Abstract
Oxidative refolding of the dimeric alkaline protease inhibitor (API) from Streptomyces sp. NCIM 5127 has been investigated. We demonstrate here that both isomerase and chaperone functions of the protein folding catalyst, protein disulfide isomerase (PDI), are essential for efficient refolding of denatured-reduced API (dr-API). Although the role of PDI as an isomerase and a chaperone has been reported for a few monomeric proteins, its role as a foldase in refolding of oligomeric proteins has not been demonstrated hitherto. Spontaneous refolding and reactivation of dr-API in redox buffer resulted in 45% to 50% reactivation. At concentrations <0.25 μM, reactivation rates and yields of dr-API are accelerated by catalytic amounts of PDI through its isomerase activity, which promotes disulfide bond formation and rearrangement. dr-API is susceptible to aggregation at concentrations >25 μM, and a large molar excess of PDI is required to enhance reactivation yields. PDI functions as a chaperone by suppressing aggregation and maintains the partially unfolded monomers in a folding-competent state, thereby assisting dimerization. Simultaneously, isomerase function of PDI brings about regeneration of native disulfides. 5-Iodoacetamidofluorescein–labeled PDI devoid of isomerase activity failed to enhance the reactivation of dr-API despite its intact chaperone activity. Our results on the requirement of a stoichiometric excess of PDI and of presence of PDI in redox buffer right from the initiation of refolding corroborate that both the functions of PDI are essential for efficient reassociation, refolding, and reactivation of dr-API.
Keywords: protein folding catalyst, foldase, aggregation, chemoaffinity labeling
Understanding the nature of events by which an unfolded protein attains its functional three-dimensional structure is the most challenging problem in structural biology today. One of the major complications encountered in both in vivo and in vitro protein folding is that of protein misfolding and aggregation (Goldberg et al. 1991; Dobson and Ellis 1998). Efficiency of folding depends on the kinetic competition between pathways leading to correct folding and aggregation (Goldberg et al. 1991). It is now generally well accepted that, in most cases, folding and assembly of nascent polypeptides to functional proteins in the cell are assisted by a whole array of molecular chaperones (Gething and Sambrook 1992; Hartl 1996) and folding catalysts (Gilbert 1997). Chaperones increase the folding efficiency by suppressing the tendency of unfolded proteins to aggregate and generally do not increase overall rate of folding. Folding catalysts, on the other hand, act on covalent bonds and accelerate the rate of slow chemical steps such as proline isomerization, disulfide bond formation, and rearrangement. Correct formation of disulfide bonds is essential for proper folding of a large number of proteins. Although folding of small, single domain proteins devoid of disulfide bonds or with intact native disulfide bonds has been studied extensively (Kim and Baldwin 1990), there is a relative lack of data on oxidative refolding of proteins. Only a few disulfide containing proteins such as bovine pancreatic trypsin inhibitor (van Mierlo et al. 1992), lysozyme (van den Berg et al. 1999a), α-lactalbumin (Lindner et al. 1997), and ribonuclease A (Rothwarf et al. 1998) have been investigated in detail.
Protein disulfide isomerase (PDI), a multifunctional 55-kDa protein present in near millimolar concentrations in the lumen of the endoplasmic reticulum, assists in disulfide bond formation and rearrangement of secreted proteins (LaMantia and Lennarz 1993; Freedman et al. 1994). PDI has two domains with homology to the small, redox-active protein, thioredoxin. The thiol/disulfide centers of the two thioredoxin-like domains function as two independent active sites (Freedman et al. 1994). PDI-catalyzed oxidative refolding of proteins has been investigated (Tang et al. 1994; van den Berg et al. 1999b; Vinci et al. 2000). In concert with its isomerase activity, PDI is also known to display molecular chaperone-like activity by suppressing aggregation and increasing reactivation and, therefore, is referred to as a “foldase” that promotes protein folding and catalyses formation of native disulfide bonds (Wang 1998). The role of PDI as a foldase has been demonstrated during the oxidative refolding of disulfide containing proteins such as lysozyme (Puig and Gilbert 1994) and acidic phospholipase A2 (APLA2; Yao et al. 1997). The chaperone function of PDI has been shown in refolding of proteins without disulfide bonds such as d-glyceraldehyde-3-phoshate dehydrogenase (GAPDH; Cai et al. 1994) and rhodanese (Song and Wang 1995). Although the role of PDI as a foldase has been reported for a few monomeric proteins, there are no reports in refolding of oligomeric proteins. Available reports on the oxidative refolding of oligomeric immunoglobulins have focused mainly on the participation of PDI in the disulfide bond formation (Roth and Pierce 1987; Lilie et al. 1994). The influence of isomerase function of PDI in cooperation with BiP (immunoglobulin heavy-chain binding protein) as a chaperone was shown during the oxidative folding of antibodies (Mayer et al. 2000).
In an attempt to decipher whether PDI can also function as a folding catalyst in the refolding of oligomeric proteins that require an additional step of regeneration of native quaternary structure, we have investigated the oxidative refolding of the dimeric alkaline protease inhibitor (API). API isolated from a Streptomyces sp. (NCIM 5127) is a dimeric protein of 28 kDa containing disulfide linkages that are vital for its biologically active conformation (Vernekar et al. 1999, 2001). The spontaneous refolding of denatured and reduced API (dr-API) is only 45%–50% due to lack of proper reassociation of unfolded monomers and correct formation of essential disulfide linkages and also due to its propensity to aggregate. Our results demonstrate that both chaperone and isomerase functions of PDI are essential for the successful refolding and reactivation of dimeric API.
Results
API from the Streptomyces sp. is a dimeric protein containing five disulfide linkages and two cysteine residues (Vernekar et al. 1999). The denaturation and reduction of API in presence of 6 M GdmHCl and 20 mM DTT for 4 h at 37°C led to its complete inactivation and resulted in the dissociation of the dimer with total disruption of native disulfide bonds. On SDS-PAGE, under reducing conditions API migrated at a molecular weight of ~13.5 kDa whereas on a nonreducing SDS-PAGE, API migrated at a distance corresponding to a molecular weight of 28 kDa, indicating that in the absence of reducing agent the two subunits are linked by a disulfide linkage (data not shown).
