, showing paramagnetically shifted peaks. (B) The temperature dependencies of peaks 1–4 demonstrate anti-Curie behavior. The filled circles are data points, and the lines represent the best fits to the data using nonlinear regressions of equation 1.
Activity assays
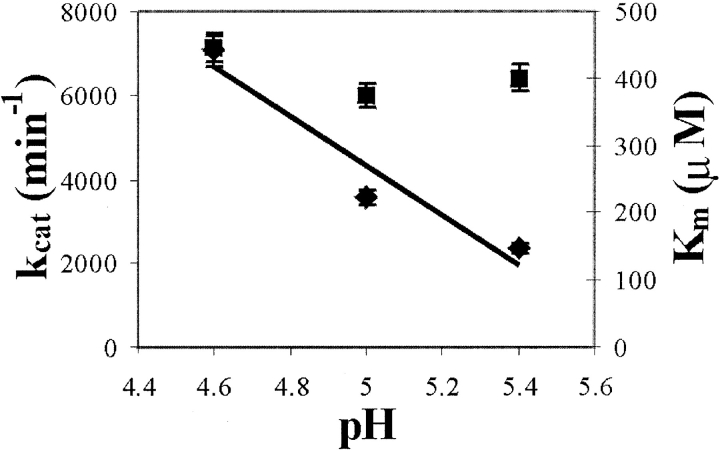
We observed that SLAC has activity against a wide range of substrates, including t-butyl catechol, 3-(3,4-dihydroxyphenyl)-L-alanine (DOPA), syringaldazine (SGZ), DMP, and Fe(CN)64−. The reaction velocities are linear with respect to enzyme concentration. Interactions with DMP and Fe(CN)64− were selected for further characterization; DMP is a member of the phenolic class of substrates that is particularly stable against autooxidation at high pH (because of its high pKa value), and Fe(CN)64− represents nonphenolic substrates. This distinction is important, as the reduction potentials of phenols have a marked pH-dependence. Aqueous iron(II), a substrate of the iron oxidoreductase-type laccases, does not react with SLAC. In contrast, the negatively charged ferrocyanide complex is rapidly oxidized. The Km for ferrocyanide is constant (~400 μM) over the short pH range (4.5 < pH < 5.5) studied. The pH range is limited by protein instability in the acidic regime and substrate auto-oxidation at high concentrations of ferrocyanide at more alkaline pHs. The kcat of Fe(CN)64− decreases with an increase in pH, similar to that observed in all other laccases (Fig. 6 ▶).
Figure 6.
Plots of the Michaelis-Menten constant, Km (squares, right axis), and the kcat (diamonds, left axis) for the reaction of SLAC with ferrocyanide vs. pH. The line represents a linear regression of the data points for kcat.
