Abstract
Computational analysis of the bonding, geometric, and topological relationships within proteins typically takes on the order of hours, mainly devoted to the writing of scripts and code to correctly parse the data. The Structured Query Language (SQL) built into modern database management systems eliminates the need for data parsing, effectively reducing the analysis time to seconds. To this end, we have formulated a conceptual data model (CDM) for proteins based on the IUPAC recommendations for biochemical nomenclature. This conceptual data model makes explicit the inherent bonding relationships between the atoms of a protein, as well as the geometric (bond angle and torsion angle) and topological (chirality) relationships between the bonds. The validity of the CDM has been tested with a reduced implementation using commercial database software. The ease in both populating the database with data from the Protein Data Bank and formulating/executing queries supports the correctness of the model. The ability to conduct truly interactive analyses of protein structure is essential to fully capitalize on the explosion in postgenomic protein structure data.
Keywords: IUPAC, relational database, protein structure, conceptual data model
Relational databases fill two important roles. Superficially, they act as repositories of information, allowing database management systems (DBMS) to provide archival and retrieval functions. At a more profound level, they also define explicitly the inherent relationships between data elements, allowing important subsets of the data to be collected, extracted, and analyzed efficiently. It is through this second, often-neglected, role that relational databases show their power in the analysis of scientific data, allowing new hypotheses to be generated through data mining (Piatetsky-Shapiro and Frawley 1991). It is our goal to develop and implement a relational database containing structural data of proteins, which will assist in the search and cataloguing of correlations hidden within the data.
It is axiomatic in the database engineering community that explicit specification of a high-level conceptual data model is required before implementation of any database. The structural biology community has long relied upon the well-defined IUPAC nomenclature for describing the conformation of polypeptide chains (IUPAC-IUB Commission on Biochemical Nomenclature, 1970, IUPAC-IUB Commission on Biochemical Nomenclature, 1984; Markley et al. 1998). From a database engineering perspective, this notation represents an implied data model. In this article we make the IUPAC model explicit. The conceptual data model implied by the IUPAC nomenclature may serve as a starting point for various software implementations, thereby ensuring that any such implementation is based on the scientific principles of the system rather than on technological concerns of parsing the data. An explicit data model also serves the goal of IUPAC, namely to enhance communication by facilitating data exchange between different software and databases.
It is the object of this article to outline and critique the explicit data model implied by the nomenclature recommended by the IUPAC commissions. Incompatibilities between the various recommendations are illustrated, as well as recommended compromises where differences exist. Finally, it is shown how this molecular data model can be used as a core for other expanded data models. The inherently hierarchical model has convenient tiers on which to integrate data with the Protein Data Bank (PDB), which houses atomic coordinates of biological macromolecules (Berman et al. 2000); the BioMagResBank (BMRB), which stores NMR-specific data, including chemical shifts, coupling constants, and relaxation data (Seavey et al. 1991); as well as with structure computation software such as XPLOR (Brünger 1992) or CYANA (Güntert et al. 1997). In that sense, the recommendations introduced by IUPAC transcend nomenclature by providing a referential interface between the fields of chemistry, biochemistry, bioinformatics and structural biology. To test the validity of our model, we have provided a database implementation of the conceptual data model that has been populated with a limited subset of the PDB.
