Abstract
The oxidative folding, particularly the arrangement of disulfide bonds of recombinant extracellular N-terminal domains of the corticotropin-releasing factor receptor type 2a bearing five cysteines (C2 to C6), was investigated. Depending on the position of a His-tag, two types of disulfide patterns were found. In the case of an N-terminal His-tag, the disulfide bonds C2–C3 and C4–C6 were found, leaving C5 free, whereas the C-terminal position of the His-tag led to the disulfide pattern C2–C5 and C4–C6, and leaving C3 free. The latter pattern is consistent with the disulfide arrangement of the extracellular N-terminal domain of the corticotropin-releasing factor (CRF) receptor type 1, which has six cysteines (C1 to C6) and in which C1 is paired with C3. However, binding data of the two differently disulfide-bridged domains show no significant differences in binding affinities to selected ligands, indicating the importance of the C-terminal portion of the N-terminal receptor domains, particularly the disulfide bond C4–C6 for ligand binding.
Keywords: protein folding, affinity tag, corticotropin-releasing factor receptor
Affinity tags of fusion proteins are widely used in recombinant protein expression techniques. The most common affinity tags, such as poly-histidine (Bornhorst and Falke 2000), glutathione S-transferase (GST; Smith 2000), maltose binding protein (MBP; Sachdev and Chirgwin 2000), and streptavidin tags (Skerra and Schmidt 2000), are routinely used for increasing expression, enhancement of solubility, and protein purification. In novel developments of high-throughput screening techniques, such as scintillation proximity assay (SPA) and alphaScreen (PerkinElmer Life and Analytical Sciences, Inc.), one or two proteins, respectively, are attached to beads via affinity tags. It is known that large affinity tags (MBP, GST) influence the conformational homogeneity and hence the three-dimensional structure of in vivo folded target proteins (Smyth et al. 2003). To date, however, there are no data about the impact of small affinity tags, such as the (His)6-tag, on the protein conformation and, especially, the disulfide pattern when generating cysteine-containing proteins by recombinant expression.
The large N-terminal domains of G protein–coupled receptor (GPCR) class B, such as the receptor for the peptide hormone corticotropin-releasing factor (CRF; Vale et al. 1981), contain several cysteines. There are two receptor subtypes for CRF in mammals, the CRF1 and CRF2 receptor (Chang et al. 1993; Chen et al. 1993; Sklenar et al. 1993; Vita et al. 1993; Lovenberg et al. 1995; Stenzel et al. 1995; Smith 2000), for the latter three splice variants, CRF2(a), CRF2(b) and CRF2(c), exist. The CRF1 and CRF2(b) receptor N terminus (NT) without their putative signal sequence contain six cysteines (C1 through C6) and form three disulfide bridges. In contrast, the CRF2(a) and the CRF2(c) receptor NT without signal peptides exhibit only five cysteines (C2 through C6) and therefore forms only two disulfide bridges, leaving one cysteine unbound (Fig. 1 ▶). Recent studies on the N-terminal domains of the CRF receptors (CRF1 receptor [Hofmann et al. 2001; Perrin et al. 2001], CRF2(b) receptor [Perrin et al. 2003]), as well as other receptors of this class, such as PTHR1 (Grauschopf et al. 2000) and GLPR1 (Bazarsuren et al. 2002), showed that the N-terminal domains serve as the major ligand binding site. For the CRF1 receptor, it is also known that the disulfide pattern of the N-terminal domain is critical for binding of the natural ligand CRF (Qi et al. 1997). Thus, a defined conformation of the N-terminal domains of CRF receptors is suggested to be important for interactions with the ligands.
Figure 1.
Alignment of the expressed CRF2(a) receptor N termini with an N- and C-terminal (His)6-tag, respectively.
In this study, we investigated the disulfide patterns of recombinant, in vitro, folded N-terminal domains of the CRF2(a) receptor in dependence on the position of a (His)6-tag. The binding behavior of several ligands (Table 1) to the refolded, C- or N-terminally, (His)6-tagged receptor NT was studied by ligand binding assay by using the SPA technique.
Table 1.
