Abstract
The transcription factor, Runx2, is known to play crucial roles in skeletal and tooth morphogenesis. Here we document that Runx2 has a regulatory role in skin and hair follicle development. The expression of Runx2 is restricted to hair follicles and is dynamic, pari passu with follicle development. Follicle maturation is delayed in the absence of Runx2 and overall skin and epidermal thickness of Runx2 null embryos is significantly reduced. The Runx2 null epidermis is hypoplastic, displaying reduced expression of Keratin 14, Keratin 1 and markers of proliferation. The expression pattern of Runx2 in the bulb epithelium of mature hair follicles is asymmetric and strikingly similar to that of Sonic hedgehog. This suggests that Runx2 may be a regulator of hedgehog signaling in skin as it is in bones and teeth. Supporting this possibility, we demonstrate that Sonic hedgehog, Patched1 and Gli1 transcripts are reduced in the skin of Runx2 null embryos. Moreover, we document Patched1 expression in epidermal basal cells and show that the skin of Sonic+/− embryos is thinner than that of wild-type littermates. These observations suggest that Runx2 and hedgehog signaling are involved in the well known, but unexplained, coupling of skin thickness to hair follicle development.
Keywords: Runx2, Skin, Hair follicles, Mouse, Sonic hedgehog, Development, Gene expression, Hair cycle, Skin allograft, Skin thickness
INTRODUCTION
Early embryonic skin consists of a single layer of proliferating cells overlying a dermis populated with undifferentiated mesenchymal cells. In hair-bearing skin, as the epithelium differentiates and stratifies (Hanson, 1947) and the dermal mesenchyme matures (Van Exan and Hardy, 1984), hair follicles begin to develop as a result of molecular interactions between the mesenchyme and the overlying epithelium. The initial signal for hair follicle development is thought to originate in clusters of committed fibroblast-like cells in the sub-epithelial mesenchyme that instruct the overlying epithelium to produce a skin appendage. The epithelium in turn signals these specialized fibroblasts to compact and form a dermal papilla which then instructs the epithelium to make a hair follicle (Hardy, 1992). As the epithelium grows downward, reciprocal molecular interactions between the papilla and epithelium continue as the follicle matures and the hair grows (Fuchs et al., 2001; Hardy, 1992; Paus and Cotsarelis, 1999). Soon after morphogenesis is completed, hair follicles begin to undergo lifelong cycles of regression (catagen) and regeneration and growth (anagen) with an intervening resting phase (telogen) (Chase, 1954). The cycling of hair follicles is also thought to be dependent upon inductive molecular interactions between the dermal papilla and the adjacent epithelium (Link et al., 1990); (Chase et al., 1951; Millar, 2002; Panteleyev et al., 1999; Paus and Cotsarelis, 1999). As hair follicles mature during morphogenesis there is a rather spectacular increase in skin thickness involving both the inter-follicular epithelium and the dermis. Similar marked increases or decreases in skin thickness occur during follicle cycling depending upon the phase of the cycle (Chase et al., 1953; Hansen et al., 1984; Muller-Rover et al., 2001; Paus et al., 1990). The molecular signals effecting these changes in skin thickness have not been identified.
During studies of Runx2 deficient mice, we noted Runx2 expression in the dermal papillae of developing hair follicles. Runx2 is a transcription factor of the Runt domain family that plays crucial roles in skeletal and tooth development (Inada et al., 1999; Kim et al., 1999; Komori et al., 1997; Otto et al., 1997; Zelzer et al., 2001). In light of the inductive role of the dermal papilla in developing and cycling hair follicles (Fuchs et al., 2001; Hardy, 1992) our observation of Runx2 expression in the dermal papilla suggested a role for Runx2 in hair follicle and skin development. Such a role would be consistent with the finding that Runx2 deficiency in both man (Mundlos et al., 1997) and mouse (Aberg et al., 2004a; Aberg et al., 2004b; D'Souza et al., 1999) has a profound impact on tooth development which is controlled by mesenchymal-epithelial interactions similar to those of developing hair follicles.