Data model
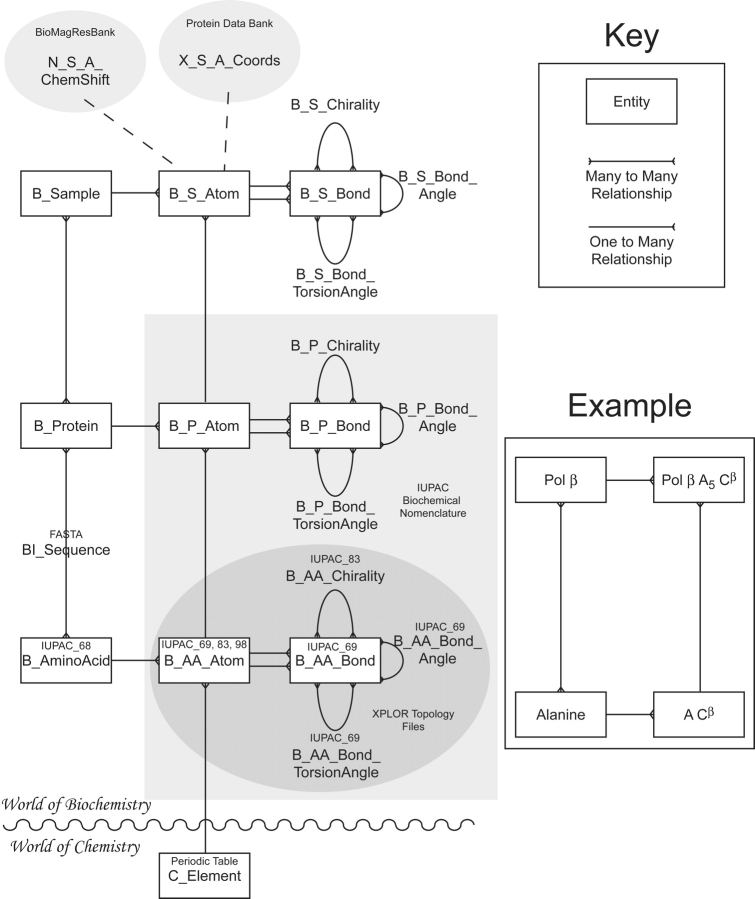
An entity-relationship diagram3 for the implied IUPAC model is shown in Figure 1 ▶. Several features of the model are important to note. First, while the IUPAC nomenclature is intended to uniquely designate atoms in a polypeptide rather than designating the chemical bond network, the bonding relationship of the atoms is partially coded in the naming convention. It is implicit that each residue shares a common framework for the main chain atoms, and that the linear organization of the side chain atoms can be inferred from the designated nomenclature (for instance, the Cβ is always covalently bound to the Cα and Cγ [if present]). However, not all bonding relationships can be inferred from the nomenclature (such as the N-Cδ bond in proline). All bonding relationships in a protein are made explicit in our proposed data model as relationships between individual atoms. Bond angle, torsion angle, and chirality are explicitly defined as the geometric relationship between bonds. It is clear that these additional concepts should not be modeled as relationships between atoms, as the absolute position of the atoms does not affect these terms (i.e., bond angle is geometrically independent of bond length). However, there are other isomorphic representations for these concepts, such as that of the torsion angle being defined as the angle between two planes. We have chosen to model each as a relationship between bonds to minimize the number of entities in the model; alternative representations and translations between them can easily be added if they provide additional utility.
Figure 1.
Conceptual data model implied from IUPAC nomenclature. Entities are noted as boxes; relationships shared between the entities as lines. Some relationships (such as bonds) are important enough to warrant entity status. The side panel shows an example of how the amino acid, alanine, and its component atoms relate to the protein Pol β. The dotted lines connected to B_S_Atom illustrate the potential mapping to structural data. The IUPAC nomenclature technically define the lower two tiers of the hierarchy, while the bottom tier is reflected in the definition of topology files for the structure refinement package, XPLOR (Brünger 1992). Our notation for entities begins with a prefix describing the scientific discipline to which the data most naturally reside: Chemistry, Biochemistry, BioInformatics, X-ray crystallography, NMR spectroscopy.
