Abstract
Grb14 is an adapter protein that is known to be overexpressed in estrogen receptor positive breast cancers, and in a number of prostate cancer cell lines. Grb14 has been demonstrated to bind to a number of activated receptor tyrosine kinases (RTKs) and to modulate signals transduced through these receptors. The RTKs to which Grb14 binds include the insulin receptor (IR), the fibroblast growth factor receptor (FGFR), the platelet-derived growth factor receptor (PDGFR), and the tunica endothelial kinase (Tek/Tie2) receptor. Grb14 has been shown to bind to these activated RTKs through its Src homology 2 (SH2) domain, with the exception of the insulin receptor, where the primary binding interaction is via a small domain adjacent to the SH2 domain (the BPS or PIR domain). Grb14 is a member of the Grb7 family of proteins, which also includes Grb7 and Grb10. We have solved the solution structure of the human Grb14–SH2 domain and compared it with the recently determined Grb7–SH2 and Grb10–SH2 domain structures.
Keywords: Src homology 2, nuclear magnetic resonance, cell signaling, protein structure
Growth factors are signaling proteins responsible for stimulating mitogenic and morphogenic events required for cellular growth and proliferation. Growth factors bind to the extracellular domains of their cognate receptors to transduce a signal across the cell membrane to carry out an intracellular response. The responses carried out by these signaling proteins include cellular proliferation (Fabian et al. 1993; Iwama et al. 1993; Reith et al. 1993; Casteran et al. 1994), differentiation (Shilo 1992; Xu et al. 1994), motility (Fridell et al. 1998; Qi et al. 1999), and apoptosis (Merlo et al. 1996; Ueno et al. 1997). Many of these transmembrane receptors are receptor tyrosine kinases (RTKs) that upon binding their cognate ligand, dimerize and become activated (autophosphorylated) on tyrosine residues within their intracellular domain. Thus activated, the intracellular domains can further transduce the signal by phosphorylating intracellular targets, or by recruiting intracellular target proteins to bind to the activated phosphotyrosine residues. Src homology 2, or SH2, domains are one of a few protein motifs that have been demonstrated to bind phosphotyrosine moieties (Sierke and Koland 1993), and have been shown to be discriminating in their specificities based upon the three to six residues immediately C-terminal to the phosphotyrosine residue (Waksman et al. 1993; Pascal et al. 1994; Songyang et al. 1994).
A family of 14 proteins, the Grb family, has been identified based on its ability to bind to the activated epidermal growth factor receptor (EGFR, also known as erbB1). A subfamily of this, the Grb7 family of proteins, has been identified based on a similar molecular topology (Ooi et al. 1995). This topology consists of an N-terminal proline rich region, a Ras association-like domain, a pleckstrin homology domain, a “between pleckstrin and Src homology” region (BPS), and a C-terminal SH2 domain (Daly 1998). The Grb7 family of proteins is comprised of Grb7, Grb10, and Grb14. The SH2 domains of the Grb7 family are unique among SH2 domains owing to a four residue insertion at the juncture of the βE strand and the EF loop. The members of the Grb7 family may carry out a more specialized signaling pathway than other ubiquitously expressed SH2 domain-containing proteins as their expression is tissue-specific. Grb7 is expressed in the liver, kidney, and gonads (Margolis et al. 1992), while Grb14 is expressed in the liver, kidney, pancreas, gonads, heart, and skeletal muscle (Daly et al. 1996). In addition, the individual SH2 domains of the Grb7 family show considerable selectivity in binding to their RTK partners. Grb7 binds in vivo to the RTK erbB2 via its SH2 domain, while the Grb14–SH2 domain does not appreciably bind with this RTK (Janes et al. 1997). Correlations have been made between Grb7 protein family overexpression and cancer. Grb7 has been shown to be co-over-expressed with erbB2 in 20% to 30% of all breast cancers (Stein et al. 1994), and Grb14 has been demonstrated to be highly overexpressed in some prostate cancer cell lines (Daly et al. 1996). Grb14 has been shown to bind to a number of RTKs, including EGFR, PDGFR, FGFR, IR, and Tek/Tie2.
We have solved the solution structure of the human Grb14–SH2 domain, and compare it to the recently solved human Grb7–SH2 and Grb10–SH2 domain structures (Ivancic et al. 2003; Stein et al. 2003). In addition, we present a hypothesis that the orientation of the βE strand, EF loop, and βF strand in the SH2 domains of this protein family may play a role in determining binding specificity.