Here we show that Runx2 expression in the skin is restricted to hair follicles and displays a dynamic, stage-dependent pattern. Hair follicle development is delayed in Runx2 null skin, but the most striking feature of the null skin phenotype is a marked decrease in epidermal and overall skin thickness. The thin inter-follicular epidermis is hypoplastic but undergoes normal differentiation. We demonstrate substantial reductions in Sonic hedgehog, Patched1 and Gli1 transcript levels suggesting that Runx2 (directly or indirectly) regulates hedgehog signaling in the skin. These findings, together with our findings that Sonic hedgehog deficient skin is also abnormally thin and that Patched1 is expressed in epithelial basal cells, suggest that decreased hedgehog signaling is a part of the molecular mechanism for the thin, hypoplastic skin phenotype of Runx2 null embryos. We hypothesize that Runx2 and Sonic hedgehog signaling may be players in the as yet undefined molecular connection between hair follicle development and skin thickness (Hansen et al., 1984; Muller-Rover et al., 2001); (Paus et al., 1990); (Chase et al., 1953).
Materials and Methods
Mice
Runx2 mutant mice were the gift of M. J. Owen, generated as previously described (Otto et al., 1997). The colony was maintained by outbreeding with wild type C57BL6 mice. The mothers of Sonic hedgehog +/− and their wild-type littermate E18.5 embryos were generously bred and supplied by C. Tabin and E. McGlinn. To generate these mice ShhGFPcre mice that have one inactivated Sonic hedgehog allele (Harfe et al., 2004) were mated with RC::PFwe mice (Farago et al., 2006). Patched1 heterozygous mice were purchased from Jackson Laboratory (Stock Ptch1 tm/Mps/J, Stock # 003081, (Goodrich et al., 1997). Animals were housed in a pathogen free facility and maintained in accordance with strict guidelines for humane care and treatment of animals. Embryos at desired developmental stages were obtained by timed pregnancies using vaginal plugs or overnight mating, counting noon of the day of the vaginal plug or after overnight copulation as E 0.5.
Genotyping was accomplished by PCR amplification of genomic DNA prepared from tails (Laird et al., 1991). The PCR products were analyzed by electrophoresis through 0.8% agarose (details of the PCR reactions for genotyping for Runx2, Cre recombinase and the mutant Patched1 allele upon request). For genotyping for inactivated Sonic hedgehog, Cre recombinase-expressing embryos were assumed to be heterozygous for Sonic hedgehog whereas Cre negative embryos were considered to be wild-type. The presence or absence of Lac Z staining in several embryos was used to confirm genotyping.
Expression pattern of Runx2
Expression of Runx2 in pelage hair follicles during development and follicle cycling was studied in heterozygous Runx2 null mice using staining for the Lac Z gene in the targeted allele (Otto et al., 1997) as previously validated (Kim et al., 1999). The use of heterozygous mice (which are viable in contrast to null animals) allowed the study of postnatal skin and thus all 8 stages of hair follicle development as well as the stages of cycling follicles. Skin was examined on each gestation day from E14.5 through E19.0. To study all developmental stages of follicles, skin of heterozygous mice at P0, P1, P3, P7 and P13 was also examined.
To document the expression pattern in cycling follicles, synchronized hair cycles were induced by plucking the dorsal hair (as described below) of approximately 45 day-old Runx2 heterozygous mice (Muller-Rover et al., 2001; Slominski et al., 1991). Skin was examined on days 1, 3, 5, 8, 12, 18 and 22 post-plucking, allowing study of anagen, early catagen and return to telogen.