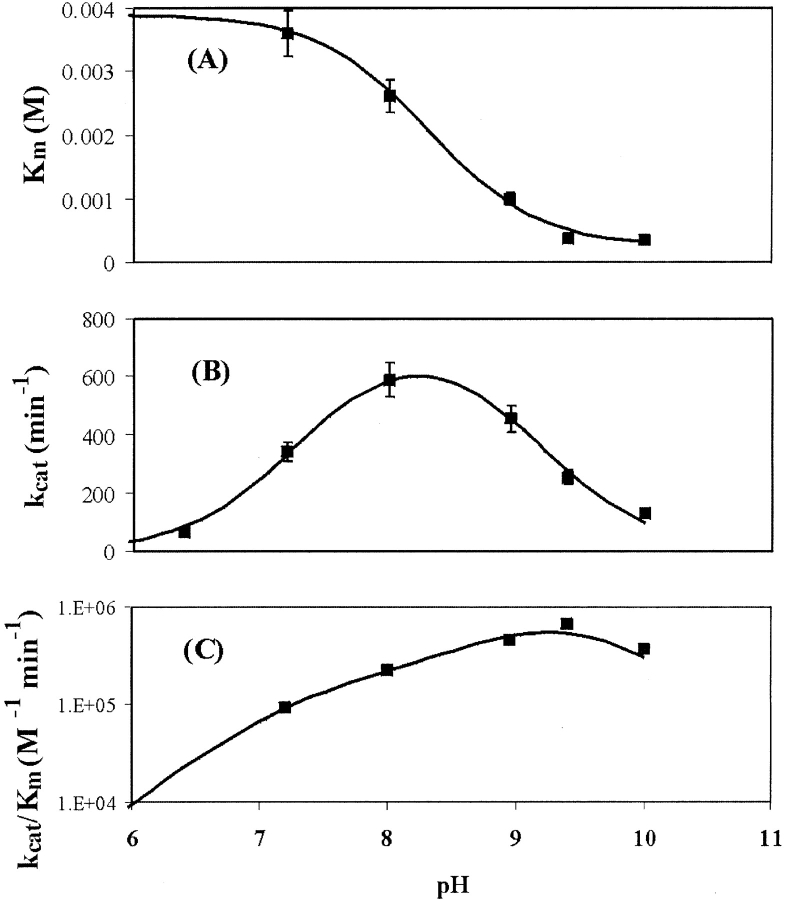
The Km for DMP is pH-dependent, decreasing as the pH increases (Fig. 7 ▶). The binding behavior of DMP is consistent with pKa values of 8.3 ± 0.1 for the bound form and 9.5 ± 0.3 for the free form. Thus, the phenolate is bound more tightly than the phenol. The pKa value of free DMP, 9.6, is in good agreement with this analysis (Xu 1996). The kcat of DMP reaches a maximum at approximately pH 8.2, with the standard bell-shaped dependence seen for other laccases. The derived pKa values are 7.3 ± 0.1 and 9.2 ± 0.1 for the enzyme–DMP complex. The maximum of kcat/Km for DMP occurs at pH 9.4, which is the most alkaline pH optimum measured for laccases.
Figure 7.
Plots of the Michaelis-Menten constant (A) Km, (B) kcat, and (C) the ratio kcat/Km for the reaction of SLAC with DMP vs. pH. The lines represent the best fits to the data using nonlinear regressions of equations 2, 3, and the quotient of equation 3 divided by equation 2.
Discussion
Multiple sequence alignment clearly shows that the ligands for the type 1 site and trinuclear cluster are present, and located in the same relative positions as found in other laccases. Secondary structure prediction and comparisons find significant alignment of the two domains of SLAC with cupredoxin folds. It has been hypothesized that the cupredoxin domain has been often recycled for various purposes, from copper nitrite reductase to the human coagulation factor VIII (Murphy et al. 1997). One of the most important similarities between these enzymes is the position and identity of the metal ligands. The alignment shows that all the copper ligands are conserved in their proper locations. The spectroscopy and copper quantitation validate the presence and structure of the three expected copper sites.
One unusual feature is the presence of 24 histidines in the full sequence. If 10 residues are the expected copper ligands, 14 are left. These 14 residues represent 4.6% of SLAC’s amino acids, about double the average histidine composition (2.26%) of proteins (Gasteiger et al. 2001). CueO from E. coli has a high percentage of methionine residues and has been suggested to play a role in resistance to copper toxicity (Roberts et al. 2002). It is possible that a secondary function of SLAC is to bind a surplus of intra-cellular copper and carry it when exported by the TAT secretory apparatus. Extensive in vivo work is currently being done to address this question.
The first 30 amino acids encoded by the SLAC gene form a twin-arginine-targeted (TAT) leader sequence that signals the protein for export via the folded-protein pathway. Many laccase proteins are transported in a similar manner, including the E. coli proteins CueO and PcoA (Lee et al. 2002). When expressed recombinantly in E. coli, SLAC is found in the cytoplasm and the N-terminal residue is one of several occurring downstream from the TAT-leader sequence. These results suggest that SLAC is degraded in the cytoplasm of E. coli. However, in cultures of S. coelicolor, laccase activity is detected in the media. Laccase activity is not present in the strain with the SLAC gene disrupted, indicating that SLAC is exported when expressed homologously and that SLAC is the primary extracellular laccase.