Results
Human Grb14–SH2 domain structure
For all the Grb7 protein family SH2 domains, the nomenclature used to describe secondary structural features follows that of Eck et al. (1993). That is, β strands and α helices are named sequentially A, B, C, etc., beginning from the N terminus (Fig. 2A ▶; see below). Loop portions of the SH2 domain are named according to the two secondary structural elements that they connect (e.g., the D′E loop is the loop between the βD′ strand and the βE strand). Sequence assignments of the human Grb14–SH2 domain have been published previously (Scharf and Lyons 2002). Of the 117 residues composing the Grb14–SH2 domain, the N-terminal glycine residue remains unassigned.
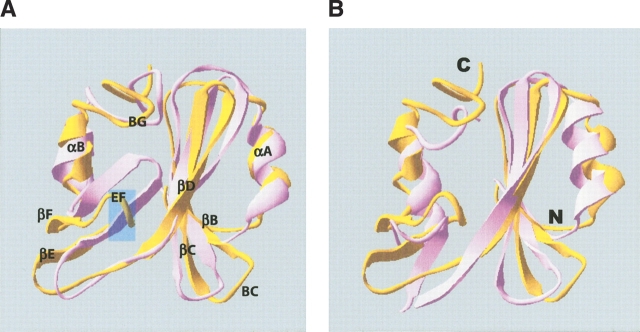
Figure 2.
(A) Ribbon representation of one of the 20 representative NMR solution structures of the human Grb14–SH2 domain (gold) aligned with a representative structure of the Grb7–SH2/erbB2 peptide complex (mauve). Secondary structural elements are labeled from the N terminus to the C terminus, with αA being the N-terminal α-helix, αB the C-terminal α-helix, etc. The approximate location of the insertion described in the text is indicated by a transparent blue box. (B) Ribbon representation of one of the 20 representative NMR solution structures of the human Grb14–SH2 domain (gold) aligned with the X-ray crystallographic structure of the Grb10–SH2 domain (mauve; Stein et al. 2003). The N and C termini are labeled.
The NMR solution structure of the SH2 domain of human Grb14 was solved using three-dimensional heteronuclear 15N- and 13C-edited NOESY NMR experiments (Figs. 1 ▶, 2 ▶). The calculations yield a family of structures with a root-mean-square deviation (RMSD) of 0.65 Å for the backbone heavy atoms and 1.03 Å for all atoms (residues 16–113). The Grb14–SH2 structure (Figs. 1 ▶, 2 ▶), with its three long antiparallel β-strands, flanked by a few smaller β-strands, and an α-helix at each end reveals the standard SH2 domain structure with the exception of the βE strand, which is two residues longer than the classic SH2 domain βE strands. This extension is part of a four amino acid insert (L80–G83) in the region that elongates the EF loop by two residues, and corresponds to a region of the domain that often interacts with the (pY) + 3 residue in ligand binding (Waksman et al. 1993). Throughout the manuscript, peptide residue positions subsequent (C-terminal) to the phosphotyrosine residue are referred to as +1 (the first residue after the pTyr), +2 (the second residue after the pTyr), etc. Residue positions prior (N-terminal) to the phosphotyrosine are referred to as −1 (the residue immediately preceding the pTyr), and so forth.
Figure 1.
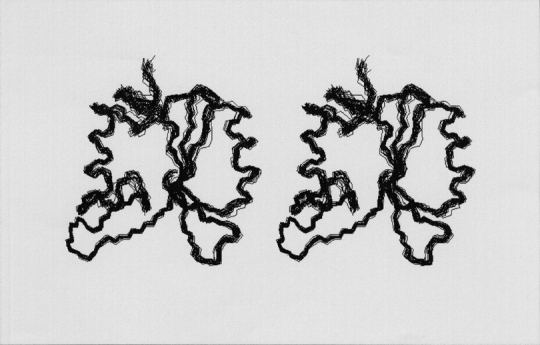
Backbone-trace stereo views of 20 representative NMR solution structures of the human Grb14–SH2 domain. Residues 16–114 are visualized. The orientation of the Grb14–SH2 domain in the figure is the same as that shown in Figure 2 ▶. The structures were selected based upon the lowest NOE violation energies out of 50 calculated structures.