Reactivation yield as a function of dr-API concentration
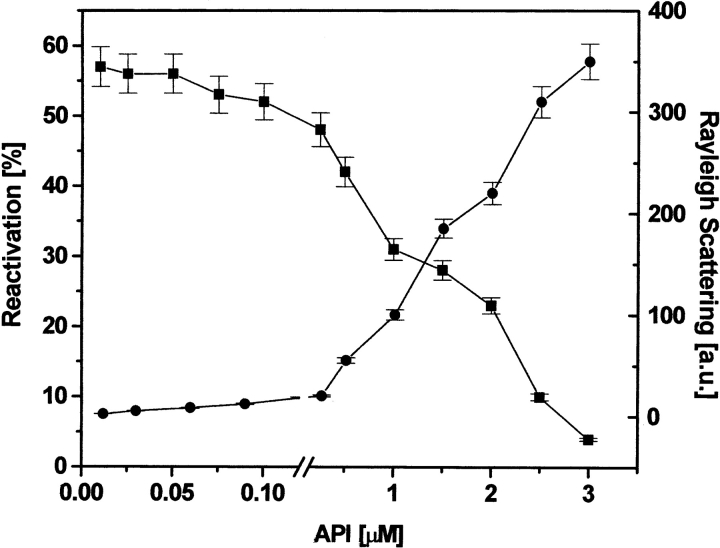
The spontaneous reactivation of dr-API (0.5 μM) initiated by a 100-fold dilution in refolding buffer in the absence of redox buffer resulted only in a 5% to 10% yield of API. In the presence of redox buffer, a reactivation yield of 45% was obtained in 3 h, which decreased to 23% when the concentration of dr-API was increased to 2 μM (Fig. 1A ▶). The kinetics of reactivation of dr-API followed a biphasic sequential reactivation. The initial rate of reactivation had a rate constant (k1) of 6.2 ± 0.8 × 10−4s−1 for 0.5 μM and 5.9 ± 0.7 × 10−4s−1 for 2 μM dr-API, respectively (Fig. 1B ▶). The second phase of reactivation was faster for both 0.5 and 2 μM dr-API, with a similar rate constant (k2) of 2.0 ± 0.3 × 10−3s−1. At lower concentrations of dr-API (0.01−0.25 μM) in redox buffer, a maximum reactivation yield of 50% to 55% was obtained. However, the reactivation yield markedly decreased from 55% to 4% as the concentration of API was increased from 0.25 to 3 μM (Fig. 2 ▶). The susceptibility of dr-API to aggregate increased with increasing protein concentration (0.25−3 μM) with a concomitant decrease in the reactivation yield (Fig. 2 ▶).
Figure 1.
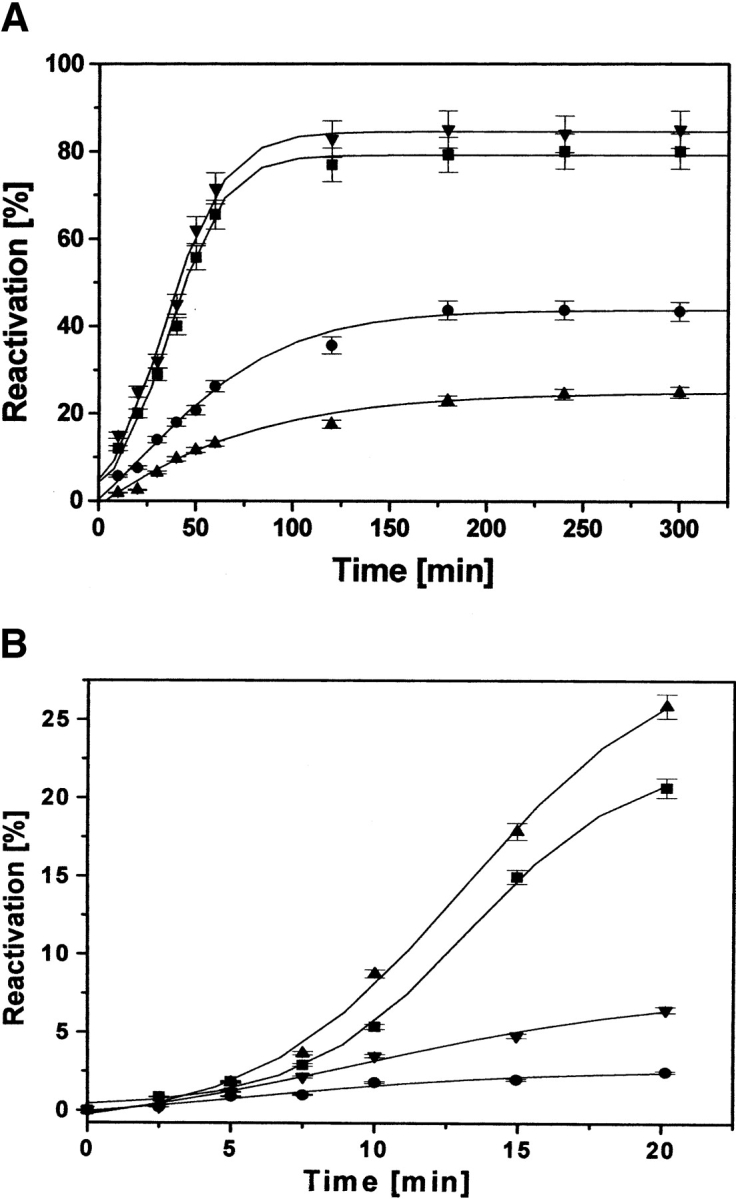
Reactivation of dr-API in presence and absence of PDI. (A) Reactivation of 0.5 μM dr-API in absence (circles) and presence (down-pointing triangles) of 5 μM PDI and 2 μM dr-API in absence (up-pointing triangles) and presence (squares) of 20 μM PDI was initiated in redox buffer as described in Materials and Methods, and the recovery of anti-proteolytic activity was determined by the caseinolytic assay. (B) The figure shows the initial time points (up to 20 min) of the curves as in A. The curves represent the best fit of the reactivation data that are expressed as mean ± SD (n = 3 to 5).
Figure 2.
Concentration-dependent reactivation and aggregation of dr-API in redox buffer. The concentration-dependent reactivation of dr-API was carried out in redox buffer for 3 h. The recovery of inhibitory activity (squares) and aggregation by light scattering at 385 nm (circles) were determined. The data are expressed as mean ± SD (n = 3 to 5).