The lack of the second domain that is normally found in laccases raises several issues. One such issue is the question of how the two domains are connected. Normally, a short loop or helix of nine amino acids connects domains one and two, but domains two and three are usually linked by a long coil of 40 or more amino acids. Laccases have very low sequence homology in this coil. It is thus not surprising that the two domains of SLAC are connected by ~60 amino acids of unknown structure. This section of SLAC also would include all of the substrate-binding features from a full-size laccase. The 60-amino-acid segment is predicted to be almost neutral at pH 7.0, and negative charges seem randomly distributed. There are two small clusters of positive residues (following R180 and R203), perhaps near a substrate-binding site.
Another issue regarding the absent domain is “Can the enzyme function without this highly conserved element?” It clearly does but only in the dimeric form, as predicted by Murphy et al. (1997). The crystal structure of the ascorbate oxidase dimer from cucumber shows that 32 of the 39 residues that form the dimer interface are in domains one and three (Messerschmidt et al. 1992). Domain two frequently contributes several residues to the substrate pocket, but other residues that form the binding cleft near the type 1 copper are provided by loops and turns between domain two and domains one and three. These comparisons show that although the majority of the surface for dimer formation is present, the machinery for binding the reducing substrate must have been reengineered. An alternative explanation could be that the copper ions of the trinuclear cluster bridge the first domain of one monomer and the second domain of a second monomer, so that the two monomers form a head-to-tail arrangement. Finally, it is possible that the monomeric, folded form has activity, but that this species is not observed in solution or on SDS-PAGE gels.
The interactions for dimer formation may still be present, but another two-domain copper enzyme, the nitrite reductase from Alcaligenes faecalis, naturally forms a homotrimer. EpoA of S. griseus, the only other characterized example of a laccase lacking the second domain, is 69% identical to SLAC. Despite this near identity, EpoA is also active as a trimer (Endo et al. 2002). It is possible that agglomeration occurs with the high percentage of histidines and copper, although such behavior is not witnessed in SLAC.
The activity of SLAC against DMP is striking in that it reaches an optimum at an alkaline pH. Quite often, phenoloxidase activity is measured at acidic pHs (4.0–5.0), and a few enzymes peak at pH 7.0–8.0 (Table 1). Mutational analyses on several laccases suggest that an acidic residue (analogous to D295 in SLAC) raises the optimum of Acremonium ascorbate oxidase by one pH unit, but the reported differences in activity are very small, that is, the top of the bell-shaped curve of activity versus pH is almost flat (Sugino et al. 2002). The increase of activity with pH has been attributed to a decrease in substrate reduction potential that is favorable for electron transfer to the type 1 copper site (Xu 1997). The other half of the bell curve, representing a decrease of activity with pH, has been attributed to hydroxide inhibition at the trinuclear cluster.
Table 1.