Hair plucking for induction of synchronous hair cycle
Synchronous hair cycles were induced by plucking the dorsal hair during the normally synchronized second post-natal telogen phase (Lavker et al., 1993; Muller-Rover et al., 2001; Paus et al., 1990; Slominski et al., 1991) Mice were anesthetized with Ketamine® (0.1 mg/gm mouse) and Xylazine® (0.02 mg/gm mouse) and the dorsal skin depilated with Surgiwax™ or with a 50/50 mixture (by weight) of Rosin gum (Sigma) and Beeswax (Aldrich) which was melted, cooled and applied to the dorsum of the mouse. After hardening the wax was removed together with the hair. Wax was reapplied 1 to 2 times as necessary to complete the depilation. The same technique was used to depilate embryonic allografts, taking care not to apply an overly strong force that would avulse the skin graft.
Tissue preparation, embedding and sectioning
For study of embryonic skin, mothers were euthanized with CO2. Whole embryos were quickly harvested, placed in cold PBS and then embedded on their sides in a mold containing cold Tissue-Tek® OCT compound. Molds were partially frozen in a dry ice-95% ethanol mixture, placed at −20° C for ∼1 hr and then stored at −80° C.
For neonatal and adult mice dorsal skins were harvested after marking the dorsal midline. Skin was first placed flat in cold OCT in a small mold superficial side up and partially frozen in 95% ETOH-dry ice mixture and then placed at −20°C for ∼1hr. To embed the skin on edge these blocks were cut along the previously marked midline and both sides were placed cut side down into cold OCT in a deeper mold, partially frozen in a 95% ETOH-dry ice mixture, complexly frozen at −20° C and then stored at −80° C until needed for sectioning.
A similar technique was used for embedding of skin for in situ hybridization. Using RNase clean conditions E18.5 embryos were harvested, the dorsal midline marked and the dorsal skin was excised and placed on 3M® paper. Specimens were fixed overnight in 4% PFA in PBS, rocking, at 4° C. After washing in cold PBS, the tissues were impregnated with 10%, 15% and 20% sucrose in PBS. The 3M® paper was removed and skin was embedded as described above.
The technique used for paraffin embedding of skin labeled with bromodeoxyuridine was similar to that used for frozen tissue. After overnight fixation in 4% paraformaldehyde in PBS, rocking, at 4° C. tissue was dehydrated and infused with paraffin using a tissue processor (Leica 2000). The paper backing was removed and the skin was embedded on edge in a mold and stored at room temperature until sectioning.
For embryonic skin 8μm and 16μm para-sagittal sections of the whole embryos were cut with a cryostat (Leica®). Similar sections of embedded postnatal skin were made. Cut sections were placed on Superfrost Plus® microscopic slides which were dried for 20 minutes, fixed in acetone or methanol at −20° C for 20−30 min, dried and stored at −20° C until stained. Paraffin embedded sections for BrdU staining were sectioned at 4 μm with a microtome (Microm®), mounted on Superfrost Plus® slides and stored at room temperature until stained.
ß-galactosidase (LacZ) staining
Slides were dried for 20 min at room temperature, post-fixed in 2% PFA, 2mM MgCl2 in PBS for 5 min at 4° C and then washed twice in PBS with 2 mM MgCl2 for 5 min at room temperature. Slides were stained for 24−48 h at 37° C in a soultion containing 1 mg/ml X-gal (Sigma), 2mM MgCl2, 0.02% NP40, 0.01% Na desoxycholate, 5mM ferricyanide, 5 mM K ferrocyanide and buffered to pH 7.3 with Na phosphate. After washing in PBS for 5 min at room temperature, slides were counterstained with Nuclear Fast Red (Vector) for 5 min, washed, rapidly dehydrated in graded ethanol and xylene and mounted with xylene-based medium.
Alkaline phosphatase staining
The method was essentially that described by Handjiski et al., 1994 (Handjiski et al., 1994). Eight μm cryosections were used. After alkaline phosphatase staining the tissues were counterstained with Gill's hematoxylin (Vector) for 45 sec, washed in H2O and mounted with Kaiser's glycerol gelatin (Merck).