PDI-accelerated refolding of dr-API
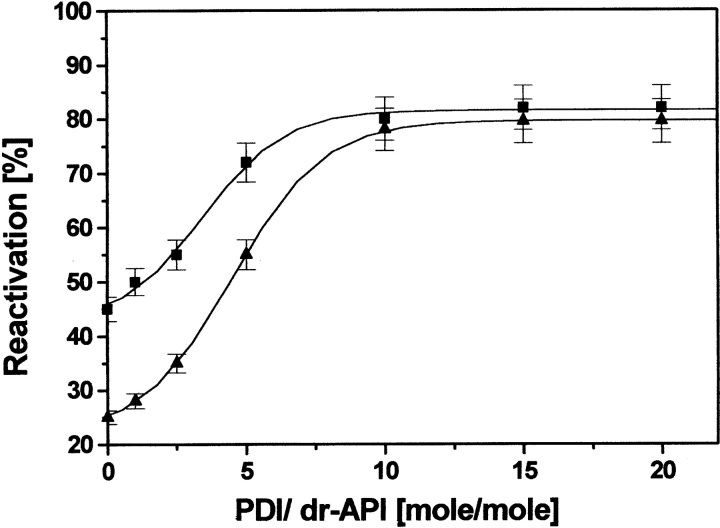
Addition of 0.05 μM dr-API to redox buffer containing only catalytic amount of PDI (0.025 μM) significantly increased its activity to 90%, indicating the participation of its isomerase activity during reactivation (data not shown). However, at higher concentrations of dr-API (0.5−3 μM), even a stoichiometric amount of PDI was unable to promote substantial reactivation. The reactivation yields increased as a function of increasing PDI concentrations (Fig. 3 ▶). A 10-fold molar excess of PDI was required to achieve maximum increase of ~85% in the recovery of 0.5 and 2 μM dr-API. The initial rate of reactivation was accelerated in the presence of PDI compared with the uncatalyzed reactivation with rate constant (k1) of 1.5 ± 0.2 × 10−3s−1 and 1.2 ± 0.2 × 10−3s−1 for 0.5 μM and 2 μM dr-API, respectively (Fig. 1B ▶). The second phase of reactivation again had similar rate constants (k2 = 2.5 ± 0.3 × 10−3s−1), which were almost similar to those in the absence of PDI (Fig. 1 ▶).
Figure 3.
Effect of PDI on the reactivation of dr-API. Reactivation of 0.5 μM (squares) and 2 μM (triangles) dr-API in the presence of increasing amounts of PDI was carried out in redox buffer for 3 h, and the recovery of inhibitory activity was determined. The data are expressed as mean ± SD (n = 3 to 5).
Effect of PDI on the suppression of aggregation
Efficiency of PDI to suppress aggregation of dr-API increased with increasing PDI/dr-API ratios (Fig. 4A ▶) and was in agreement with the increase in reactivation. Aggregation of 2 μM dr-API is markedly suppressed by the presence of 10 μM PDI and further by 20 μM PDI. The time-dependent light scattering studies revealed that the interaction between PDI and dr-API is complete within 3 to 5 min, and the amount of aggregated API remained constant thereafter (Fig. 4B ▶).
Figure 4.
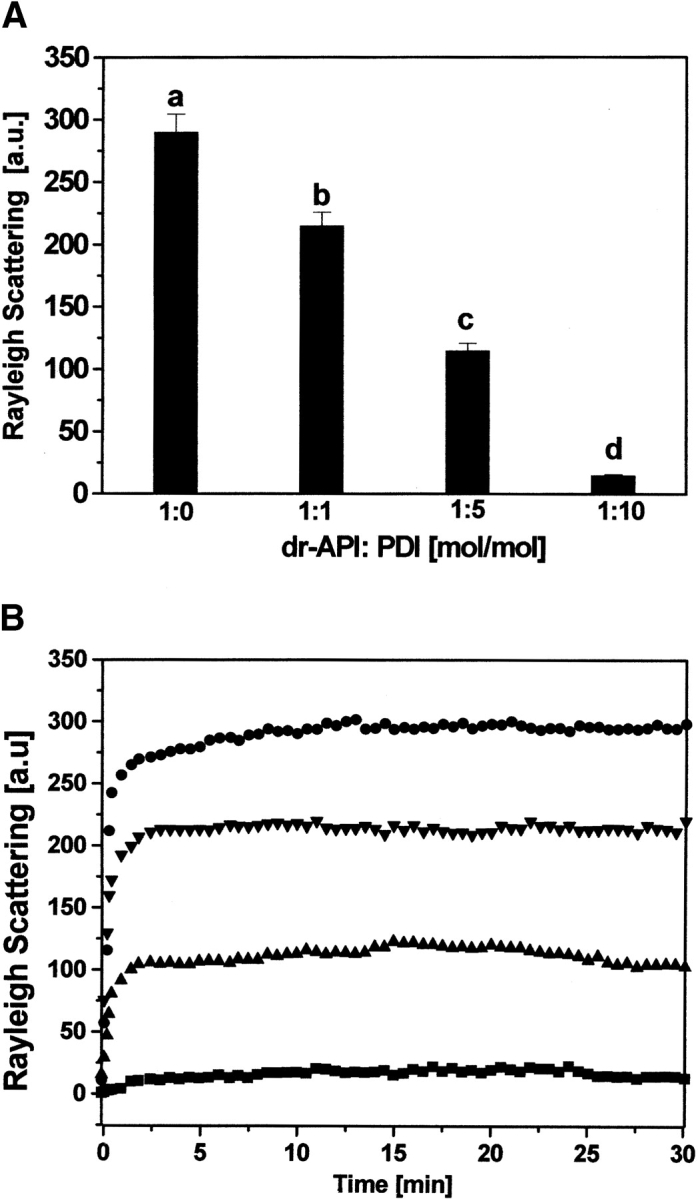
Influence of PDI on aggregation of dr-API during refolding. (A) The aggregation of 2 μM dr-API in the presence of varying dr-API/PDI ratios during refolding was monitored by light scattering at 385 nm. The molar ratios of dr-API to PDI were as follows: (a) 1:0; (b) 1:1; (c) 1:5; and (d) 1:10. The data are expressed as mean ± SD (n = 3 to 5). (B) The time-dependent suppression of aggregation of 2 μM dr-API in presence of varying amounts of PDI was monitored in redox buffer for 30 min at 25°C. The molar ratios of dr-API to PDI were 1:0 (circles), 1:1 (down-pointing triangles), 1:5 (up-pointing triangles), and 1:10 (squares).