Ramachandran plot analysis (Laskowski et al. 1993) of the secondary structure elements yields 81.1% of residues falling within the most favored region, 10.8% within the additionally allowed region, 2.7% within the generously allowed region, and 2.7% within the disallowed region. The homology of the highly conserved hydrophobic core of SH2 domains (corresponding in Grb14 to residues W16, I30, F40, L41, V42, L54, M56, I62, I67, F77, L90, L93, V94, L104, and L108) is conserved, except for the semiconservative substitution of M56 for the V/I/L/F usually found at this position (Waksman et al. 1993). The electrostatic potential of the molecule is divided, with about two-thirds of the domain being basic, and the other third being highly acidic. The highly basic phosphotyrosine-binding pocket, in conjunction with numerous exposed lysine residues at one end of the molecule forms the basic region. The acidic region of the molecule is formed by two juxtaposed loops, the D′E and EF loops. This region is well conserved among the Grb14–, Grb7–, and Grb10–SH2 domains.
Discussion
Human Grb14–SH2 domain structure
The human Grb14–SH2 domain has the same overall fold as that of the numerous other published SH2 domain structures (Waksman et al. 1993; Pascal et al. 1994; Thornton et al. 1996; Hoedemaeker et al. 1999; Ogura et al. 1999; Mallis et al. 2002; Ivancic et al. 2003; Stein et al. 2003; and others). The most notable difference in the Grb14–SH2 domain structure is the four-residue insertion at the juncture of the βE strand and EF loop, resulting in an extension of two residues in each. The Grb14–SH2 domain contains a highly acidic region consisting of the adjacent D′E and EF loops, with both loops having a D-D-G sequence, and the D′E loop also containing two additional Glu residues. This region is also acidic in both the Grb7 (a D-D-G and an E-E-G sequence, and one additional Glu in the D′E loop) and Grb10 (two D-D-G and one additional Glu in the D′E loop) SH2 domains. The functional role of these residues is unknown; one hypothesis may be that the region is involved in stabilizing interactions with other basic regions within the Grb14 protein (the Pro-rich domain, the pleckstrin homology domain, and the SH2 domain all have candidate regions). The possibility that such interactions may occur has been demonstrated in the oligomerization seen in Grb10 in which the SH2 domain was demonstrated to play a role (Dong et al. 1998). Another possibility may be the involvement of these residues in similar stabilizing interactions with such regions on its target RTK. Evidence supporting this hypothesis is the demonstration that mutation of the Grb10 EF2 aspartate residue to a valine abrogates binding of the Grb10–SH2 to activated EGFR and IR, as well as nonphosphorylated Raf1 and MEK1 (Nantel et al. 1998).
The Grb7 protein family (Grb 7, 10, and 14) SH2 domains are classified as type I SH2 domains, owing to the presence of either a phenylalanine or a tyrosine at the βD5 position. This class of SH2 domains is typified by a preference for a phosphotyrosine-hydrophilic-hydrophilic-hydrophobic recognition sequence in the pY−+1−+2−+3 positions of the peptide ligand. The observed chemical shift changes in the Grb14–SH2 domain upon binding to a FGFR-derived peptide (pYLDL) have been previously reported (Scharf and Lyons 2002). The FGFR-derived peptide matches this sequence, with the exception of the +1 residue. The only other noted preference of the Grb14–SH2 domain for its ligands appears to be for a bulky hydrophobe in the +3 position (Janes et al. 1997; Jones et al. 1999).
The Grb14–SH2 domain contains a four-residue insertion in the EF loop region as do all three members of the Grb7 protein family. Based upon sequence identity to the Grb7–SH2 domain (68%) the Grb14–SH2 domain should bind its phosphorylated peptide ligands in a turn conformation, as does the Grb7–SH2 domain (Ivancic et al. 2003). It has been experimentally shown that the Grb14–SH2 domain does not bind to erbB2 (Janes et al. 1997). In fact, as stated previously, the consensus binding site sequence for the Grb14–SH2 domain includes a hydrophobic residue in the +3 position following pY. This is the consensus binding sequence for SH2 domains that bind their phosphorylated ligands in an extended conformation (Waksman et al. 1993). These contradictory results, the Grb14–SH2 domain experimentally derived consensus binding sequence versus Grb7–SH2/Grb14–SH2 domain homology, may be explained by specificity determinants imposed by the βE strand, EF loop and βF strand regions in this family of SH2 domains.