Second, the IUPAC nomenclature implies an inherently hierarchical data model. The atoms of a given polypeptide are designated on an amino acid residue basis, each individual amino acid of the polypeptide chain being distinguished by a numerical suffix. The implied chemical bonding network is also designated on an amino acid residue basis, with successive amino acid networks linked by the Ci′-Ni + 1 peptide bond. This hierarchical approach is clearly beneficial for its simplicity in uniquely defining all the atoms of an arbitrary polypeptide sequence. This benefit is mirrored in the database implementation of the model in that instances for each atom in an arbitrary polypeptide can be generated through a table join4 between the polypeptide sequence and the atoms in the generic amino acid. The generic amino acid in this context is an idealized abstraction of residues occurring in polypeptides, rather than an actualization of the free amino acid. In this sense, the IUPAC nomenclature refers to an idealized abstraction of a polypeptide chain, applicable to theoretical modeling, and molecular dynamics studies, as well as practical modeling of structural data obtained from X-ray crystallography, NMR spectroscopy or cryo-electron microscopy. Because the structural characteristics of a polypeptide are likely to differ between these various studies, we have chosen to add a tier to the hierarchy to include instances of the polypeptide in various real or virtual samples. This is the level at which scientific data from crystallography or NMR is most appropriately mapped, and is the level to which the PDB and BMRB public databanks relate (Fig. 1 ▶).
Results and Discussion
To demonstrate the appropriateness and utility of this data model, we have implemented a reduced form of it as a database in Microsoft Access. (The utility of the model is independent of the software implementation.) All atom type designations from the IUPAC recommendations were included in the table B_AA_Atom as well as the implied chemical bonding network in the table B_AA_Bond. An instance for the protein DNA polymerase β (Swiss-Prot ID: P06746) was included in the table B_Protein and the individual atoms and bonds created for this protein. The table B_S_Atom was populated with the X-ray crystallographic data available from the PDB (1BPY) (Sawaya et al. 1997). Finally, the table B_S_Bond was populated using a single SQL query that calculated all individual bond lengths from the X-ray coordinates in B_S_Atom. The database implementation is freely available for download at http://sbtools.uchc.edu/.
Once populated in this fashion, the Structured Query Language (SQL) built into the commercial database product allowed the facile search for correlations in the PDB data. The dataset was queried to determine the average observed bond lengths for (1) all bonds, (2) only C—N bonds, (3) only C—O bonds; and (4) only C—C bonds. The results are shown in Table 1. More complex queries were also run, including determining the average bond length of all C—C bonds where one C is bonded to an O, and all C—C bonds where neither C is bonded to an O (Table 1). While the first three queries could be efficiently reproduced by parsing the database for specific atom types, the latter two would be far more complicated to determine without the explicit designation of the bonding relationships in the database.
Table 1.
SQL queries run on a test data set containing the crystallographic coordinates of 1BPY (Sawaya et al. 1997)
| Bonds selected | Avg. bond len. | Selection criteriaa |
| All Heavy | 1.423 ± 0.1131 | None |
| C—N | 1.386 ± 0.065 | WHERE (Atom1.B_AA_A_Element= ‘N’ AND Atom2.B_AA_A_Element= ‘C’) OR (Atom1.B_AA_A_Element= ‘C’ AND Atom2.B_AA_A_Element= ‘N’); |
| C—O | 1.256 ± 0.057 | WHERE (Atom1.B_AA_A_Element= ‘O’ AND Atom2.B_AA_A_Element= ‘C’) OR (Atom1.B_AA_A_Element= ‘C’ AND Atom2.B_AA_A_Element= ‘O’); |
| C—C | 1.505 ± 0.048 | WHERE (Atom1.B_AA_A_Element= ‘C’ AND Atom2.B_AA_A_Element= ‘C’); |
| C—C(—O) | 1.511 ± 0.036 | INNER JOIN [C-O_Bonds_Subset] |
| ON ([C-C_Bonds_Subset].Atom1.B_P_A_ID= [C-O_Bonds_Subset].Atom1.B_P_A_ID) | ||
| Or ([C-C_Bonds_Subset].Atom1.B_P_A_ID= [C-O_Bonds_Subset].Atom2.B_P_A_ID) | ||
| Or ([C-C_Bonds_Subset].Atom2.B_P_A_ID= [C-O_Bonds_Subset].Atom1.B_P_A_ID) | ||
| Or ([C-C_Bonds_Subset].Atom2.B_P_A_ID= [C-O_Bonds_Subset].Atom2.B_P_A_ID); | ||
| C—C(—X) | 1.501 ± 0.053 | RIGHT JOIN [C-C_Bonds_Subset] |
| X: Not O | ON [C-C′_Bonds_Subset].[B_P_B_ID]= [C-C_Bonds_Subset].[B_P_B_ID] | |
| WHERE [C-C′_Bonds_Subset].[B_P_B_ID] is Null; | ||
| Atoms selected | Avg. Debye-Waller factor | Selection criteria |
| Backbone | 48.98 ± 10.58 | WHERE B_AA_A_Serial<=1; |
| Side chain | 53.26 ± 15.52 | WHERE B_AA_A_Serial>=2; |
| ɛ and greater | 58.60 ± 18.51 | WHERE B_AA_A_Serial>=5; |
a The bond selection shown is based on the chemical element of the atoms in the bond. The element type is labeled with the identifier, B_AA_A_Element. The atom selection shown is based on how many bonds separate the atom from the protein backbone. This degree of separation is labeled with the identifier, B_AA_A_Serial.