Effect of delayed addition of PDI and redox buffer on reactivation of dr-API
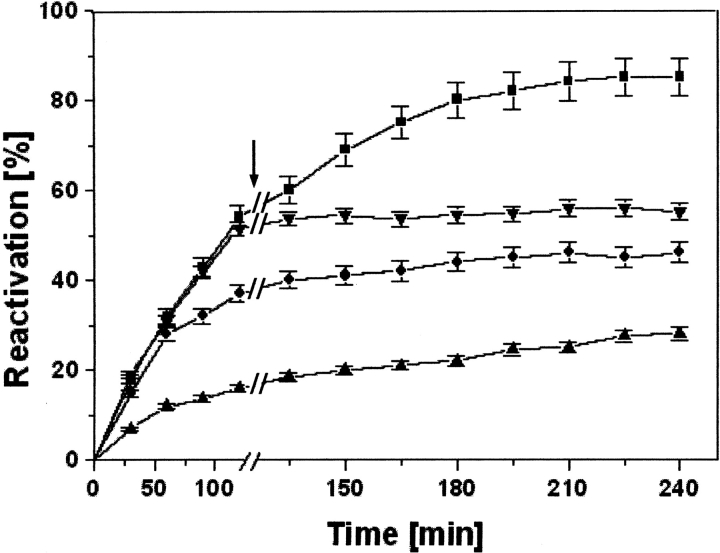
dr-API was allowed to refold in redox buffer for 2 h before the addition of PDI, and the reactivation was monitored. A time period of 2 h was used as a representative time. The delayed addition of only 0.025 μM PDI to a lower concentration of 0.05 μM dr-API after 2 h increased the reactivation to 85% (Fig. 5 ▶). However, at higher concentrations of 0.5 and 2 μM dr-API, the reactivation yields of only 45% and 25%, respectively, were obtained upon the delayed addition of a 10-fold molar concentration of PDI. These yields were similar to the uncatalyzed reactivation. Thus, at higher concentrations of dr-API, the delayed addition of PDI failed to enhance its reactivation.
Figure 5.
Effect of delayed addition of PDI on reactivation of dr-API in redox buffer. Refolding of dr-API 0.05 μM (squares), 0.5 μM (circles), and 2 μM (up-pointing triangles) was initiated in redox buffer followed by an addition of 10-fold molar PDI after 2 h. As a control, 0.05 μM dr-API was refolded in the redox buffer without addition of PDI (down-pointing triangles). The arrow indicates the time of addition of PDI. The data are expressed as mean ± SD (n = 3 to 5).
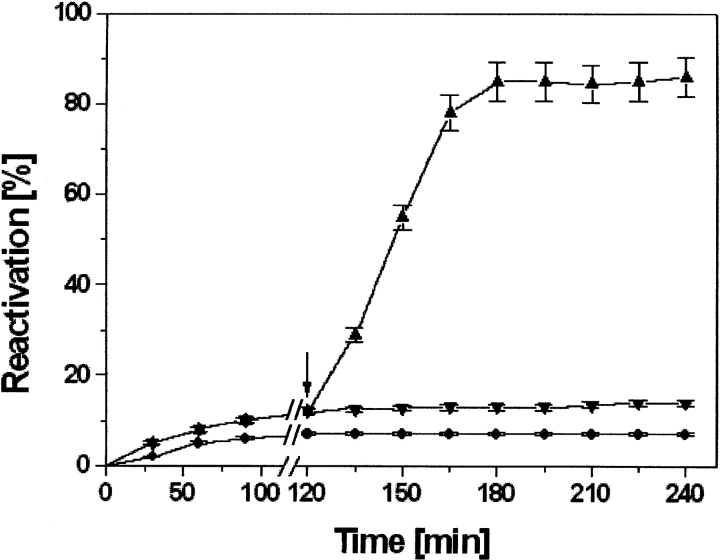
The incubation of 1 μM dr-API with 10 μM PDI in the absence of redox buffer resulted in a reactivation yield of only 14% to 15% (Fig. 6 ▶). However, addition of redox buffer after 120 min accelerated the reactivation to 85%. Thus, only PDI in the absence of redox buffer was unable to enhance the reactivation of dr-API.
Figure 6.
Effect of delayed addition of redox buffer on the reactivation of dr-API in presence of PDI. Reactivation of 1 μM dr-API in the absence of PDI and redox buffer (circles), in the presence of 10 μM PDI (down-pointing triangles), and after the delayed addition of redox buffer (up-pointing triangles). The arrow indicates the time of addition of redox buffer. The data are expressed as mean ± SD (n = 3 to 5).
Effect of modified PDI on the refolding of dr-API
In an attempt to decipher the influence of modified PDI during refolding of dr-API on its isomerase and chaperone functions, we have labeled PDI by using the chemoaffinity fluorescent label 5-iodoacetamidofluorescein (5-IAF). PDI contains two cysteine residues in its active site. They are known to function in thiol/disulfide exchange for the regeneration of native disulfide linkages (Freedman et al. 1994). The a and a′ domains of PDI involved in the isomerase activity are highly homologous to thioredoxin. In thioredoxin, Cys32, the more amino-terminal of the active-site Cys residues, has an anomalously low pK of 6.7, so that at neutral pH, the vicinal dithiol formed by the two Cys residues comprises one −S− and one −SH group (Freedman et al. 1994). The fluorescent probe, 5-IAF, is known to react specifically with the free SH groups of proteins (Tanksale et al. 2000). The labeling ratio indicated that two Cys residues were labeled by 5-IAF. The reactivation of 0.5 μM dr-API in the presence of 5 μM IAF-PDI was only 45% similar to the uncatalyzed reactivation (Fig. 7A ▶). Thus, the labeling of PDI with 5-IAF abolished the isomerase activity of PDI. The simultaneous addition of a 10-fold molar concentration of PDI and IAF-labeled PDI in a ratio of 1:9, respectively, gave an increase in the reactivation yield of dr-API (0.5 μM; Fig. 7A ▶). The increase in reactivation by the presence of only a catalytic amount of PDI indicated the essential nature of the isomerase function of PDI and also showed the intact chaperone activity of labeled-PDI because the IAF-labeled PDI could maintain dr-API in a folding-competent state. The integrity of the chaperone activity of IAF-labeled PDI and the requirement of the isomerase activity of PDI for the reactivation of dr-API was further corroborated as observed by the increase in the reactivation to ~80% after the delayed addition of 0.5 μM PDI to 0.5 μM dr-API already refolded in the presence of 4.5 μM IAF-PDI in redox buffer for 2 h (Fig. 7B ▶).
Figure 7.