Structure comparison between the Grb14–SH2 domain, the Grb10–SH2 domain, and the Grb7–SH2/pY1139 complex
Alignment of the secondary structural elements (αA helix, βB strand, βC strand, βD strand, and αB helix) of a representative human Grb14–SH2 domain structure with the X-ray diffraction structure of the human Grb10–SH2 domain (Stein et al. 2003) gives a RMSD of 3.7 Å. One noticeable difference between the Grb14–SH2 and Grb10–SH2 domain structures is the relative orientation of the C-terminal helix (Fig. 2B ▶). The C-terminal helix orientation in the Grb14–SH2 domain more closely resembles the orientation of this helix in the Grb7–SH2/pY1139 complex (Fig. 2A ▶; Brescia et al. 2002; Ivancic et al. 2003). In contrast, the conformation of the EF loop region in the Grb14 and Grb10 SH2 domain structures are more similar to each other than to the EF loop conformation of the Grb7–SH2/pY1139 complex structure (Fig. 2A ▶). Alignment of representative Grb14–SH2 and Grb7–SH2/pY1139 structures reveals close similarities between the two domains. The backbone RMSD between the two structures for the N-terminal αA helix, βB strand, βC strand, βD strand, and αB helix is 3.4 Å. The majority of the differences between the Grb14–SH2 and Grb7–SH2 domain structures reside in the orientation of the βE strand, EF loop, and βF strand.
The structure of the Grb14–SH2 domain in comparison to the Grb7–SH2/pY1139 complex structure has provided clues to a potential functional difference related to the conformation of the EF loop region in these proteins. The βE strand, EF loop, and βF strand conformations between Grb7 and Grb14 are substantially different. In the Grb7–SH2 domain this region is rigidly extended and packed against the remainder of the SH2 domain structure (i.e., “closed”; Fig. 2A ▶). This is similar to the EF loop conformation in the Grb2–SH2 domain (Ogura et al. 1999). In contrast, the EF loop conformation in the Grb14–SH2 and Grb10–SH2 domain structures are similar, which is in a curved downward and outward position relative to the rest of the SH2 domain structure (Fig. 2B ▶). The rigidly extended and closed EF loop conformation of the Grb7–SH2 and Grb2–SH2 domains may facilitate occlusion of the +3 binding pocket in these domains. The more curved-outward and open conformation of the EF loop in the Grb14–SH2 and Grb10–SH2 domains may leave the +3 binding pocket available for binding a ligand in an extended conformation.
The Grb14–SH2 domain structure reveals one residue (E75) in the disallowed region of the Ramachandran plot. E75 is located in the D′E loop. The residue in the corresponding position in the Grb7–SH2 domain is R78, and falls into the generously allowed region of the Ramachandran plot (Ivancic et al. 2003). The corresponding residue in the Grb10–SH2 domain falls into a region of the structure lacking electron density (residues 490–494; Stein et al. 2003). Preliminary backbone nuclear relaxation measurements performed on the Grb7–SH2 domain indicate this region of the SH2 domain may contain sites of increased mobility (B. Lyons, unpubl.). In addition, sequence specific assignments for the Grb14–SH2 domain were initially difficult to obtain in the D′E loop region because some resonances appear exchanged broadened. Increased mobility in this “hinge” region of the SH2 domain may be a characteristic found in all three members of the Grb7 protein family, and could play a role in determining the overall conformation (and thus SH2 domain specificity) of the βE strand, EF loop, and βF strand regions. Evidence for mobility in the D′E region has been observed in at least one other SH2 domain. The order parameter values (S2) for the D′E transition and loop region of the phospholipase C-γ1 C-terminal SH2 domain display the lowest stretch of values for the entire molecule in both the unliganded and liganded states (Farrow et al. 1994).
There are subtle sequence differences in the βE strands and EF loops of these two SH2 domains that may affect the type of stabilizing interactions formed upon ligand binding, thus directing specificity. A thorough investigation of the affect of mutations in the βE strand and EF loop is the next step in confirming this hypothesis. Solving the structure of the Grb14–SH2 domain in complex with a phosphorylated peptide ligand representative of a natural Grb14 RTK target is another immediate goal within our laboratory.
Atomic coordinates for the human Grb14–SH2 domain have been deposited with the protein database (PDB) at Rutgers University. Sequence specific assignments for the human Grb14 SH2 domain have been deposited at the BioMagRes Bank (http://www.bmrb.wisc.edu) under BMRB accession number 5314.
Materials and methods
Sample preparation
Expression and purification of the human Grb14–SH2 domain has been described in detail previously (Scharf and Lyons 2002). Sample conditions for NMR spectroscopy were 0.6–1.0 mM protein, 50 mM sodium acetate, 100 mM NaCl, 5 mM DTT, 1 mM EDTA, and pH 5.5.