pH optima of DMP and SGZ oxidase activity for several laccases
| Organism | Enzyme | pH | Substrate | Reference |
| E. coli | CueO | 5.5, 8 | DMP | Roberts et al. 2003 |
| T. pubescens | Laccase | 3–4.5 | DMP, SGZ | Galhaup et al. 2002 |
| B. subtilis | CotA | 7.0 | SGZ | Martins et al. 2002 |
| P. cinnabarinus | Laccase | 4 | SGZ | Record et al. 2002 |
| P. pinsitus | Laccase | 5 | SGZ | Xu et al. 1996 |
| R. solani | Laccase | 7 | SGZ | Xu et al. 1996 |
| M. thermophila | Laccase | 6 | SGZ | Xu et al. 1996 |
| S. thermophilium | Laccase | 7 | SGZ | Xu et al. 1996 |
| R. vernicifera | Laccase | 9 | SGZ | Xu et al. 1996 |
| M. verrucaria | Bilirubin oxidase | 8 | SGZ | Xu et al. 1996 |
The dependencies of kcat and Km on pH in this work form a different understanding for the reactivity of SLAC. The decrease of Km with increasing pH reflects the binding of the anionic form of DMP, with a commensurate correlation to the substrate’s pKa value. That ferrocyanide and not aqueous iron(II) is bound by and reacts with SLAC confirms that the enzyme prefers to bind negative substrates over uncharged or positive ones. It is possible that the two positive clusters of residues in the interdomain linker are responsible for this behavior. Also, it is usually the situation that the variation in kcat/Km mirrors the trend in kcat near the pH of optimum activity. Here, in contrast, the binding of substrate to SLAC dominates kcat/Km until the pKa value of the free substrate is reached and the pH dependency of Km levels off.
The catalysis of phenol oxidation mirrors the results of three-domain laccases. From pH 6.4 to pH 10.0, the catalytic rate constant takes the shape of a bell curve (Xu 1997). Optimum activity is consistent with a deprotonation at pKa 7.3 and a protonation at pKa 9.2. It is important to note that these pKa values correspond to the bound enzyme–substrate complex (the species responsible for catalytic activity), and not to the parameters of the free substrate that are calculated from fitting Km. Also, because neither of these pKa values corresponds to the pKa value of the enzyme–substrate complex (8.3) that is derived from the substrate-binding behavior, they are likely to involve changes in a separate part of the protein: the trinuclear cluster. This hypothesis is in concordance with previous work that ascribes a reduction in activity to hydroxide inhibition at the trinuclear cluster.
Rough correlations have also been made between the pI of laccases and their optimal pH for SGZ oxidation (Xu et al. 1996). From the measured pI of 8.2, the optimal pH should be above 8.0, second only to R. vernicifera laccase. The fact that a high pH optimum is seen in SLAC for DMP, a similar substrate to SGZ, is further support for this correlation. Similar correlations between the kcat and the reduction potentials of laccases have been made. The values of kcat for DMP oxidation measured in this study suggest that SLAC is a “low-potential” laccase, with the reduction potential of the type 1 site near 0.5 V versus normal hydrogen electrode.
Paramagnetic NMR of copper proteins, primarily blue copper proteins, has been studied for more than two decades. A mononuclear copper(II) center typically exhibits Curie behavior, or an increase in chemical shift with a decrease in temperature. Recently, this lab began to characterize tyrosinase, a protein with a type 3 dinuclear copper center, which shows anti-Curie behavior as a result of antiferromagnetic coupling between the two copper ions (Bubacco et al. 1999; Tepper et al. 2002). Herein, we have reported the paramagnetic NMR spectrum of a laccase, which has recently been also reported by Battistuzzi et al. (2003). The resonances in the NMR spectrum of that work are assigned to the mononuclear type 1 site; in contrast, the anti-Curie behavior of several of the peaks in SLAC shows that, for the first time, the trinuclear cluster of a laccase has been observed with NMR. The characterization of these spectra is the subject of another manuscript.
A novel laccase enzyme has been cloned and characterized, and is denoted “small laccase” (SLAC) because it lacks a domain and is thus much smaller than other laccases. Despite the lack of this domain, the enzyme maintains the normal complement of four copper ions and is active against the common range of substrates, preferring to bind those that are negatively charged. A group of ~60 amino acids takes the place of the substrate-binding loops that are found in three-domain laccases. The protein is functional in solution and denaturing gels as a dimer, and converts phenols effectively at unprecedentedly high pHs. It also appears to be the primary extracellular laccase in its host organism, S. coelicolor. These many features, coupled with the ease of preparation of SLAC, suggest that the enzyme will be a useful implement for understanding the properties of laccases.