Embryonic skin allografts
Male and female heterozygous Runx2 null mice were time mated. Pregnant females were sacrificed at 18.5 dpc using an overdose of isofluorane and cervical dislocation. Using clean surgical technique embryos were harvested and the full thickness dorsal skin was removed and placed in ampicillin 5mg/ml in PBS. Embryo tails were taken for later PCR genotyping. Nude (nu/nu) BALBc outbred female mice (Harlan or Charles River) were anesthetized by intra-peritoneal injection of Ketamine® (0.1mg/gm mouse) and Xylazine® (0.02mg/gm mouse) and a 1 cm2 full thickness piece of skin was excised with curved scissors leaving intact the panniculus carnosus. A 1 cm2 portion of the embryo skin from mid-dorsal area was cut and carefully placed on this bed without sutures or glue and a sterile, non-adherent elastic dressing was applied (details upon request). The recipient mice were caged separately. Dressings usually remained in place until removed 6−8 days after the procedure.
The genotypes of the embryos corresponded roughly to the expected Mendelian ratios. Operative mortality was nil and 94% of grafts were satisfactory for study.
Morphometric staging of hair follicle development
Near mid-sagittal 8μm cryosections of whole embryos were cut at several levels and stained for alkaline phosphatase to define the dermal papillae. The stage (Paus et al., 1999) was recorded of each stageable (i.e. properly oriented) hair follicle on the dorsal skin from the level of the thoracic inlet to the bottom of the peritoneal cavity. At least 200 follicles per animal were staged taking care during cutting that each section was at least 100μ distant from the previous to avoid staging the same follicle more than once. For each animal the number of follicles at each stage was tabulated and then expressed as the percent of the total number of follicles examined for that animal.
The mean percent follicles at each stage of all Runx2 null and wild-type embryos along with the standard deviations were calculated and the significance of the differences in the means was determined using an unpaired, 1 tailed T-test.
Skin thickness measurements
Whole embryo 8 μm cryosections of E 18.5 embryos stained for alkaline phosphatase and counterstained with Gill's hematoxylin were used. For each animal 4 near mid sagittal sections, cut at least 100 microns apart, were evaluated. Digital photomicrographs of the dorsal skin of a standard area at the level of the diaphragm were taken with a 4× objective. Using an image-processing program (Adobe Photoshop® 5.5 or 8.0), colored lines at right angles to the surface were drawn (at fixed intervals determined by crossing gridlines and thus random for the section) traversing the thickness of the epidermis and the epidermis and dermis together. Epidermal thickness was from the top of the granular layer to the bottom of the basal layer and total thickness was from the top of the granular layer to the top of the panniculus carnosus. The length of these lines was measured on color prints of photomicrographs using as a ruler a photomicrograph of a 1 micron stage micrometer also taken with the 4× objective to give the actual values of thickness in microns at each point. For each Runx2 null or wild-type embryo there were 4 sections with 10 measurements (11 for Sonic hedgehog) on each section for epidermis and total thickness. Therefore, there were 40 thickness measurements per mouse (44 for Sonic hedgehog). The means and standard deviations of the individual measurements for wild-type and mutant animals were compared and the significance of the differences was determined using an unpaired, 1 tailed T-test.
Immunohistochemistry
Para-sagittal cryosections (8μm) of OCT embedded whole embryos were used. The protocols for filaggrin, K1 and K14 were horseradish peroxidase based with detection with diaminobenzidine (DAB, Bio Genex). Slides were post-fixed in 4% PFA, washed in H20 and PBS and then quenched in 3% H2O2 for 5 min, Blocking was with 5% Normal Goat Serum (Vector) for 30 min at room temperature. The following primary antibodies and concentrations, all diluted in PBS, were applied: Filaggrin 1:250 (AF111, Covance); K1 1:500 (MK1, Covance); K14 1:1000 (MK14, Covance). Incubation was at room temperature for ∼16 hr in a moisturized chamber. Secondary antibody (peroxidase conjugated goat anti-rabbit IgG [F(ab')fragment] Jackson Immunology 1:500 in PBS) was applied with incubation for 2 hr at room temperature in a moisturized container. After washing, DAB substrate was applied for 2 min or more, time determined under microscopic observation. After washing the slides were counterstained with 0.25% methyl green for ∼1−2 sec and immediately washed with H20. The tissue was rapidly dehydrated in graded ethanol and xylene and slides mounted with xylene-based mounting medium.