NMR spectroscopy
All NMR experiments were carried out using a Varian INOVA 500 MHz NMR Spectrometer. Data sets were processed using the software program AZARA v2.0 (Boucher 1993). Processing in all dimensions sequentially involved multiplication with a phase-shifted sine or sine2 bell curve followed by zero-filling to the next power of two (De Marco and Wüthrich 1976). The NMR experiments employed are all found in the suite of triple resonance experiments available in the pulse sequence library ProteinPack (Kay 1995). Amide nitrogen and hydrogen, and α-carbon assignments were obtained using the two- and three-dimensional double and triple resonance experiments 2D NHSQC (Bax et al. 1990a,b), 3D HNCA (Clore et al. 1990), and 3D HN(CO)CA (Bax and Ikura 1991). For the triple resonance experiments, a 13C(F1) carrier frequency at 50.66 ppm, a spectral width of 4400 Hz, and 64 and 32 transients for each increment were used for the HNCA and HN(CO)CA experiments, respectively. α-Carbon resonances were assigned using the 3D CBCANH (Bax and Grzesiek 1993) and CBCA(CO)NH (Grzesiek and Bax 1992) experiments at the same carrier frequency, a spectral width of 9050 Hz, and 16 (CBCANH) and 32 (CBCA(CO)NH) transients for each increment. α-Carbon and farther side-chain carbon resonance assignments were made using the 3D C(CO)NH experiment (Kuboniwa et al. 1994) at the same carrier frequency and spectral width as for the CBCANH and with eight transients per increment. Nonexchangeable hydrogen assignments and NOE constraints were made using a combination of the NHSQC-TOCSY and NHSQC-NOESY (60- and 150-msec mixing times, respectively) for amide to aliphatic resonances. For aliphatic to aliphatic resonances, the HCCH-TOCSY and 13C-edited NOESY (100-msec mixing times) experiments were employed (Bax et al. 1990b; Driscoll et al. 1990).
Structure determination
All spectral analysis was accomplished using the software Ansig v3.3 (Kraulis 1989). NOE cross-peak intensities were classified into three categories: strong (1.8 Å to 2.5 Å), medium (2.6 Å to 3.5 Å), and weak (3.6 Å to 6.0 Å). A total of 55 hydrogen bond restraints were employed, assigned based upon evaluation of candidate hydrogen bonds calculated in the absence of hydrogen bond restraints, coupled with backbone NOE patterns and chemical shifts. Structure calculations used a total of 1870 NOE constraints divided into 473 intraresidue constraints, 689 sequential constraints, 335 medium-range constraints (1 < |i−j| ≤4), and 373 long-range constraints (|i−j| >4) as listed in Table 1. NOEs were tabulated and quantified manually using the intraresidue amide proton:α proton distance at 2.8 Å as the reference for distance calibrations. Pseudo-atom corrections were made for methyl groups and degenerate methylene groups. Structure calculations were performed using CNS software version 1.1 beginning with extended structures in torsion angle space dynamics followed by Cartesian minimization (Brunger et al. 1998). Dihedral, improper, and nonbonded bond and angle parameters for nonhydrogen atoms were derived from the Cambridge database protein parameter and topology files (Engh and Huber 1991) parallhdg5.2.pro and topallhdg5.2.pro, obtained from ftp://ftp.pasteur.fr/pub/BIS/nilges/parameters/dg_version5.2. The initial high-temperature stage was computed at 50,000 K with 1000 molecular dynamic steps of 15 fsec and NOE and dihedral scaling factors of 150 and 200, respectively. The first slow-cooling stage was comprised of 1000 steps of 15 fsec each with the annealing temperature stepping from 50,000 K to 0 K in 250-K increments. The second slow-cooling stage consisted of 3000 steps of 3 fsec each, cooling from 2000 K to 0 K. The Cartesian energy minimization consisted of 200 steps with the dihedral angle and NOE energy force constants set to 400 kcal−1 rad−2 and 75 kcal mole−1 Å−4, respectively.
Table 1.