Materials and methods
Recombinant expression and purification of SLAC
A 1.4-kb fragment containing the gene encoding SLAC was generated by PCR with genomic DNA of S. coelicolor M145 as template and the following primers: CGCGAATTCATATGGACAG GCGAGGCTTTAACCGACG (MCOF) and CGCAAGCTTCTC CGGTTCCGCGGCGACG (MCOR). The forward primer, MCOF, introduced an NdeI site (underlined) on the start codon (italics) and an EcoRI site (bold), whereas the reverse primer, MCOR, introduced a HindIII (underlined) site 420 bp after the stop codon. The NdeI/HindIII-digested PCR fragment was cloned into pET 20b (Novagen), generating pSLAC1, which was transformed into E. coli BL21(DE3). Transformants were selected on LB media containing 100 μg/mL ampicillin. DNA sequencing (Baseclear) confirmed the sequence of the cloned gene.
For medium-scale production of SLAC, 3 L of 2xYT media was inoculated with 30 mL of an overnight culture. The cells were grown at 30°C until reaching OD600 ~ 1.5, when the temperature was reduced to 25°C and the cells were induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (Eurogentec). After 20 h of growth, the cells were collected by centrifugation for 10 min at 5000g. The pellet was resuspended in 150 mL of 10 mM NaPi (pH 7.3), and the resulting solution was sonicated to lyse the cells. The soluble fraction was obtained by centrifuging for 20 min at 27,000g. The soluble fraction was incubated for several hours with 1 mM Cu(SO4), at which time 150 μg of DNAase I (Roche) and10 μg of RNAase (Eurogentec) were added.
The solution was then dialyzed four times against 5 L of the same buffer that was used to suspend the cell pellets. Then 1 mM EDTA was added to the second dialysis to remove excess copper from the protein. The solution was then applied to a diethylaminoethyl (DEAE) column (100 mm long by 50 mm diameter; Amersham Pharmacia) that had been equilibrated with the same phosphate buffer. A step gradient elutes the pure SLAC protein at 100 mM NaCl. The blue fractions were pooled and concentrated, and the purity was determined by SDS-PAGE and UV-visible absorbance measurements (A280 nm/A590 nm < 11 indicates >97% purity). Protein concentration was initially assessed using both the Bradford technique and the absorbance at 280 nm of the unfolded protein in 6 M guanidinium hydrochloride (Gill and von Hippel 1989); these concentrations were then used to determine the extinction coefficients for the 280 nm, 330 nm, and 590 nm peaks of the folded protein. The folded-protein extinction coefficients were used in all other measurements. When testing for export of the protein, the periplasmic fraction of the E. coli was isolated by osmotic shock.
Other methods
The amount of copper bound to the protein was typically determined through the trichloroacetic acid/bicinchoninic acid (BCA) method of Brenner and Harris (1995) and confirmed by atomic absorption (Department of Toxicology, Leiden University Medical Center). The isoelectric point of the protein was determined using IEF gels on a Phast System (Amersham Biosciences) with broad pI standards. Reduced protein was prepared by incubation with ascorbic acid or sodium dithionite. For gel filtration, an aliquot of protein was injected onto a Superose 12 column (Amersham Biosciences) that had been equilibrated with 10 mM NaPi (pH 7.3), and the elution volume was compared against a series of standards (Sigma): blue dextran (2000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa dimer), carbonic anhydrase (29 kDa), cytochrome c (12 kDa), and tryptophan (0.17 kDa).
Electron paramagnetic resonance (EPR)
EPR measurements were made at 40 K on a Bruker Elexsys 680 (Bruker Biospin Gmbh Rheinstetten) spectrometer operating at 9.73 GHz (X-band). The protein sample contained 1.5 mM SLAC in 30% glycerol, 70% 100 mM NaPi buffer (pH 7.2). The instrument parameters were microwave power, 19.75 μW; modulation frequency, 100 kHz; modulation amplitude, 0.5 mT; time constant, 2.56 msec; sweep rate, 19.1 mT/sec.