The ease with which the database was populated with data from the PDB gives strong evidence that the model is consistent with that representation. The advantage of our proposed model is that the chemical bonding network is explicitly defined as relationships in the database rather than implicitly layered onto the nomenclature. Once explicitly defined, it is possible to use the query functionality built in to commercial database software to extract correlations inherent in the data or to test hypotheses. To prove the utility of this approach, we have limited our test queries to those that exploit the chemical bonding relationships in proteins. Other queries are also possible, which group common atoms/bonds on the basis of the type/strength of the chemical bond, the electronegativity or charge of the atoms, and other chemical attributes of the protein.
There have been three sets of recommendations that have been supported by IUPAC (1970, 1984; Markley et al. 1998). Although the bulk of the latter revisions are additions for nucleic acids and other molecular types, there have also been revisions to the original proposed nomenclature, and it is worthwhile to examine the consequences with respect to the implied data model. From a notational standpoint, the first recommendations set the stage by defining unique identifiers for each of the peptide backbone atoms, as well as an algorithm for defining unique identifiers for any arbitrary amino acid side chain. All backbone atoms of the peptide bond are noted by their element type alone (H, N, C, and O). The side chain atoms as well as the backbone Cα, Hα atoms are treated differently than the four peptide bond atoms by labeling the heavy atoms sequentially (α, β, γ, etc.) from the backbone outward. Branches to the heavy atom chain are numbered sequentially by defined priority rules. H atoms are labeled with the same Greek symbol as the heavy atom to which they are attached. Multiple H atoms attached to the same heavy atom are numbered sequentially according to priority rules. H atoms can be treated differently from the heavy atoms because hydrogen is only able to form one covalent bond, effectively terminating the chain at that point.
This aforementioned algorithm for defining side chain atom identifiers has several unfortunate consequences. First, the Greek notation is limited to 20 identifiers, a notation that fails when giving unique identifiers to large nonnatural side chains or side-chain modifications. Second, the notation requires the use of a mixture of Greek symbols for the sequential labels and Roman for the element types. It is recommended in the 1969 article that the Greek symbols can be substituted for their Roman counterparts when convenient (Cα becoming CA); however, this results in the additional consequence that atom identifiers that were sequential in the Greek system (Cβ, Cγ, Cδ) are no longer sequential in the Roman system (CB, CG, CD).5 The 1983 recommendation to forgo the Greek notation in favor of numerical notation rectified this problem; however, it has never been widely adopted (Markley et al. 1998). Maintaining sequential labels is imperative to search the database for correlations based on an atom’s remoteness from the backbone. For these reasons, we have chosen to model the side chains with sequential numeric identifiers rather than Greek symbols, although the legacy notation will be retained where appropriate.6 Once modeled sequentially, it was easy to demonstrate that in our test data set, the observed Debye-Waller factor is larger, as more bonds separate the atom from the peptide backbone (Table 1).