Summary of statistics for the 20 final structures
| NOE upper distance limitsa | 1870 |
| Intraresidue | 473 |
| Sequential | 689 |
| Medium range | 335 |
| Long range | 373 |
| Hydrogen bonds | 55 |
| RMSD from experimental constraints | |
| Distances (Å) | 0.0478 ± 0.0019 |
| Dihedrals (°) | 0.00 ± 0.00 |
| Average number of NOE distance constraint violations | |
| >0.5 Å | 0.00 ± 0.00 |
| >0.2 Å | 19.4 ± 2.2 |
| RMSD from idealized covalent geometry | |
| Bonds (Å) | 0.0038 ± 0.00016 |
| Angles (deg) | 0.5710 ± 0.0221 |
| Improper angles (deg) | 0.3733 ± 0.0116 |
| Atomic RMSD values (Å)b | |
| Backbone atoms | 0.65 |
| All atoms | 1.03 |
| Ramachandran plotc,d | |
| Most favored regions | 81.1% |
| Additional allowed regions | 10.8% |
| Generously allowed regions | 2.7% |
| Disallowed regions | 2.7% |
Structure calculation statistics are reported as averages of the 20 structures with the lowest NOE violation energies.
a Sequential NOEs are NOEs between adjoining (i + 1, i − 1) amino acid residues. medium-range NOEs are residues separated by two to four residues. Long-range NOEs are residues separated by five or more residues.
b Residues 16–113.
c Glycines and prolines are not included.
d Residues 23–29, 41–45, 51–58, 61–67, 74–76, 86–88, and 94–97.
Acknowledgments
This work was supported by a Department of Energy grant DEFG02-00er45828 and a grant from the Lake Champlain Cancer Research Organization.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BPS, “between plekstrin and Src”
EGFR, epidermal growth factor receptor
erbB2 (a.k.a. HER2, EGFR2), Erythroblastosis B
FGFR, fibroblast growth factor receptor
Grb, growth factor receptor bound
HSQC, heteronuclear single-quantum coherence
IR, insulin receptor
NMR, nuclear magnetic resonance
NOE, nuclear Overhauser effect
NOESY, NOE spectroscopy
PDGFR, platelet-derived growth factor receptor
TEK, tunica endothelial kinase
RMSD, root-mean-square deviation
RTK, receptor tyrosine kinase
SH2, Src homology 2
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04884704.
References
- Bax, A. and Grzesiek, S. 1993. Methodological advances in protein NMR. Acc.Chem. Res. 26 131–138. [Google Scholar]
- Bax, A. and Ikura, M. 1991. An efficient three-dimensional NMR technique for correlating the proton and nitrogen backbone amide resonances with the α carbon of the preceding residue in uniformly 13C/15N enriched proteins. J Biomol. NMR 1 99–104. [DOI] [PubMed] [Google Scholar]
- Bax, A., Ikura, M., Kay, L.E., Torchia, D.A., and Tschudin, R. 1990a. Comparison of different modes of two-dimensional reverse correlation NMR for the study of proteins. J. Magn. Reson. 86 304–318. [Google Scholar]
- Bax, A., Clore, G.M., and Gronenborn A.M. 1990b. 1H-1H correlation via isotropic mixing of 13C magnetization: A new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 88 425–431. [Google Scholar]
- Boucher, W. 1993–1996. Azara v2.0. Department of Biochemistry, University of Cambridge, Cambridge, UK.
- Brescia, P.J., Ivancic, M., and Lyons, B.A. 2002. Assignment of backbone 1H, 13C, and 15N resonances of human Grb7–SH2 domain in complex with a phosphorylated peptide ligand. J. Biomol. NMR 23 77–78. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macro-molecular structure determination. Acta Crystallogr. D Biol. Crystallogr 54 905–921. [DOI] [PubMed] [Google Scholar]
- Casteran, N., Rottapel, R., Beslu, N., Lecocq, E., Birnbaum, D., and Dubreuil, P. 1994. Analysis of the mitogenic pathway of the FLT3 receptor and characterization in its C terminal region of a specific binding site for the PI3′ kinase. Cell Mol. Biol. 40 443–456. [PubMed] [Google Scholar]
- Clore, G.M., Bax, A., Driscoll, P.C., Wingfield, P.T., and Gronenborn, A.M. 1990. Assignment of the side-chain 1H and 13C resonances of interleukin-1 β using double- and triple-resonance heteronuclear three-dimensional NMR spectroscopy. Biochemistry 29 8172–8184. [DOI] [PubMed] [Google Scholar]
- Daly, R.J. 1998. The Grb7 family of signaling proteins. Cell Signal. 10 613–618. [DOI] [PubMed] [Google Scholar]