Ki 67 horseradish peroxidase-based staining required streptavidin amplification. Post-fixation, H2O2 quenching and proteinase K treatment were not required. Blocking was accomplished with 10% normal rabbit serum (Vector) in PBS for 20 min at room temperature. The primary antibody was Rat monoclonal anti-mouse Ki 67 (DAKO) 1:25 in PBS with incubation for ∼16 hr at room temperature. Secondary antibody, biotinylated rabbit anti-rat IgG (Vector BA 4001) 1:200 in PBS, was applied for 20 min at room temperature. After washing in PBS, strepavidin peroxidase (Biogenex) 1:50 in diluent was applied with incubation for 20 min at room temperature. After washing the DAB protocol was utilized with counter-staining with methyl green, rapid dehydration and mounting as described above.
Bromodeoxyuridine (BrdU) immuno-staining
Pregnant mothers at 18.5 days post-coitus were injected intra-peritoneally with 0.5 ml undiluted Labeling Reagent (Roche) and sacrificed 2 hr later. Embryos and skin were harvested and paraffin blocks were prepared. Sections of skin cut at 4 μm were used. A BrdU staining kit (Zymed Laboratories) was employed, using the protocol and control slide supplied by the manufacturer.
Immunofluorescence histochemistry for Ki 67
Cryosections (8μm) prepared as described above were used. The protocol was slightly modified from one kindly provided by Jennifer Zhang. All steps were carried out at room temperature. Care was taken to avoid drying at any point. Blocking was accomplished with 10% normal goat serum (Vector) for 20 min. After shake off, slides were incubated for 2 hr with the primary antibody (Rat monoclonal anti-mouse Ki 67 [DAKO] 1:25 in 1.5% normal goat serum in PBS with 0.1% Tween 20). Slides were washed three times in PBS with 0.1% Tween-20 and then incubated for 45 min with the secondary antibody (Alexafluor 488- labeled goat anti-rat IgG [Molecular Probes)] 1:250 in 1.5% normal goat serum in PBS with 0.1% Tween 20). Slides were then washed three times in 0.1% Tween-20 and counterstained with Hoechst 33342 1 mg/L for 30 sec with prompt wash in PBS. Mounting was with ProLong Gold® (Invitrogen).
Quantitation of fluorescence
As soon as possible after staining sections were examined with a Nikon® 80i microscope under UV light from a metal halide lamp, and optimally stained regions of the dorsal skin were identified. Using Metamorph® software gray scale photomicrographs using blue and green filters of these areas were taken with a 10× objective using a Hamamatsu Cool CCD® camera. Photomicrographs using several exposure times with each filter were taken so that images with the same exposure time and with a maximum pixel value of <4095 would be available for all specimens. Using Metamorph® software the basal layer of the skin was outlined on the blue image and the identical area transferred to the green image. Each area was “auto-thresholded” and the ratios of green-stained and blue-stained areas above an arbitrary baseline level above auto-fluorescence was determined. These ratios were used as surrogates for the percent of basal cells expressing Ki 67 for each animal. The means and standard deviations of all values for wild-type and null embryos were determined and the significance of the difference between the means was determined using an unpaired, 1 tailed T test.
In situ hybridization
The protocol used for in situ hybridization using cryosections was modified from one kindly provided by Bruce Morgan. Digoxigenin-11-UTP-labeled single strand ribo-probes were prepared essentially as previously described (Huang et al., 1997). Plasmids were generously supplied by P. Mill (Sonic hedgehog), A. McMahon (Patched1) and C. Tabin (Gli and Gli2).
Each protocol step was carried out at room temperature unless otherwise specified. Hybridization steps were performed using clean gloves and RNase-free containers. Solutions were DEPC treated when feasible.