Nuclear magnetic resonance
NMR samples of fresh protein (1 mM) were prepared in 100 mM NaPi buffer (pH 7.3) in H2O/D2O (90/10 v/v). 1H NMR spectra were recorded on a Bruker DMX-600 NMR spectrometer using a super-WEFT pulse sequence (interpulse delay, 100 msec; repetition rate, 6 sec−1; spectral width, 150 ppm). In all, 32,000 free induction decays were acquired and Fourier-transformed using a Gaussian window (LB = −10 Hz, GB = 0.02) and baseline-corrected using Bruker-provided software. The temperature-dependence of the paramagnetic shift of the peaks was fit to the following equation, which assumes that two copper ions are antiferro-magnetically coupled with an exchange coupling constant J, yielding a diamagnetic ground state with an excited paramagnetic (S = 1) state -2J in energy above the ground state (Bubacco et al. 1999):
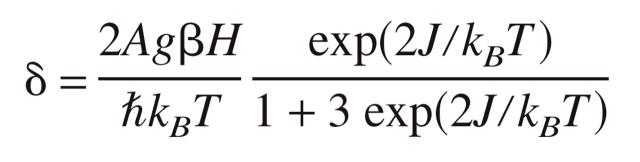 |
(1) |
In this equation, β is the Bohr magneton, ħ is Planck’s constant divided by 2π, kB is Boltzmann’s constant, T is the absolute temperature, H is the externally applied magnetic field, and A is the isotropic dipolar interaction energy between the magnetic moment of the electrons and the nuclear magnetic moment.
Activity assays
Activity assays were typically carried out in 100 mM acetate (pH 4.0–6.0), NaPi (pH 6.0–8.0), Tris (pH 7.0–9.0), or carbonate (pH 9.0–11.0) buffers. DMP turnover was monitored at 468 nm (ɛ = 14,800 M−1 cm−1 for the dimeric product; Solano et al. 2001), Fe(CN)64− at 405 nm (ɛ = 900 M−1 cm−1), and Feaq2+ at 315 nm (ɛ = 2200 M−1 cm−1). All UV-visible absorption measurements were obtained using a Shimadzu UV-2101PC spectrophotometer. The method of initial rates was used to determine the substrate-dependent reaction velocities, which were then fit with the Michaelis-Menten equation. The pH-dependencies of the catalytic rate constant (kcat) and Michaelis-Menten constant (Km) were fit using equations 2 and 3 (Segel 1975):
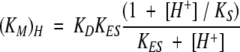 |
(2) |
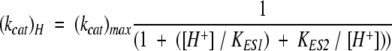 |
(3) |
In equation 2, KD is the dissociation constant of the substrate in the deprotonated form, KES is an acid dissociation constant for the enzyme–substrate complex, and KS is the acid dissociation constant for the free substrate. In equation 3, KES1 and KES2 are acid dissociation constants for the enzyme–substrate complex (these constants are unrelated to KES in equation 2).
Streptomyces strains and growth
M145 (SCP1-, SCP2-) was used for all Streptomyces experiments, except where the disruption mutant was used (vide infra). Spores, mycelia, and protoplasts were all manipulated by established protocols (Kieser et al. 2000). Disruption of the SLAC gene in M145 by the insertion of an apramycin-resistance cassette was accomplished by PCR-targeted gene replacement (Gust et al. 2003). The cosmid 4C6 was used for recombination, and the PCR primers used were TCTGCGCACATTCCGTATCGGCGTGGGGAGTTC GGCATGATTCCGGGGATCCGTCGACC (MCOKOF) and CC GGCGGCGCCGGTGGCGGCGCCGGCCGGCCCCGGCTCAT GTAGGCTGGAGCTGCTTC (MCOKOR). Disruption was confirmed by Southern blot.