- Daly, R.J., Sanderson, G.M., Janes, P.W., and Sutherland, R.L. 1996. Cloning and characterization of GRB14, a novel member of the GRB7 gene family. J. Biol. Chem. 271 12502–12510. [DOI] [PubMed] [Google Scholar]
- De Marco, A. and Wüthrich, K. 1976. Digital filtering with a sinusoidal window function: An alternative technique for resolution enhancement in FT NMR. J. Magn. Reson. 24 201–204. [Google Scholar]
- Dong, L.Q., Porter, S., Hu, D., and Liu, F. 1998. Inhibition of hGrb10 binding to the insulin receptor by functional domain-mediated oligomerization. J. Biol. Chem. 273 17720–17725. [DOI] [PubMed] [Google Scholar]
- Driscoll, P.C., Clore, G.M., Marion, D., Wingfield, P.T., and Gronenborn, A.M. 1990. Complete resonance assignment for the polypeptide backbone of interleukin 1 β using three-dimensional heteronuclear NMR spectroscopy. Biochemistry 29 3542–3556. [DOI] [PubMed] [Google Scholar]
- Eck, M.J., Shoelson, S.E., and Harrison, S.C. 1993. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362 87–91. [DOI] [PubMed] [Google Scholar]
- Engh, R.A. and Huber, R. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47 392–400. [Google Scholar]
- Fabian, J.R., Morrison, D.K., and Daar, I.O. 1993. Requirement for Raf and MAP kinase function during the meiotic maturation of Xenopus oocytes. J. Cell Biol. 122 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., and Kay, L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33 5984–6003. [DOI] [PubMed] [Google Scholar]
- Fridell, Y.W., Villa Jr., J., Attar, E.C., and Liu, E.T. 1998. GAS6 induces Axl-mediated chemotaxis of vascular smooth muscle cells. J. Biol. Chem. 273 7123–7126. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax, A. 1992. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am. Chem. Soc. 114 6291–6293. [Google Scholar]
- Hoedemaeker, F.J., Siegal, G., Roe, S.M., Driscoll, P.C., and Abrahams, J.P. 1999. Crystal structure of the C-terminal SH2 domain of the p85α regulatory subunit of phosphoinositide 3-kinase: An SH2 domain mimicking its own substrate. J. Mol. Biol. 292 763–770. [DOI] [PubMed] [Google Scholar]
- Ivancic, M., Daly, R.J., and Lyons, B.A. 2003. Solution structure of the human Grb7–SH2 domain/erbB2 peptide complex and structural basis for Grb7 binding to ErbB2. J. Biomol. NMR 27 205–219. [DOI] [PubMed] [Google Scholar]
- Iwama, A., Hamaguchi, I., Hashiyama, M., Murayama, Y., Yasunaga, K., and Suda, T. 1993. Molecular cloning and characterization of mouse TIE and TEK receptor tyrosine kinase genes and their expression in hematopoietic stem cells. Biochem. Biophys. Res. Commun. 195 301–309. [DOI] [PubMed] [Google Scholar]
- Janes, P.W., Lackmann, M., Church, W.B., Sanderson, G.M., Sutherland, R.L., and Daly, R.J. 1997. Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J. Biol. Chem. 272 8490–8497. [DOI] [PubMed] [Google Scholar]
- Jones, N., Master, Z., Jones, J., Bouchard, D., Gunji, Y., Sasaki, H., Daly, R., Alitalo, K., and Dumont, D.J. 1999. Identification of Tek/Tie2 binding partners. Binding to a multifunctional docking site mediates cell survival and migration. J. Biol. Chem. 274 30896–30905. [DOI] [PubMed] [Google Scholar]
- Kay, L.E. 1995. Pulsed field gradient multi-dimensional NMR methods for the study of protein structure and dynamics in solution. Prog. Biophys. Mol. Biol. 63 277–299. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1989. ANSIG: A program for the assignment of protein 1H 2D NMR spectra by interactive computer graphics. J. Magn. Reson. 84 627–633. [Google Scholar]
- Kuboniwa, H., Grzesiek, S., Delaglio, F., and Bax, A. 1994. Measurement of HN-H α J couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR 4 871–878. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Moss, D.S., and Thornton, J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231 1049–1067. [DOI] [PubMed] [Google Scholar]
- Mallis, R.J., Brazin, K.N., Fulton, D.B., and Andreotti, A.H. 2002. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat. Struct. Biol. 9 900–905. [DOI] [PubMed] [Google Scholar]