Primary structure of CRF analog
| Peptide | Sequence |
| Rat urocortin 1 (1–40) | D D P P L S I D L T F H L L R T L L E L A R T Q S Q R E R A E Q N R I I F D S V |
| Rat urocortin 1 (8–40) | - - - - - - - D L T F H L L R T L L E L A R T Q S Q R E R A E Q N R I I F D S V f H L L R E V L E B A R A E Q L A Q E A H K N R K L B E I I |
| Astressina | * * |
All peptides were synthesized as C-terminal amides. The truncated rat urocortin 1 (8–40) is N-terminally acetylated.
a f indicates D-phenylalanine; B, norleucine.
* Lactam bridge connecting the side chains of glutamic acid and lysine.
Results and Discussion
The recombinant production of cysteine-containing proteins in Escherichia coli in inclusion bodies requires an in vitro folding step. The in vitro folding efficiency of disulfide bond–containing proteins is adjustable by several parameters, such as pH, protein concentration, ratio of low-molecular-weight thiols in reduced and oxidized form (Saxena and Wetlaufer 1970), and the concentration of non-denaturating reagents. The influence of other impacts, such as fusion tags on disulfide bridging, has to be elucidated for each individual protein.
Overexpression and in vitro folding
To investigate the influence of an N- and C-terminal (His)6-tag, respectively, on the in vitro folding and resulting disulfide pattern of the five cysteine containing CRF2(a) receptor NT, both recombinant proteins were produced in E. coli (Fig. 1 ▶). According to different algorithms (Persson and Argos 1996; Cserzo et al. 1997; Nielsen et al. 1997; Reczko et al. 2002), amino acids Ala19-Arg114 form the extracellular N-terminal domain of the rat CRF2(a) receptor without signal peptide. Overexpression of the CRF2(a) receptor NT with an N- and C-terminal (His)6-tag, respectively, gave 40 to 60 mg N-(His)6-CRF2(a) receptor NT and 30 to 60 mg CRF2(a)-C-(His)6 receptor NT per liter cell culture. Here, the position of the (His)6-tag had no remarkable influence on the expression levels. Sodium dodecylsulfate–polyacrylamide gel electrophoresis of soluble and insoluble protein fractions showed that both CRF2(a) receptor NTs were deposited in inclusion bodies almost exclusively, and that the apparent molecular size of both CRF2(a) receptor NT is ~12.5 kDa according to the predicted molecular size. It should be noted that the presence of the (His)6-tag on both termini led to increased expression levels in comparison to the untagged protein.
For subsequent protein renaturation, an in vitro refolding protocol according to procedures described elsewhere (Buchner and Rudolph 1991; Rudolph and Lilie 1996; Lilie et al. 1998) was applied. The optimized protocol led to a rapid reshuffling of improper disulfide bonds and to an enrichment of one folding species for the N- and C-terminally (His)6-tagged protein, respectively, with yields of renaturation in a range of 40% to 50%. Because of the requirement of a high resolution for the separation of folding intermediates, preparative reversed-phase high-performance liquid chromatography (HPLC) was used to purify the oxidized products, with an overall yield of 10% for the entire folding and purification process.
Assignment of disulfide bridges
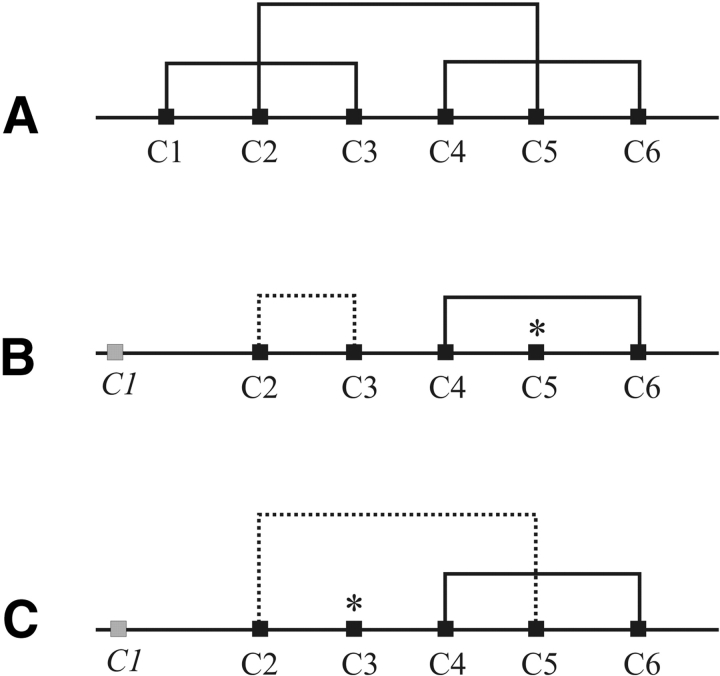