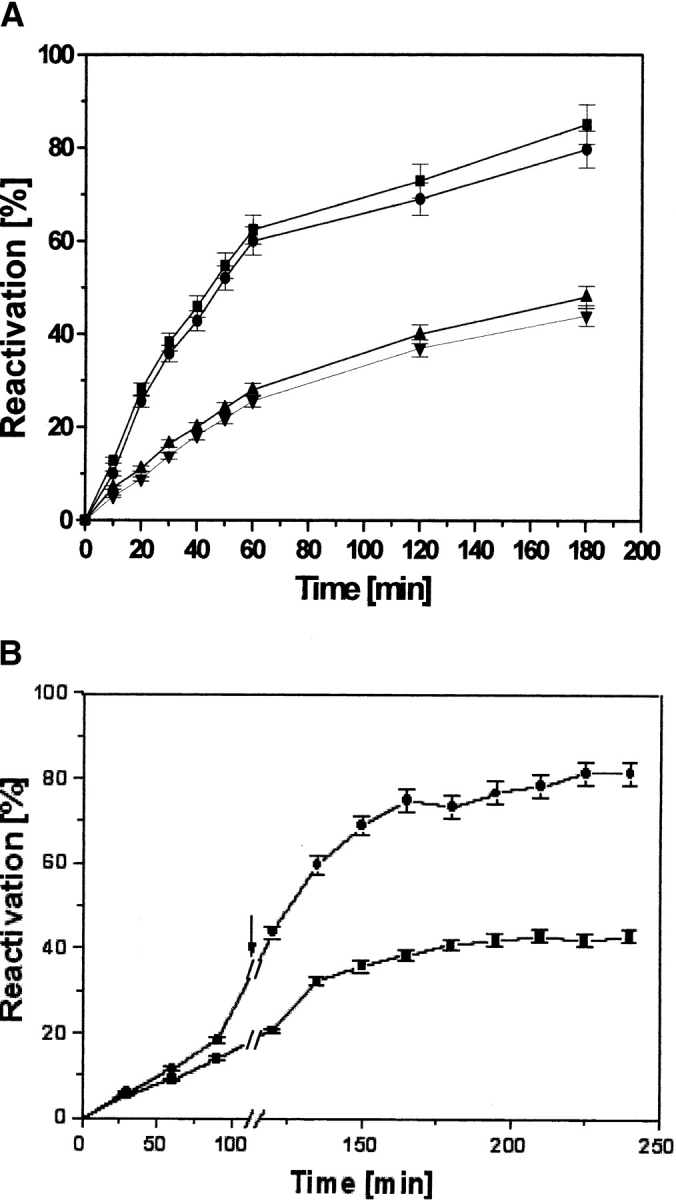
Reactivation of dr-API in the simultaneous presence of PDI and IAF-labeled PDI. (A) Reactivation of 0.5 μM dr-API in redox buffer in the absence (down-pointing triangles) and presence (squares) of 5 μM PDI, the presence of 5 μM IAF-PDI (up-pointing triangles), and the simultaneous presence of 0.5 μM PDI and 4.5 μM IAF-PDI (circles). The data are expressed as mean ± SD (n = 3 to 5). (B) Reactivation of 0.5 μM dr-API in the presence of 4.5 μM IAF-PDI (squares) and after the delayed addition (2 h) of 0.5 μM unlabeled PDI and redox buffer (circles). The arrow indicates the time of addition. The data are expressed as mean ± SD (n = 3 to 5).
To further confirm our results showing that modification of PDI by IAF does not affect the chaperone activity of PDI, we have used fluorescence spectroscopy. The IAF-PDI displays a λmax at 527 nm, which does not interfere with that of dr-API because dr-API does not emit at this wavelength. Therefore, the changes in the emission spectra of IAF-PDI during its interaction with dr-API will exclusively reflect the conformational changes due to its binding with the substrate protein. Incubation of 5 μM IAF-PDI with 0.5 μM dr-API resulted in a rapid quenching in the fluorescence yield at 527 nm with time (Fig. 8A ▶), indicative of their effective binding and the intactness of the chaperone activity of labeled PDI. The binding interaction of IAF-PDI and dr-API was complete within 3 to 5 min, which is similar to the time required for suppression of aggregation of dr-API by PDI. The functional integrity of the chaperone activity was further confirmed by the time-dependent light scattering studies that revealed the suppression of aggregation of 1 μM dr-API by a 10-fold molar excess of IAF-PDI (Fig. 8B ▶). Thus, the IAF-labeled PDI suppresses aggregation and maintains dr-API in a folding-competent state.
Figure 8.
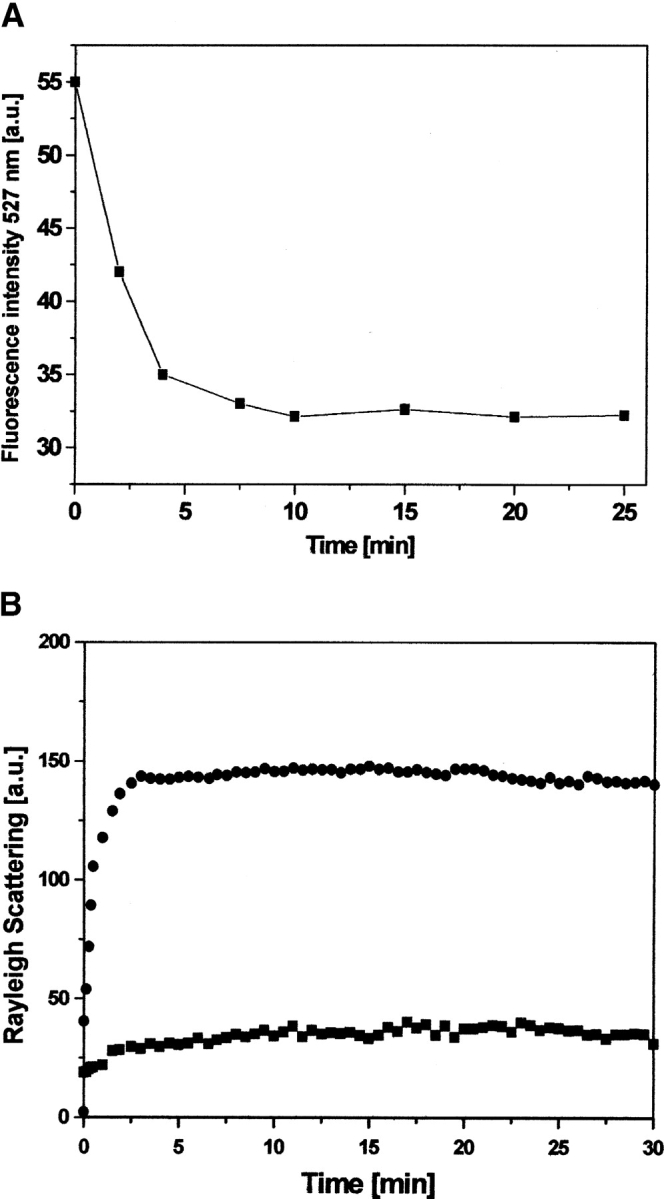
Interaction of chemoaffinity-labeled PDI with dr-API. (A) Five micromolar IAF-PDI was excited at a wavelength of 490 nm, and the time-dependent change in fluorescence emission at 527 nm (square) was monitored during its interaction with 0.5 μM dr-API. (B) The aggregation of 1 μM dr-API during refolding in the presence (squares) and absence (circles) of 10 μM IAF-PDI was monitored by light scattering at 385 nm.