All three sets of recommendations treat the terminal groups as unique entities, such that the C-terminal oxygen atoms are labeled O′ and O″ rather than O, and the N-terminal hydrogen atoms are labeled H1, H2, and H3 rather than H/HN. This is conceptually dissatisfying, for even though the terminal O′ would be unaffected by addition of another amino acid, it would be required to change its identifier. We therefore have chosen to model all backbone oxygen atoms as O1 with the additional terminal hydroxyl as O2-H2, and all backbone amides as H1 with the additional terminal protons as H2 and H3. (In keeping with the 1998 recommendations, H1 can also be referred to as HN or HN1 if desirable.)
A final note regarding the notation is that of the priority rules themselves. Rule 2.1.5 (IUPAC-IUB, 1970) recommends that in the absence of other distinguishing characteristics, atoms are given notational preference based on isotopic composition. This rule is inappropriate in the context of conveying the geometric relationships between chemically bonded atoms, which is not altered through isotopic labeling. This rule would be particularly troublesome applied to the field of NMR spectroscopy, as it is common to use isotopic enrichment to distinguish the branches of prochiral centers. The unfortunate consequence of this priority rule would be that if the Cγ2 was selectively enriched with 13C it would become the Cγ1. Therefore, we propose that isotopic composition should not be used in determining priority.
In summary, we believe that IUPAC recommendations for biochemical nomenclature provide a solid framework from which to build a conceptual data model for protein structure. In providing this implied data model, the IUPAC recommendations transcend the mere standardization of nomenclature and allow for the abstraction of structural data types. Implementing the conceptual data model in a commercial database provides powerful computational tools for structural data analysis. In providing an explicit representation of the implied model, we hope to stimulate expansion and refinement of the proposed model.
Acknowledgments
We thank Drs. Jeffrey C. Hoch, Mark W. Maciejewski, and Scott A. Robson for thoughtful discussion. This research was supported by the NIH grant ES09847 (M.R.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04810204.
Footnotes
Entity-relationship models define an abstraction of a given system.“Entities” are any part of the system worthy of an identity. They are essentially the components of the system and are described by any number of “attributes.” “Relationships” define the associations between the entities, some relationships being important enough to be treated as entities themselves.
In database terminology, a “table join” combines all of the entities common to two tables. Therefore, a table containing all the atoms in a peptide sequence is easily created by joining the atoms of the amino acids with the amino acids of the peptide sequence.
The 1998 recommendation to refer to the backbone H as HN may also be problematic for nonnatural side chains containing Nv and Hv.
Commercial database management systems provide the facile conversion between opposing nomenclatures using a CREATE VIEW statement.
References
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T. 1992. X-PLOR: A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Güntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- IUPAC-IUB Commission on Biochemical Nomenclature. 1970. Abbreviations and symbols for the description of the conformation of polypeptide chains. Tentative rules (1969). Biochemistry 9 3471–3479. [DOI] [PubMed] [Google Scholar]
- IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). 1984. Nomenclature and symbolism for amino acids and peptides. Recommendations 1983. Biochem. J. 219 345–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley, J.L., Bax, A., Arata, Y., Hilbers, C.W., Kaptein, R., Sykes, B.D., Wright, P.E., and Wüthrich, K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 280 933–952. [DOI] [PubMed] [Google Scholar]
- Piatetsky-Shapiro, G. and Frawley, W. 1991. Knowledge discovery in databases. AAAI Press/The MIT Press, Cambridge, MA.
- Sawaya, M.R., Prasad, R., Wilson, S.H., Kraut, J., and Pelletier, H. 1997. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry 36 11205–11215. [DOI] [PubMed] [Google Scholar]
- Seavey, B.R., Farr, E.A., Westler, W.M., and Markley, J.L. 1991. A relational database for sequence-specific protein NMR data. J. Biomol. NMR 1 217–236. [DOI] [PubMed] [Google Scholar]