Homologous expression and localization
Cultures of M145 and the ΔSLAC strain were seeded with freshly harvested spores and grown for 5 d at 30°C in flasks containing metal springs and liquid R5 media without antibiotics. Mycelia were harvested by centrifugation (3000g for 10 min). DMP oxidase activity of the media was determined under the following conditions: 50 μL of growth medium was added to 950 μL of a 2.5 mM solution of DMP with 100 mM Tris buffer at pH 8.0. Absorption at 468 nm was monitored with time.
Acknowledgments
We acknowledge M. Ubbink for assistance with the NMR, M. Huber and M. Finiguerra for their EPR measurements, and R. van der Heijden for mass spectrometry measurements. We thank J. van Beeumen (Gent) for valuable comments concerning the amino acid sequencing of the SLAC. B.S. is a postdoctoral fellow of the Fund for Scientific-Research-Flanders (F.W.O.-Vlaanderen, Belgium).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04759104.
References
- Battistuzzi, G., Di Rocco, G., Leonardi, A., and Sola, M. 2003. 1H NMR of native and azide-inhibited laccase from Rhus vernicifera. J. Inorg. Biochem. 96 503–506. [DOI] [PubMed] [Google Scholar]
- Berks, B.C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22 393–404. [DOI] [PubMed] [Google Scholar]
- Brenner, A.J. and Harris, E.D. 1995. A quantitative test for copper using bicinchoninic acid. Anal. Biochem. 226 80–84. [DOI] [PubMed] [Google Scholar]
- Bubacco, L., Salgado, J., Tepper, A.W.J.W., Vijgenboom, E., and Canters, G.W. 1999. 1H NMR spectroscopy of the binuclear Cu(II) active site of Streptomyces antibioticus tyrosinase. FEBS Lett. 442 215–220. [DOI] [PubMed] [Google Scholar]
- Claus, H. 2003. Laccases and their occurrence in prokaryotes. Arch. Microbiol. 179 145–150. [DOI] [PubMed] [Google Scholar]
- Edens, W.A., Goins, T.Q., Dooley, D., and Henson, J.M. 1999. Purification and characterization of a secreted laccase of Gaeumannomyces gramminis var. tritici. Appl. Environ. Microbiol. 65 3071–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, K., Hosono, K., Beppu, T., and Ueda, K. 2002. A novel extracytoplasmic phenol oxidase of Streptomyces: Its possible involvement in the onset of morphogenesis. Microbiology 148 1767–1776. [DOI] [PubMed] [Google Scholar]
- Galhaup, C., Goller, S., Peterbauer, C.K., Strauss, J., and Haltrich, D. 2002. Characterization of the major laccase isoenzyme from Trametes pubescens and regulation of its synthesis by metal ions. Microbiology 148 2159–2169. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., Jung, E., and Bairoch, A. 2001. SWISS-PROT: Connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 3 47–55. [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Gust, B., Challis, G.L., Fowler, K., Kieser, T., and Chater, K.F. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. 100 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakulinen, N., Kiiskinen, L., Kruus, K., Saloheimo, M., Paananen, A., Koivula, A., and Rouvinen, J. 2002. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear site. Nat. Struct. Biol. 9 601–605. [DOI] [PubMed] [Google Scholar]
- Jones, D.T. 1999a. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287 797–815. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kieser, T., Bibb, M.J., Buttner, M.J., Chater, K.F., and Hopwood, D.A. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich UK.