Discussion
Disulfide bonds confer conformational and thermodynamic stability to proteins and are essential for their biological activity (Woycechowsky and Raines 2000). Formation/isomerization of disulfide bonds is the rate-limiting step and is, therefore, critical for the proper folding of proteins. We present here, the oxidative refolding of a dimeric disulfide containing API from a Streptomyces sp. During its refolding, the association of monomers and the regeneration of native disulfide linkages are the main essential prerequisites for regaining its biological function. The spontaneous refolding of dr-API upon dilution in redox buffer resulted in 45% reactivation, which is remarkably high for a disulfide containing protein. Low concentrations of 0.01 to 0.1 μM dr-API that are not prone to aggregation also result in incomplete reactivation, suggesting the formation of nonnative disulfides. Refolding yields decreased with increasing concentrations of dr-API concomitant to its propensity to aggregate. Denatured and reduced disulfide containing proteins have a tendency to aggregate during refolding due to nonproductive hydrophobic interactions and incorrect non-native disulfide bond formation (Yao et al. 1997). Thus, the partial recovery of API may be attributed to the formation of misfolded and misoxidised monomers and inactive oligomeric aggregates that may require the assistance of chaperones and folding catalysts for complete recovery. To verify this notion, we have studied the refolding of dr-API in the presence of PDI. Because dr-API is not susceptible to aggregation at lower concentrations, catalytic amounts of PDI effected complete reactivation, indicating that its isomerase activity was sufficient for efficient refolding. However, with increasing concentrations of dr-API, only a large stoichiometric excess of PDI was able to enhance the refolding efficiency, revealing that only isomerase activity of PDI was inadequate. Interestingly, the refolding yields of API increased as a function of PDI concentration concomitant with the suppression of aggregation, suggesting that PDI also functions as a chaperone. Requirement of a large molar excess of PDI for suppression of aggregation of dr-API implied the possibility that multiple PDI molecules must bind each substrate molecule in order to prevent aggregation. The interaction between PDI and dr-API at all PDI concentrations used is complete within 3 to 5 min after which the amount of aggregated material remains constant.
The reactivation of dr-API followed a biphasic reactivation; in the initial phase the rate of reaction was slow in the absence of PDI, and the presence of PDI brings about acceleration in the reaction rate, thereby decreasing the number of misoxidized/misfolded monomers and resulting in increased reactivation. Once there is formation of properly folded and oxidized monomers, in the second phase they become committed to dimerization; therefore, the rate of dimerization is almost the same in the presence/absence of PDI. Thus, the initial phase of native disulfide bond formation and rearrangement is the rate-limiting step in the reactivation process. Only catalytic amounts of PDI are required to enhance reactivation in the absence of aggregation; however, at higher concentration of dr-API, though the formation of native disulfide linkages is the rate-limiting step, a large molar excess of PDI is required to maintain the monomers in a folding-competent state, which will prevent formation of undesirable aggregates and allow their correct isomerization.
The delayed addition of PDI during the refolding of lower concentrations of dr-API (0.05 μM) promoted the reactivation, validating that its isomerase activity is sufficient for efficient reactivation in the absence of aggregation. However, the delayed addition of PDI to higher concentrations of dr-API (2 μM) failed to promote the refolding of dr-API that was already aggregated, suggesting the requirement of the chaperone function of PDI in the refolding of dr-API. The presence of PDI was required right from the initiation of refolding to maintain the unfolded monomers in a folding competent state and to prevent the incorrect association of the partially folded monomers that are prone to aggregation. In the absence of redox buffer, although PDI displayed chaperone activity by suppressing aggregation, it was unable to promote reactivation because under these conditions only oxygen from air can be used as an oxidant. Similar results were obtained during the refolding of lysozyme in the absence of oxidized glutathione (GSSG) (Song et al. 1997). The isomerase activity of PDI requires the presence of redox conditions for efficient catalysis of disulfide bond formation and rearrangement. In the absence of redox buffer, although PDI was able to maintain dr-API in an assembly competent state for 2 h, it did not enhance the reactivation yield, indicating that the chaperone activity alone is not adequate for correct refolding and complete reactivation. The isomerase activity in presence of redox buffer brings about the proper disulfide bond formation and enables complete reactivation. The delayed addition of redox buffer to the mixture of PDI and dr-API enhanced the reactivation, indicating that both the isomerase and chaperone activities are required to work in cooperation for the efficient reactivation of dr-API. The interaction of PDI with the partially unfolded monomers promotes their correct folding and keeps the cysteine residues accessible for PDI to function subsequently as an isomerase in the presence of redox buffer that results in maximum reactivation of API. The binding of IAF-PDI to dr-API was unaffected by labeling, as revealed by the quenching of fluorescence. However, IAF-PDI, even in a 10-fold molar excess, failed to enhance the reactivation of dr-API, signifying that the isomerase activity of PDI is indispensable for reactivation of dr-API. Furthermore, the replacement by a catalytic amount of PDI instead of IAF-labeled PDI enabled reactivation. The simultaneous presence of native PDI (conferring isomerase activity) and a molar excess of labeled PDI (conferring chaperone activity) can increase reactivation of dr-API. Although a 10-fold molar excess of IAF-labeled PDI did not increase reactivation, it was able to retain dr-API in a folding-competent state for 2 h as a chaperone as revealed by the reactivation of dr-API after 2 h upon addition of native PDI. The integrity of the chaperone activity of IAF-labeled PDI was further validated by the suppression of aggregation of dr-API. The labeled PDI suppresses aggregation and the unlabeled PDI brings about the correct disulfide formation, thus cooperatively promoting refolding and reactivation. The essential nature of the chaperone and isomerase functions of PDI was thus confirmed by fluorescence chemoaffinity labeling of PDI, substantiating its role as a foldase in the refolding and reactivation of dr-API.
PDI is a resident protein of the secretory pathway present in near millimolar concentrations in the lumen of the endoplasmic reticulum and is known to interact with nascent and newly translocated secretory proteins. Our results reinforce the role of PDI as a protein folding catalyst not only for small single domain proteins but also for the reactivation of oligomeric proteins such as API. The extension of these studies to other oligomeric proteins and the exploration of the molecular mechanisms involved in the isomerase and chaperone functions in the folding of these proteins will further strengthen our understanding of the functions of PDI.
Materials and methods
Reagents
Dithiothreitol (DTT), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), guanidinium hydrochloride (GdmHCl), glutathione reduced (GSH) and oxidized (GSSG), and protein disulfide isomerase (PDI) from bovine liver were obtained from Sigma Chemical Co. All other reagents used were of analytical grade.