The characterization of disulfide patterns in the extracellular N-terminal domains of the N- and C-terminally (His)6-tagged CRF2(a) receptors was done by a combination of enzymatic/chemical cleavage and mass spectrometry (MS) of cleaved fragments. The homogeneity of the digested, final, purified refolding products was confirmed by the lack of differently connected species. It should be noted that CRF2(a) receptor NT carries only five cysteines (C2 through C6); consequently, only two disulfide bridges and one unlinked cysteine can be expected. Moreover, a chemical cleavage at Asn69-Gly70 by hydroxylamine (Bornstei and Balian 1970) before the Glu-C digestion was necessary to increase the susceptibility of N-(His)6-CRF2(a) receptor NT for the enzyme. The following analysis of digested fragments revealed the linkages Cys64(C4)–Cys98(C6) and Cys40(C2)–Cys50(C3) leaving Cys83(C5) as unbound (Table 2). For the CRF2(a)-C-(His)6 receptor NT, we faced the problem of disulfide scrambling (Glocker et al. 1995) due to the uneven number of cysteines when performing standard digestion at pH 7.5. For this case, we used the strategy for blocking the free cysteine applying alkylation (Sechi and Chait 1998) or cyanylation (Wakselman et al. 1976) at pH 5.7 with iodoacetamide (IAA) and cyanodimethylaminopyridinium tetrafluoroborate (CDAP), respectively. After enzymatic digestion of the alkylated or cyanylated NT using chymotrypsin, the found fragments clearly show disulfide bridging between Cys40(C2) and Cys83(C5) as well as Cys64(C4) and Cys83(C6) (Table 2). We concluded from these unambiguously obtained disulfide bridges that Cys50(C3) is not involved in disulfide bonds. Thus, the disulfide pattern of N-(His)6-CRF2(a) receptor NT is not analog with disulfide bridges C1–C3, C2–C5, and C4–C6 found in both, CRF1 receptor NT (Hofmann et al. 2001; Perrin et al. 2001) and CRF2(b) receptor NT (Perrin et al. 2003), respectively, which were expressed as soluble, in vivo, folded proteins. In opposite, the CRF2(a)-C-(His)6 receptor NT shows a disulfide pattern in accordance with the described ones for CRF1 receptor NT and CRF2(b) receptor NT, respectively (Fig. 2 ▶).
Table 2.
Disulfide analysis of CRF2(a) receptor NT determined by MALDI-MS of cleaved fragments
| Fragment | Cysteine | Cysteine pattern | Calc. [M+H]+ | Found [M+H]+ |
| N-(His)6-CRF2(a) receptor NT, hydroxylamine, GluC | ||||
| 35-61 | Cys40-Cys50 | C2-C3 | 2869.30 | 2869.40 |
| 62-66+87-103 | Cys64-Cys98 | C4-C6 | 2546.20 | 2548.81 |
| CRF2(a)-C-(His)6 receptor NT, chymotrypsin, cyanylation | ||||
| 40-44+81-89 | Cys40-Cys83 | C2-C5 | 1655.71 | 1655.74 |
| 38-59+81-84a | Cys40,50-Cys83 | C2,3-C5 | 2868.42 | 2868.21 |
| 45-68+96-102a | Cys50,64-Cys98 | C3,4-C6 | 3486.93 | 3486.90 |
| CRF2(a)-C-(His)6 receptor NT, chymotrypsin, alkylation | ||||
| 39-43+80-88 | Cys40-Cys83 | C2-C5 | 1655.70 | 1655.50 |
| 59-67+95-108 | C64-Cys98 | C4-C6 | 2868.31 | 2868.30 |
| 39-58+80-88b | Cys40,50-Cys83 | C2,3-C5 | 3486.80 | 3485.48 |
a Cyanylation adduct.
b Glutathione adduct.
Figure 2.
Disulfide pattern of the expressed, in vitro refolded CRF2(a) receptor N termini with an N- and C-terminal (His)6-tag (B,C), respectively, in comparison to the in vivo folded CRF1 receptor N terminus (A). Unbound cysteines are marked with *. Cysteine 1 in CRF2(a) receptor N termini is located in the signal sequence and labeled in italics.