- Margolis, B., Silvennoinen, O., Comoglio, F., Roonprapunt, C., Skolnik, E., Ullrich, A., and Schlessinger, J. 1992. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc. Natl. Acad. Sci. 89 8894–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo, G.R., Graus-Porta, D., Cella, N., Marte, B.M., Taverna, D., and Hynes, N.E. 1996. Growth, differentiation and survival of HC11 mammary epithelial cells: Diverse effects of receptor tyrosine kinase-activating peptide growth factors. Eur. J. Cell Biol. 70 97–105. [PubMed] [Google Scholar]
- Nantel, A., Mohammad-Ali, K., Sherk, J., Posner, B.I., and Thomas, D.Y. 1998. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J. Biol. Chem. 273 10475–10484. [DOI] [PubMed] [Google Scholar]
- Ogura, K., Tsuchiya, S., Terasawa, H., Yuzawa, S., Hatanaka, H., Mandiyan, V., Schlessinger, J., and Inagaki, F. 1999. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J. Mol. Biol. 289 439–445. [DOI] [PubMed] [Google Scholar]
- Ooi, J., Yajnik, V., Immanuel, D., Gordon, M., Moskow, J.J., Buchberg, A.M., and Margolis, B. 1995. The cloning of Grb10 reveals a new family of SH2 domain proteins. Oncogene 10 1621–1630. [PubMed] [Google Scholar]
- Pascal, S.M., Singer, A.U., Gish, G., Yamazaki, T., Shoelson, S.E., Pawson, T., Kay, L.E., and Forman-Kay, J.D. 1994. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-γ1 complexed with a high affinity binding peptide. Cell 77 461–472. [DOI] [PubMed] [Google Scholar]
- Qi, J.H., Ito, N., and Claesson-Welsh, L. 1999. Tyrosine phosphatase SHP-2 is involved in regulation of platelet-derived growth factor-induced migration. J. Biol. Chem. 274 14455–14463. [DOI] [PubMed] [Google Scholar]
- Reith, A.D., Ellis, C., Maroc, N., Pawson, T., Bernstein, A., and Dubreuil, P. 1993. “W” mutant forms of the Fms receptor tyrosine kinase act in a dominant manner to suppress CSF-1 dependent cellular transformation. Oncogene 8 45–53. [PubMed] [Google Scholar]
- Scharf, P.J. and Lyons, B.A. 2002. Assignment of backbone 1H, 13C, and 15N resonances of the SH2 domain of human Grb14. J. Biomol. NMR 24 275–276. [DOI] [PubMed] [Google Scholar]
- Shilo, B.Z. 1992. Roles of receptor tyrosine kinases in Drosophila development. FASEB J. 6 2915–2922. [DOI] [PubMed] [Google Scholar]
- Sierke, S.L. and Koland, J.G. 1993. SH2 domain proteins as high-affinity receptor tyrosine kinase substrates. Biochemistry 32 10102–10108. [DOI] [PubMed] [Google Scholar]
- Songyang, Z., Shoelson, S.E., McGlade, J., Olivier, P., Pawson, T., Bustelo, X.R., Barbacid, M., Hanafusa, H., Yi, T., Ren, R., et al. 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, D., Wu, J., Fuqua, S.A., Roonprapunt, C., Yajnik, V., D’Eustachio, P., Moskow, J.J., Buchberg, A.M., Osborne, C.K., and Margolis, B. 1994. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 13 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, E.G., Ghirlando, R., and Hubbard, S.R. 2003. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J. Biol. Chem. 278 13257–13264. [DOI] [PubMed] [Google Scholar]
- Thornton, K.H., Mueller, W.T., McConnell, P., Zhu, G., Saltiel, A.R., and Thanabal, V. 1996. Nuclear magnetic resonance solution structure of the growth factor receptor-bound protein 2 Src homology 2 domain. Biochemistry 35 11852–11864. [DOI] [PubMed] [Google Scholar]
- Ueno, H., Honda, H., Nakamoto, T., Yamagata, T., Sasaki, K., Miyagawa, K., Mitani, K., Yazaki, Y., and Hirai, H. 1997. The phosphatidylinositol 3′ kinase pathway is required for the survival signal of leukocyte tyrosine kinase. Oncogene 14 3067–3072. [DOI] [PubMed] [Google Scholar]
- Waksman, G., Shoelson, S.E., Pant, N., Cowburn, D., and Kuriyan, J. 1993. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell 72 779–790. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Holder, N., Patient, R., and Wilson, S.W. 1994. Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development 120 287–299. [DOI] [PubMed] [Google Scholar]