Eight μM sections were air dried, hydrated in PBS and then post-fixed in 4% PFA in PBS for 15 min. After washing in PBS slides were placed in Proteinase K, (4 μg/ml. in 10mM Tris buffer [pH 7.5] with 1mM CaCl2) for 20 min. After washing in 0.2% glycine and then in PBS, further fixation with was carried out in 4% PFA in PBS for 10 min. After washing slides were acetylated for 15 min in a solution containing 0.2% acetic anhydride and 0.2M triethanolamine. After washing tissues were quickly dehydrated in graded ethanol and air dried for ∼30 min. The hybridization solution (details furnished upon request) was heated to 85° C and riboprobes were added (∼1μl/100μl hybridization solution). After vortexing the solution was heated to 85° C and placed on the slides (∼100μL/slide) and a cover-slip made from Parafilm® was applied. Incubation was carried out for ∼16 hrs. at 65° C in a chamber moisturized with 50/50 formamide/water.
After hybridization the following washes were carried out at 65° C in increasingly dilute SSC (20× SSC: 3M NaCl, 300mM Na Citrate, pH 7.0). Cover-slips were removed in 6× SSC and washed in 6× SSC for 5 min, 50% formamide 2× SSC 10mM EDTA for 30 min, 2× SSC for 30 min (2 times), and 0.2× SSC for 30 min (2 times).
The steps for the color reaction were carried out at room temperature. Slides were placed in Dig 1 buffer (100mM Tris, 150mM NaCl, pH 7.5, filtered before use) for 5 min. Blocking agent (Roche Cat # 1 096 176) 1.5% in Dig 1 buffer was applied with incubation in a moisture chamber with H2O for 1 hr. The blocking reagent was poured off and, after brief wash in Dig 1 buffer, alkaline phosphatase-conjugated anti-digoxigenin antibody (anti-digoxigenin-AP Roche Cat #11 093 274 910) 1:500 in 1.5% blocking agent was applied. Incubation was for 2 hr in a moisturized chamber. The slides were then washed in Dig 1 buffer and placed in filtered Dig 3 buffer (100mM Tris, 100mM NaCl, pH 9.5 before adding MgCl2 to a concentration of 50mM) for 5 min. The slides were then covered with alkaline phosphatase substrate (4-Nitro blue tetrazolium chloride 4.5 μl [Roche 11 383 213 001] and BCIP 3.5 μl [Roche 1 383 221] per 1 ml Dig 3 buffer) and incubated (without a coverslip) overnight at 4° C in a moisture chamber. When sufficiently developed the reaction was stopped in TE buffer for 10 min at room temperature (Tris 10mM, EDTA, 0.5mM pH 7.5) after brief wash-off in tap water. Further fixation in 8% formalin in de-ionized water was carried out for 1 hr in a fume hood. After washing slides were were counterstained with methyl green 0.25% for ∼1−2 sec. Rapid dehydration in graded ethanol and impregnation with xylene was carried out followed by mounting with xylene-based medium.
Real-time PCR for mRNA quantitation
Total RNA was prepared from the dorsal skin of E18.5 embryos. After the pregnant mother was killed with an overdose of ethyl ether the embryos were rapidly harvested with instruments soaked in 4M guanidine hydrothiocyanate and put in cold DEPC-treated PBS. Using instruments treated with 4M guanidine the tails were removed for genotyping and the dorsal skin was removed and placed in an RNase-free Eppendorf® tube, quickly frozen in liquid nitrogen and stored at −80° C pending genotyping and later preparation of RNA.
RNA was extracted using Tri Reagent™ (Sigma) according to the protocol recommended by the manufacturer including an extra centrifugation step recommended to minimize genomic DNA contamination. The purified RNA was re-dissolved in ∼50 μl DEPC-treated water and stored at −80° C for later cDNA preparation. Single stranded cDNA was prepared with the iScript™ Select cDNA Synthesis Kit (Bio-Rad) and oligo(dT) primers, using the manufacturer's protocol. The cDNA was stored at −20° C until needed for RT-PCR.