The results unambiguously show that depending on the position of the (His)6-tag, two dissimilar patterns were assigned for the two differently (His)6-tagged CRF2(a) receptor NT (Fig. 2 ▶). This unexpected result can only be explained by an unfavorable influence of the (His)6-tag on the folding behavior of the corresponding protein due to its close proximity to cysteine C2 and, second, a proposed less-stabilized conformation in this region of the CRF2(a) receptor NT. The latter aspect would consequently suggest a higher conformational stability in the region involving cysteines C4 and C6, where independence of cysteine-linkage on the position of the (His)6-tag was observed.
Ligand binding
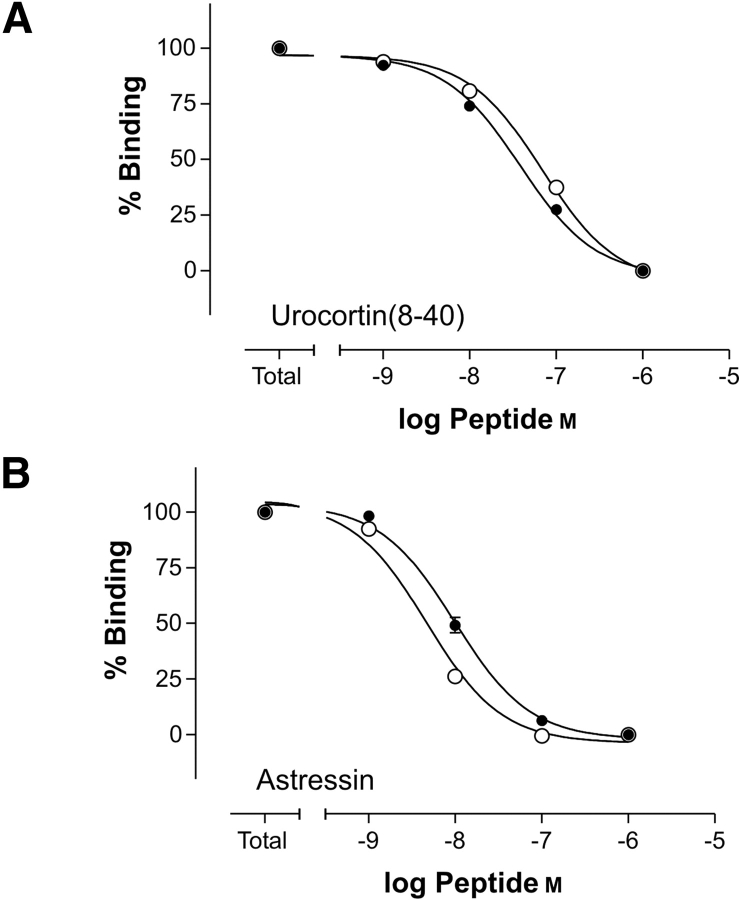
The binding characteristics of selected ligands (Table 1) to the two N-terminal domains of the CRF2(a) receptor, exhibiting different disulfide patterns due to the position of the (His)6-tag, were estimated by competitive displacement studies of radiolabeled rat urocortin 1 using SPA technique. Here, the anchorage of the extracellular N-terminal domains on copper-loaded SPA beads was mediated via the (His)6-tag. The binding data (Table 3) show no significant differences in binding behavior of the two N-terminal domains of the CRF2(a) receptor (Fig. 3 ▶). Both domains show high affinities for the antagonist astressin (~5 nM) and moderate affinities for the N-terminally truncated rat urocortin 1(8–40; 50 to 70 nM), which in this assay is equipotent to the full-length rat urocortin 1(1–40). The completely reduced and ensuing alkylated CRF2(a) receptor NT (C-terminal (His)6-tag) did not show any binding.
Table 3.
Results of ligand binding studies by SPA
| CRF2(a)-C-(His)6 receptor NT | N-(His)6-CRF2(a) receptor NT | |
| Ligand | EC50 [nM] | EC50 [nM] |
| Rat urocortin 1 (8–40) | 45.6 ± 4.9 | 71.0 ± 10.1 |
| Astressin | 6.8 ± 1.5 | 4.6 ± 0.5 |
The data are means of at least three different experiments performed in triplicates using [125I]Tyr0-urocortin as radioligand.
Figure 3.