Purification and assay of API
API was purified according as described previously (Vernekar et al. 1999). Briefly, the culture filtrate of Streptomyces sp. containing API was concentrated by ultra filtration and purified by poly-acrylamide gel electrophoresis (PAGE) using gel-X-ray film contact print technique for its detection and extraction. In the gel-X-ray film contact print technique, after electrophoretic resolution of the protein, a vertical strip of the gel was cut and incubated for 10 min in 0.1 M carbonate-bicarbonate buffer (pH 10.0) containing 0.5 mg/mL of the bacterial alkaline protease, subtilisin. The gel was overlaid on an equal sized X-ray film, and the hydrolysis of gelatin was followed for 20 min at 37°C. The X-ray film was removed and washed with warm water, and the zone where the hydrolysis was inhibited was detected. The band corresponding to the inhibitory activity was excised and eluted by homogenization followed by vacuum filtration and was purified further by DEAE cellulose chromatography.
The protease inhibitory activity of API against subtilisin was determined by the caseinolytic assay in 0.1 M carbonate-bicarbonate buffer (pH 10) at 37°C by incubating with casein (10 mg) for 20 min (Kunitz 1947). One unit of protease activity was defined as the amount of enzyme that causes an increase of one absorbance unit at 280 nm/mL reaction mixture per minute. One unit of protease inhibitor was defined as the amount of inhibitor required for inhibiting one unit of protease activity. The protein quantification was done according to Bradford method using BSA as a standard (Bradford 1976).
Unfolding and refolding of API
API was completely denatured and reduced by incubating for 4 h at 37°C in 0.05 M phosphate buffer (pH 7.5) containing 6 M GdmHCl and 20 mM DTT. It was thoroughly dialyzed against 0.1 M KCl-HCl buffer (pH 2.0) for 3 h followed by dialysis against 0.01 M of the same buffer for 16 h at 4°C. The denatured and reduced API (dr-API) was distributed in aliquots and stored at −20°C and was used for all the refolding experiments. The number of thiols in the API was determined by using DTNB (Habeeb 1972). Refolding of dr-API at concentrations in the range of 0.01–3 μM was initiated by rapid dilution to 100-fold in a redox buffer consisting of 100 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA (pH 8.5), and 1 mM GSH/0.5 mM GSSG at 25°C unless otherwise specified. The stock solutions of 50 mM GSH/GSSG in water were made freshly prior to use. Aliquots of the refolding mixture were removed at different time intervals and assayed for antiproteolytic activity. The reactivation yields are the averages of at least three different experiments. The kinetics of the reactivation reaction were modeled by using a biphasic second-order fit, and the rate constants were calculated by fitting the data into Kaliedagraph.
Refolding of dr-API in presence of PDI
The influence of PDI on the reactivation of dr-API during refolding was studied by addition of dr-API (0.01–3 μM) to varying concentrations of PDI in redox buffer. The time-dependent regain of activity of 0.5 and 2 μM dr-API in the presence of a 10-fold molar ratio of PDI to dr-API was estimated, and the rate constants were calculated as described above. The effect of delayed addition of PDI on the refolding and reactivation of dr-API was studied by the addition of PDI to refolding dr-API solutions in redox buffer after specific time intervals, and the regain in activity was monitored. The effect of PDI on the reactivation of dr-API in the absence of redox buffer was assessed by the incubation of 1 μM dr-API with 10 μM PDI for 2 h followed by the addition of redox buffer, and the time-dependent regain in activity was measured. In the text, molar concentrations of PDI are calculated by taking into account its molecular weight as 55 kDa. The specific activity of PDI was confirmed by the insulin reduction assay (Gilbert 1998). The reactivation yields are the averages of at least three different experiments.
Fluorescent chemoaffinity labeling of protein disulfide isomerase
Fifty micromolar PDI was reduced by incubation with 5 mM DTT for 30 min at room temperature followed by treatment with 100-fold molar excess of 5-IAF in 50 mM potassium phosphate buffer (pH 7.5) for 24 h in the dark. Iodoacetamide specifically reacts with free −SH groups at pH 7.5. The labeled protein was separated from the free reagent by passage through a column of Sephadex G-10. Fractions showing absorbance at 280 and 490 nm were pooled and concentrated by lyophilization. IAF-labeled PDI was homogeneous as determined by native and SDS-PAGE. The number of Cys residues labeled was determined by using the molar absorptivity for 5-IAF of 80,000 cm−1 M−1. The reactivation of dr-API in presence of IAF-labeled PDI was carried out by addition of 0.5 μM of dr-API to 5 μM IAF-labeled PDI in redox buffer for 1 h.
Fluorescence measurements
Steady-state fluorescence was recorded on a Perkin-Elmer luminescence spectrofluorometer LS50B at 25°C by using an excitation and emission slit width of 5 nm in a 1-cm path-length quartz cuvette. Measurements were performed in triplicate, and suitable baseline corrections were made. Corrections for the inner filter effect were performed as described by the following formula (Lakowicz 1983):
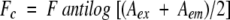 |
where Fc and F are the corrected and measured fluorescence intensities, respectively, and Aex and Aem are the solution absorbances at the excitation and emission wavelengths, respectively. The fluorescence spectrum of IAF-labeled PDI was monitored at an excitation wavelength of 490 nm and an emission wavelength of 527 nm. The magnitudes of Aex and Aem of labeled PDI were 0.08 and 0.0015, respectively. Interaction of labeled PDI (5 μM) with dr-API (0.5 μM) was monitored by addition of dr-API to labeled PDI in redox buffer, and the time-dependent quenching of fluorescence upon complex formation was monitored. Background buffer spectra were subtracted to eliminate the contribution from Raman scattering.
Measurement of protein aggregation
The aggregation during refolding of dr-API in a concentration range of 0.01 to 3 μM was followed by Rayleigh light scattering experiments with the spectrofluorometer at 25°C. Both excitation and emission wavelengths were set at 385 nm, and the change in scattering intensity was determined.
The effect of stoichiometric amounts of PDI on aggregation of dr-API was analyzed by using time-dependent scattering measurements. To 2 μM of dr-API in redox buffer, varying amounts of PDI were added, and the time-dependent scattering was monitored for 30 min. For each PDI concentration used, a new dr-API solution was used. All measurements were performed in triplicate, and suitable baseline corrections were carried out.