A Bio-Rad and Icycler iQ detection system, 96 well plates and optical tape were used for RT-PCR using SYBR Green™. The primer pairs for Patched1, Gli1 and Glyceraldehyde-3-phosphate dehydrogenase were those used by Wang et al., (2005) and those for Sonic hedgehog as used by Turner et al., (2004). Primer concentrations, cDNA concentrations and PCR conditions were optimized using melting curves and analysis of the PCR products by electrophoresis (composition of 25μl PCR reactions and conditions furnished upon request). Melting curves were used to check the specificity of the product in each well. Standard curves for each set of primers were determined using wild-type cDNA from an E16.5 embryo using 2-fold serial dilutions since the differences observed were within this range. The efficiencies were found to be 1.9 or greater for each primer pair.
For skin keratin mRNA quantification primers for Keratin 14, Keratin 1 and Glyceraldehyde-3-phosphate dehydrogenase, and Sybr Green/Fluorescein Master Mix (formulated for use in the Bio-Rad iCycler) were purchased from SuperArray™. The PCR protocol was that recommended by the manufacturer.
Each determination was done in triplicate and each experiment was performed three times. In a given experiment the mean of the triplicate crossing thresholds (cycle number at which the threshold of detection was reached) for each gene of interest was calculated. These values were normalized by subtracting the mean crossing thresholds of the Glyceraldehyde-3-phosphate dehydrogenase reference gene run simultaneously as a loading control for the amount of mRNA. The mean value of all wild-type and all null animals was then determined. The negative logarithm to the base 2 of the difference between wild-type and null gave the relative expression of null to wild-type animals. The standard deviations of all crossing values from all 3 experiments for wild-type and null animals were determined for each gene of interest and the significance of the differences between wild-type and null was calculated by an unpaired, 1 tailed T-test.
RESULTS
Runx2 expression in the skin is restricted to hair follicles in a dynamic pattern during morphogenesis and cycling
The expression of Runx2 was documented by staining for the Lac Z gene in the targeting vector (Otto et al., 1997). Runx2 expression in the skin is confined to the hair follicles. Individual follicles at a given stage vary greatly in the amount of staining (Figs. 1A, B); the amount of LacZ staining is less than that typically found in bone and cartilage (Fig. 1C). Additional representative photomicrographs of Lac Z expression during morphogenesis may be seen in the Supplementary Figure. The pattern of expression of Runx2 in hair follicles is stage-dependent irrespective of the type of pelage follicle or the age of the embryo or pup. Therefore, the expression is described mainly in terms of stages in both developing and cycling follicles.
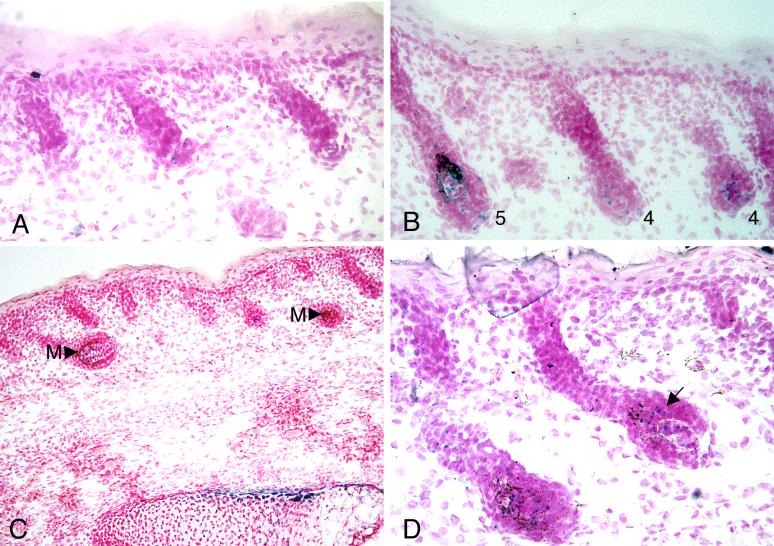
Fig. 1.