Inhibition of specific [125I-Tyr0]-urocortin 1 binding to CRF-rNT by the unlabeled CRF-like ligands urocortin 1 (8–40) and astressin by using the SPA technology. All values are given as mean ± SEM. (A) Binding of urocortin 1 (8–40) to the CRF2(a) receptor N terminus with C-terminal (His)6-tag (filled circles) and N-terminal (His)6-tag (open circles). (B) Binding of astressin to the CRF2(a) receptor N terminus with C-terminal (His)6-tag (filled circles) and N-terminal (His)6-tag (open circles).
In summary, our results show that a “locking in place” of disulfide bonds in the course of oxidative in vitro folding of proteins can be influenced by the position of the (His)6-tag. The isolated N-terminal domain of the CRF2(a) receptor containing an uneven number of cysteines exhibits two dissimilar disulfide patterns depending on the tag position, with varied C2/C3/C5 bonding but a constant C4–C6 disulfide bond. On the other hand, as shown for the CRF2(a) receptor NT, the occurrence of different disulfide patterns does not necessarily result in an altered binding behavior. Obviously, the conformation of the N-terminal portion of extracellular N-terminal domains of CRF receptors, especially the correctness of the disulfide bonds in this region, is not essential for ligand binding. In accordance with this finding are mutation studies of the CRF1 receptor (Qi et al. 1997): Replacement of the first cysteine (C1) by serine did not affect the function. However, a completely reduced and ensuing alkylated CRF2(a) receptor NT [C-terminal (His)6-tag] did not show any binding anymore, indicating the importance of the disulfide bridge between C4 and C6 and hence stability of this C-terminal region within the extra-cellular N-terminal receptor domain.
Materials and methods
Construction of expression plasmids
N-(His)6-CRF2(a) receptor NT
A PCR-amplified cDNA (312 bp, primers 5′-AGCTTGGGATC CGCCGAAGAGCTGCTTTTG-3′ and 5′-GATGAGCTCTTAT CGGTAATGCAGGTC-3′) coding for Ala19-Arg114 of the rat CRF2(a) receptor was initially inserted into pQE-30 (Qiagen) via BamHI and SacI restriction sites. For expression reasons, the resulting vector was used as template for recloning a cDNA (348 bp) coding for rat CRF2(a) receptor NT, including an N-terminal (His)6-tag (primers 5′-TTAACCATGGGAGGATCGCATCACCAT-3′ and 5′-CGGGGTCTCGAGCTCTTATCGGTAATG-3′) into pET-15b (Novagen), using NcoI and XhoI restriction sites to yield the N-terminally (His)6-tagged protein.
CRF2(a)-C-(His)6 receptor NT
To yield the C-terminally (His)6-tagged protein, a cDNA (310 bp, primers 5′-CAGCTTGCATATGGCCGAAGAGCTGCTTTTGG-3′ and 5′-GACCTCGAGTCGGTAATGCAGGTCATACTTCC-3′) coding for Ala19-Arg114 of the rat CRF2(a) receptor was inserted into pET-21a (Novagen) by using NdeI and XhoI restriction sites.
The authenticity of the resulting recombinant expression vectors was confirmed by DNA sequence analysis.
Expression in E. coli and isolation of inclusion bodies
E. coli BL21(DE3) (Novagen) was transformed with the respective plasmids and grown in LB medium supplemented with ampicillin (100 μg/mL). For expression, 1 L LB medium was inoculated with 15 to 50 mL overnight culture and grown at 37°C to an optical density of OD600nm = 0.5 to 0.7. Expression of the proteins was induced with 1 mM IPTG for 3.0 to 3.5 h at 37°C. Cells were harvested by centrifugation, and the cell pellets were resuspended in 20 mM Tris, 2% (v/v) Triton, and protease inhibitor cocktail (EDTA-free at pH 8; 15 mL/pellet from 1 L cell culture). The cells were disrupted by high-pressure dispersion and subjected to lysis, and the inclusion bodies were collected and stored at −20°C.
The sequences of the expressed proteins are shown in Figure 1 ▶.