Acknowledgments
The senior research fellowship to J.P. by the Council of Scientific and Industrial Research, India is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
API, alkaline protease inhibitor
dr-API, denatured and reduced API
PDI, protein disulfide isomerase
DTT, dithiothreitol
GdmHCl, guanidinium hydrochloride
DTNB, 5,5′-dithiobis (2-nitrobenzoic acid)
5-IAF, 5-iodoacetamidofluorescein
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03552004.
References
- Bradford, M.N. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cai, H., Wang, C.C., and Tsou, C.L. 1994. Chaperone activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J. Biol. Chem. 269 24550–24552. [PubMed] [Google Scholar]
- Dobson, C.M. and Ellis, R. J. 1998. Protein folding and misfolding inside and outside the cell. EMBO J. 17 5251–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, R.B., Hirst, T.R., and Tuite, M.F. 1994. Protein disulfide isomerase: Building bridges in protein folding. Trends. Biochem. Sci. 19 331–336. [DOI] [PubMed] [Google Scholar]
- Gething, M.J. and Sambrook, J. 1992. Protein folding in the cell. Nature 355 33–45. [DOI] [PubMed] [Google Scholar]
- Gilbert, H.F. 1997. Protein disulfide isomerase and assisted protein folding. J. Biol. Chem. 272 29399–29402. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Protein disulfide isomerase. Methods Enzymol. 290 26–50. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.E., Rudolph, R., and Jaenicke, R.A. 1991. A kinetic study of the competition between renaturation and aggregation during the refolding of denatured-reduced hen egg white lysozyme. Biochemistry 30 2790–2797. [DOI] [PubMed] [Google Scholar]
- Habeeb, A.F.S.A. 1972. Reaction of protein sulfhydryl groups with Ellman’s reagent. Methods Enzymol. 25 457–464. [DOI] [PubMed] [Google Scholar]
- Hartl, F.U. 1996. Molecular chaperones in cellular protein folding. Nature 381 571–580. [DOI] [PubMed] [Google Scholar]
- Kim, P.S. and Baldwin, R.L. 1990. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 59 631–660. [DOI] [PubMed] [Google Scholar]
- Kunitz, M. 1947. Crystalline soyabean trypsin inhibitor II: General properties. J. Gen. Physiol. 30 291–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1983. Principles of fluorescence spectroscopy. Plenum Press, New York.
- LaMantia, M. and Lennarz, W.J. 1993. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell 74 899–908. [DOI] [PubMed] [Google Scholar]
- Lilie, H., McLaughlin, S., Freedman, R., and Buchner, J. 1994. Influence of protein disulfide isomerase (PDI) on antibody folding in vitro. J. Biol. Chem. 269 14290–14296. [PubMed] [Google Scholar]
- Lindner, R.A., Kapur, A., and Carver, J.A. 1997. The interaction of the molecular chaperone, α-crystallin, with molten globule states of bovine μ-lact-albumin. J. Biol. Chem. 272 27722–27729. [DOI] [PubMed] [Google Scholar]
- Mayer, M., Kies, U., Kammermeier, R., and Buchner, J. 2000. BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J. Biol. Chem. 275 29421–29425. [DOI] [PubMed] [Google Scholar]
- Puig, A. and Gilbert, H. F. 1994. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative folding of lysozyme. J. Biol. Chem. 269 7764–7771. [PubMed] [Google Scholar]
- Roth, R.A. and Pierce, S.B. 1987. In vivo cross-linking of protein disulfide isomerase to immunoglobulins. Biochemistry 26 4179–4182. [DOI] [PubMed] [Google Scholar]
- Rothwarf, D.M., Li, Y.J., and Scheraga, H.A. 1998. Regeneration of bovine pancreatic ribonuclease A: Detailed kinetic analysis of two independent folding pathways. Biochemistry 37 3767–3776. [DOI] [PubMed] [Google Scholar]
- Song, J.L. and Wang, C.C. 1995. Chaperone-like activity of protein disulfide isomerase in the refolding of rhodanese. Eur. J. Biochem. 231 312–316. [DOI] [PubMed] [Google Scholar]
- Song, J., Quan, H., and Wang, C.C. 1997. Dependence of the anti-chaperone activity of protein disulfide isomerase on its chaperone activity. Biochem. J. 328 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B., Zhang, S.Z., and Yang, K.Y. 1994. Assisted refolding of recombinant prochymosin with the aid of protein disulfide isomerase. Biochem. J. 301 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksale, A.M., Vernekar, J., Ghatge, M.S., and Deshpande, V.V. 2000. Evidence for tryptophan in proximity to histidine and cysteine as essential to the active site of an alkaline protease. Biochem. Biophys. Res. Commun. 270 910–917. [DOI] [PubMed] [Google Scholar]
- van den Berg, B., Chung, E.W., Robinson, C.V., and Dobson, C.M. 1999a. Characterization of the dominant oxidative folding intermediate of hen lysozyme. J. Mol. Biol. 290 781–796. [DOI] [PubMed] [Google Scholar]
- van den Berg, B., Chung, E.W., Robinson, C.V., Mateo, P.L., and Dobson, C.M. 1999b. The oxidative refolding of hen lysozyme and its catalysis by protein disulfide isomerase. EMBO J. 18 4794–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo, C.P.M., Darby, N.J., and Creighton, T.E. 1992. The partially folded conformation of the Cys30-Cys51 intermediate in the disulfide folding pathway of bovine pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci. 89 6775–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernekar, J.V., Ghatge, M.S., and Deshpande, V.V. 1999. Alkaline Protease Inhibitor: A novel class of antifungal proteins against phytopathogenic fungi. Biochem. Biophys. Res. Commun. 262 702–707. [DOI] [PubMed] [Google Scholar]
- Vernekar, J.V., Tanksale, A.M., Ghatge, M.S., and Deshpande, V.V. 2001. Novel bifunctional alkaline protease inhibitor: Protease inhibitory activity as the biochemical basis of antifungal activity. Biochem. Biophys. Res. Commun. 285 1018–1024. [DOI] [PubMed] [Google Scholar]
- Vinci, M., Ruoppolo, M., Pucci, P., Freedman, R.B., and Marino, G. 2000. Early intermediates in the PDI-assisted folding of ribonuclease A. Protein Sci. 9 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.C. 1998. Isomerase and chaperone activities of protein disulfide isomerase are both required for its function as a foldase. Biochemistry (Moscow) 63 407–412. [PubMed] [Google Scholar]
- Woycechowsky, K.J. and Raines, R.T. 2000. Native disulfide bond formation in proteins. Curr. Opin. Chem. Biol. 4 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y., Zhou, Y., and Wang, C.C. 1997. Both the isomerase and chaperone activities of protein disulfide isomerase are required for the reactivation of reduced and denatured acidic phospholipase A2. EMBO J. 16 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]