- Lee, S.M., Grass, G., Rensing, C., Barrett, S.R., Yates, C.J., Stoyanov, J.V., and Brown, N.L. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295 616–620. [DOI] [PubMed] [Google Scholar]
- Martins, L.O., Soares, C.M., Pereira, M.M., Teixeira, M., Costa, T., Jones, G.H., and Henriques, A.O. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277 18849–18859. [DOI] [PubMed] [Google Scholar]
- Mayer, A.M. and Staples, R.C. 2002. Laccase: New functions for an old enzyme. Phytochemistry 60 551–565. [DOI] [PubMed] [Google Scholar]
- Messerschmidt, A., ed. 1997. Multi-copper oxidases. World Scientific, Singapore.
- Messerschmidt, A. and Huber, R. 1990. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur. J. Biochem. 187 341–352. [DOI] [PubMed] [Google Scholar]
- Messerschmidt, A., Ladenstein, R., Huber, R., Bolognesi, M., Avigliano, L., Petruzzelli, R., Rossi, A., and Finazzi-Agro, A. 1992. Refined crystal structure of ascorbate oxidase at 1.9 Å resolution. J. Mol. Biol. 224 179–205. [DOI] [PubMed] [Google Scholar]
- Murphy, M.E.P., Lindley, P.F., and Adman, E.T. 1997. Structural comparison of cupredoxin domains: Domain recycling to construct proteins with novel functions. Protein Sci. 6 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek, K., Antorini, M., and Choinowski, T. 2002. Crystal structure of a laccase from the fungus Trametes vericolor at 1.90-Å resolution containing a full complement of coppers. J. Biol. Chem. 277 37663–37669. [DOI] [PubMed] [Google Scholar]
- Record, E., Punt, P.J., Chamkha, M., Labat, M., van den Hondel, C.A.M.J.J., and Asther, M. 2002. Expression of the Pycnoporus cinnabarinus laccase gene in Aspergillus nigerand characterization of the recombinant enzyme. Eur. J. Biochem. 269 602–609. [DOI] [PubMed] [Google Scholar]
- Rensing, C. and Grass, G. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27 197–213. [DOI] [PubMed] [Google Scholar]
- Roberts, S.A., Weichsel, A., Grass, G., Thakali, K., and Hazzard, J.T. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. 99 2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.A., Wildner, G.F., Grass, G., Weichsel, A., Ambrus, A., Rensing, C., and Montfort, W.R. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 278 31958–31963. [DOI] [PubMed] [Google Scholar]
- Segel, I.H. 1975. Enzyme kinetics. John Wiley & Sons, New York.
- Solano, F., Lucas-Elio, P., López-Serrano, D., Fernández, E., and Sanchez-Amat, A. 2001. Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiol. Lett. 204 175–181. [DOI] [PubMed] [Google Scholar]
- Solomon, E.I., Sundaram, U.M., and Machonkin, T.E. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96 2563–2605. [DOI] [PubMed] [Google Scholar]
- Sugino, M., Kajita, S., Banno, K., Shirai, T., Yamane, T., Kato, M., Kobayashi, T., and Tsukagoshi, N. 2002. Upward shift of the pH optimum of Acremonium ascorbate oxidase. Biochim. Biophys. Acta 1596 36–46. [DOI] [PubMed] [Google Scholar]
- Tepper, A.W.J.W., Bubacco, L., and Canters, G.W. 2002. Structural basis and mechanism of the inhibition of the type-3 copper protein tyrosinase from Streptomyces antibioticus by halide ions. J. Biol. Chem. 277 30436–30444 [DOI] [PubMed] [Google Scholar]
- Xu, F. 1996. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: Correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35 7608–7614. [DOI] [PubMed] [Google Scholar]
- ———. 1997. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 272 924–928. [DOI] [PubMed] [Google Scholar]
- Xu, F., Shin, W., Brown, S.H., Wahleithner, J.A., Sundaram, U.M., and Solomon, E.I. 1996. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta 1292 303–311. [DOI] [PubMed] [Google Scholar]
- Yoshida, H. 1883. Chemistry of lacquer (Urushi) part 1. J. Chem. Soc. 43 472–486. [Google Scholar]