Renaturation and purification
The inclusion body pellet was solubilized in denaturation buffer (5 M guanidinium hydrochloride [GuHCl], 20 mM Tris at pH 7.5) by shaking and sonication. The proteins were purified by immobilized-metal affinity chromatography by using a chelating Sepharose FF column (Amersham Pharmacia Biotech AB) with immobilized Ni2+ ions (loading and washing buffer: 5 M GuHCl, 20 mM Tris at pH 7.5, elution buffer: 5 M GuHCl, 20 mM Tris, 0.5 M imidazole at pH 7.5). Following purification, the proteins were reduced by addition of dithiothreitol (DTT; 100 mM, 1 to 2 mg protein/mL, room temperature for 2 h). After reduction of the proteins, DTT and Ni2+ ions were removed by dialysis against denaturation buffer (pH 3.0) at 10°C. After readjusting the pH to 7.5, renaturation was achieved by dialysis against 0.5 M L-arginine, 100 mM Tris, 1 mM EDTA, 1 mM reduced glutathione (GSH), 1 mM oxidized glutathione (pH 7.5), and 0.5 to 0.7 mg protein/mL for 3 d at 10°C. After renaturation, a final dialysis step against 10% (v/v) glycerol, 50 mM NaCl, and 20 mM Tris (pH 6.5) overnight at 10°C was accomplished, and insoluble material was removed by centrifugation. The refolded proteins were purified by reversed phase (RP)–HPLC by using a Vydac C4 column (10 × 250 mm, 5-μm particle size, 300 Å pore size, number 214TP510), run in 0.1% (v/v) trifluoroacetic acid in water with increasing concentrations of acetonitrile (ACN) as mobile phase, and lyophilized.
Peptides
Astressin was purchased from Bachem AG. [125I-Tyr0]-urocortin 1 (2200 Ci/mmole) was obtained from Amersham Biosciences Europe GmbH. Other peptides were synthesized following the procedure described in reference (Beyermann et al. 2000).
Disulfide pattern analysis
Disulfide pattern analysis of all CRF receptor NTs was carried out by standard procedures of enzymatic digestion using trypsin, chymotrypsin, or Glu-C followed by matrix-assisted laser desorption/ ionization (MALDI)–MS.
N-(His)6-CRF2(a) receptor NT
Before analyzing the disulfide pattern in N-(His)6-CRF2(a) receptor NT, the protein was treated with 1.8 M hydroxylamine (Bornstei and Balian 1970) and incubated for 5 h at 45°C for cleavage of the Asn69-Gly70 and the Asn86-Gly87 peptide bonds. Following cleavage, the protein was purified by RP-HPLC (see Renaturation and Purification).
CRF2(a)-C-(His)6 receptor NT
For analyzing the disulfide pattern of CRF2(a)-C-(His)6 receptor NT, free cysteines were blocked by alkylation with IAA (phosphate buffer at pH 5.7, 25 μM protein, 5 mg/mL IAA, 2 M GuHCl, overnight at 37°C) or cyanylation with CDAP (phosphate buffer at pH 5.7, 25 μM protein, 0.25 mM CDAP, 1 mM EDTA, overnight at 40°C). Removal of excess reactants was performed by dialysis against 2 M GuHCl, 20 mM Tris, and 1 mM EDTA (pH 7.5).
MALDI-MS measurements were performed on a Voyager-DE STR BioSpectrometry Workstation MALDI–time of flight (TOF) mass spectrometer (Perseptive Biosystems, Inc.). As matrices for analyses of peptides and proteins, α-cyano-4-hydroxycinnamic acid and sinapinic acid, respectively, were used. The program SearchXLinks (www.caesar.de/searchxlinks/) was used to analyze the mass spectra of protein digests with regard to the presence of disulfide-linked fragments.
Ligand binding assay: SPA
The competitive binding assays were performed in triplicates in 2.0-mL colorless reaction tubes (Biozym Diagnostik GmbH) at room temperature by using a PVT copper His-tag SPA bead suspension (Amersham Biosciences Europe GmbH) in assay buffer containing 0.1% bovine serum albumin (BSA). The following reagents, diluted in assay buffer, were added in the order: 100 μL of unlabeled peptide with increasing peptide concentrations or buffer, 75 μL [125I-Tyr0]-urocortin (120 pM final concentration), and 75 μL of the respective CRF2(a) receptor NT (20 ng/tube). After incubation of the reaction mixture for 2 h, 50 μL of 10 mg/mL bead suspension (500 μg beads/tube) were added. The final reaction mixture was shaken and then incubated for 4 h. Finally, the tubes were counted in a Wallac 1410 set up in a 3H cpm-mode (SPA-cpm). Total binding observed and normalized at 100 pM; final concentration of tracer was ~13,000 SPA-cpm for both CRF2(a) receptor NT, with a nonspecific signal of ~3000 SPA-cpm determined in the presence of unlabeled 1 μM astressin. Binding data were analyzed by using the GraphPad Prism software.
Acknowledgments
We thank Prof. U.B. Kaupp for the gift of the CRF2(a) receptor cDNAs and G. Vogelreiter for technical assistance. This project was supported by the Sonderforschungbereich (SFB) 449.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ACN, acetonitrile
BSA, bovine serum albumin
CDAP, cyanodimethylaminopyridinium tetrafluoroborate
CRF, corticotropin-releasing factor
DTT, dithiothreitol
GPCR, G protein–coupled receptor
GST, glutathione S-transferase
GuHCl, guanidinium hydrochloride
HPLC, high-performance liquid chromatography
IAA, iodoacetamide
MALDI, matrix-assisted laser desorption/ionization
MBP, maltose binding protein
MS, mass spectrometry
NT, N terminus
RP, reversed-phase
SPA, scintillation proximity assay
TOF, time of flight
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04835904.
References
- Bazarsuren, A., Grauschopf, U., Wozny, M., Reusch, D., Hoffmann, E., Schaefer, W., Panzner, S., and Rudolph, R. 2002. In vitro folding, functional characterization, and disulfide pattern of the extracellular domain of human GLP-1 receptor. Biophys. Chem. 96 305–318. [DOI] [PubMed] [Google Scholar]
- Beyermann, M., Rothemund, S., Heinrich, N., Fechner, K., Furkert, J., Dathe, M., Winter, R., Krause, E., and Bienert, M. 2000. A role for a helical connector between two receptor binding sites of a long-chain peptide hormone. J. Biol. Chem. 275 5702–5709. [DOI] [PubMed] [Google Scholar]
- Bornhorst, J.A. and Falke, J.J. 2000. Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 326 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstei, P. and Balian, G. 1970. Specific nonenzymatic cleavage of bovine ribonuclease with hydroxylamine. J. Biol. Chem. 245 4854. [PubMed] [Google Scholar]
- Buchner, J. and Rudolph, R. 1991. Renaturation, purification and characterization of recombinant Fab-fragments produced in Escherichia coli. BioTechnology 9 157–162. [DOI] [PubMed] [Google Scholar]
- Chang, C.P., Pearse, R.V., O’Connell, S., and Rosenfeld, M.G. 1993. Identification of A 7 transmembrane helix receptor for corticotropin-releasing factor and sauvagine in mammalian brain. Neuron 11 1187–1195. [DOI] [PubMed] [Google Scholar]
- Chen, R.P., Lewis, K.A., Perrin, M.H., and Vale, W.W. 1993. Expression cloning of a human corticotropin-releasing-factor receptor. Proc. Natl. Acad. Sci. 90 8967–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo, M., Wallin, E., Simon, I., von Heijne, G., and Elofsson, A. 1997. Prediction of transmembrane α-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 10 673–676. [DOI] [PubMed] [Google Scholar]
- Glocker, M.O., Arbogast, B., and Deinzer, M.L. 1995. Characterization of disulfide linkages and disulfide bond scrambling in recombinant human macrophage-colony-stimulating factor by fast-atom-bombardment mass-spectrometry of enzymatic digests. J. Am. Soc. Mass Spektrom. 6 638–643. [DOI] [PubMed] [Google Scholar]
- Grauschopf, U., Lilie, H., Honold, K., Wozny, M., Reusch, D., Esswein, A., Schafer, W., Rucknagel, K.P., and Rudolph, R. 2000. The N-terminal fragment of human parathyroid hormone receptor 1 constitutes a hormone binding domain and reveals a distinct disulfide pattern. Biochemistry 39 8878–8887. [DOI] [PubMed] [Google Scholar]
- Hofmann, B.A., Sydow, S., Jahn, O., Van Werven, L., Liepold, T., Eckart, K., and Spiess, J. 2001. Functional and protein chemical characterization of the N-terminal domain of the rat corticotropin-releasing factor receptor 1. Protein Sci. 10 2050–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilie, H., Schwarz, E., and Rudolph, R. 1998. Advances in refolding of proteins produced in E. coli. Curr. Opin. Biotechnol. 9 497–501. [DOI] [PubMed] [Google Scholar]
- Lovenberg, T.W., Liaw, C.W., Grigoriadis, D.E., Clevenger, W., Chalmers, D.T., Desouza, E.B., and Oltersdorf, T. 1995. Cloning and characterization of a functionally distinct corticotropin-releasing factor-receptor subtype from rat-brain. Proc. Natl. Acad. Sci. 92 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Perrin, M.H., Fischer, W.H., Kunitake, K.S., Craig, A.G., Koerber, S.C., Cervini, L.A., Rivier, J.E., Groppe, J.C., Greenwald, J., Nielsen, S.M., et al. 2001. Expression, purification, and characterization of a soluble form of the first extracellular domain of the human type 1 corticotropin releasing factor receptor. J. Biol. Chem. 276 31528–31534. [DOI] [PubMed] [Google Scholar]
- Perrin, M.H., DiGruccio, M.R., Koerber, S.C., Rivier, J.E., Kunitake, K.S., Bain, D.L., Fischer, W.H., and Vale, W.W. 2003. A soluble form of the first extracellular domain of mouse type 2 β corticotropin-releasing factor receptor reveals differential ligand specificity. J. Biol. Chem. 278 15595–15600. [DOI] [PubMed] [Google Scholar]
- Persson, B. and Argos, P. 1996. Topology prediction of membrane proteins. Protein Sci. 5 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L.J., Leung, A.T., Xiong, Y.T., Marx, K.A., and Abousamra, A.B. 1997. Extracellular cysteines of the corticotropin-releasing factor receptor are critical for ligand interaction. Biochemistry 36 12442–12448. [DOI] [PubMed] [Google Scholar]
- Reczko, M., Staub, E., Fiziev, P., and Hatzigeorgiou, A. 2002. Algorithms in bioinformatics. Algorithms in bioinformatics (eds. R. Guigo and D. Gusfield), pp. 60–67. Springer Verlag, Berlin.
- Rudolph, R. and Lilie, H. 1996. In vitro folding of inclusion body proteins. FASEB J. 10 49–56. [PubMed] [Google Scholar]
- Sachdev, D. and Chirgwin, J.M. 2000. Fusions to maltose-binding protein: Control of folding and solubility in protein purification. 326: 312–321. [DOI] [PubMed]
- Saxena, V.P. and Wetlaufer, D.B. 1970. Formation of three-dimensional structure in proteins, 1: Rapid nonenzymic reactivation of reduced lysozyme. Biochemistry 9 5015–5023. [DOI] [PubMed] [Google Scholar]
- Sechi, S. and Chait, B.T. 1998. Modification of cysteine residues by alkylation: A tool in peptide mapping and protein identification. Anal. Chem. 70 5150–5158. [DOI] [PubMed] [Google Scholar]
- Skerra, A. and Schmidt, T.G.M. 2000. Use of the Strep-tag and streptavidin for detection and purification of recombinant proteins. 326: 271–304. [DOI] [PubMed]
- Sklenar, V., Piotto, M., Leppik, R., and Saudek, V. 1993. Gradient-tailored water suppression for H-1-N-15 Hsqc experiments optimized to retain full sensitivity. J. Magn. Res. A 102 241–245. [Google Scholar]
- Smith, D.B. 2000. Generating fusions to glutathione S-transferase for protein studies. Applications of chimeric genes and hybrid proteins: Methods in enzymology 326 254–270. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Mrozkiewicz, M.K., McGrath, W.J., Listwan, P., and Kobe, B. 2003. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 12 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, P., Kesterson, R., Yeung, W., Cone, R.D., Rittenberg, M.B., and Stenzelpoore, M.P. 1995. Identification of a novel murine receptor for cortico-tropin-releasing hormone expressed in the heart. Mol. Endocrinol. 9 637–645. [DOI] [PubMed] [Google Scholar]
- Vale, W., Spiess, J., Rivier, C., and Rivier, J. 1981. Characterization of A 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and β-endorphin. Science 213 1394–1397. [DOI] [PubMed] [Google Scholar]
- Vita, N., Laurent, P., Lefort, S., Chalon, P., Lelias, J.M., Kaghad, M., Lefur, G., Caput, D., and Ferrara, P. 1993. Primary structure and functional expression of mouse pituitary and human brain corticotropin-releasing factor receptors. FEBS Lett. 335 1–5. [DOI] [PubMed] [Google Scholar]
- Wakselman, M., Guibejampel, E., Raoult, A., and Busse, W.D. 1976. 1-Cyano-4-dimethylamino-pyridinium salts: New water-soluble reagents for cyanylation of protein sulfhydryl groups. J. Chem. Soc. 1 21–22. [Google Scholar]