Appearance of LacZ staining in developing follicles of Runx2 heterozygous embryos. (A) LacZ staining is seen in hair follicles as punctate blue dots or ovals. Individual follicles at a given stage vary greatly in the amount of staining for ß-galactosidase as shown in three Stage 2 follicles at E 17.5. (B) Differences in the amount of staining from follicle to follicle are also illustrated in two Stage 4 follicles and a Stage 5 follicle at E 18.5. Numbers indicate follicle stages. (C) The level of Runx2 transcription in hair follicles is lower than in cartilage and bone as shown by comparing the amount of Lac Z staining in two Stage 5 follicles with that in the subjacent vertebra. The arrowheads point to melanin (M) which should not be mistaken for Lac Z stain. (D) In later stages of development Runx2 is expressed in epithelium (arrow) as shown here in a Stage 5 follicle at E19.0. The dermal papilla is outlined by the dashed white line. Epithelial expression is typically asymmetrical, predominately on the superficial side of the follicle bulb.
The expression of Runx2 in developing follicles is summarized in Figure 2A. No Runx2 expression is found in stage 1 or early stage 2 follicles. In some of the more advanced Stage 2 follicles, expression is seen in the cap-like dermal papilla characteristic of this stage. Runx2 expression becomes more prominent in the dermal papillae of stage 3 and early stage 4 follicles when they appear at E16.5. As increasing numbers of mature stage 4 follicles appear at E17.5 and E18.5 staining is also found in the connective tissue sheath at the base of the dermal papilla. Stage 5 − 8 follicles are found mostly in post-natal animals. Pari passu with increasing stage, expression in the connective tissue sheath is seen progressively more distally (toward the skin surface) as far up as the bulge region of the follicle.
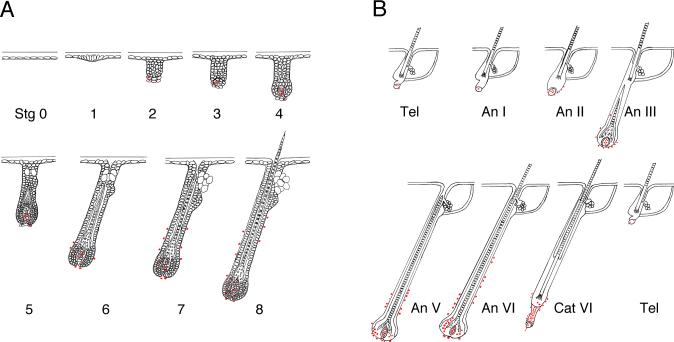
Fig. 2.
The expression pattern of Runx2 during development and cycling is dynamic. (A) During development Runx2 expression (Lac Z staining as indicated by the red dots) is first seen in the dermal papillae of Stage 2 follicles. With increasing stage, expression increases in the dermal papillae and begins to be seen in the connective tissue sheath in which it progresses distally. In later follicle stages asymmetrically distributed staining begins to be seen in the bulb epithelium and increases. (B) In an induced cycle scant Runx2 expression is found in the dermal papilla of telogen (resting) follicles. Similar to the development stages, during the anagen (growing) phases, expression increases in the dermal papilla, appears in the connective tissue sheath and then progresses distally. At later stages asymmetrical epithelial expression is seen in the bulb epithelium and increases. Comparatively strong Runx2 expression is seen in early catagen (regressing) epithelium. Stage (Stg), Telogen (Tel) Anagen (An), Catagen (Cat). Follicle diagrams after Paus, R et al, 1999 (A) and Muller-Rover et, 2001 (B).
Minimal and isolated Runx2 expression is found in follicle epithelium as early as stage 2 and in stages 3 and 4 (not depicted). However, starting with stage 5 follicles, Runx2 is increasingly expressed in the bulb epithelium (Figs. 1D, 2A, Supplementary Figure). At stages 7 and 8 there is more prominent expression in the bulb epithelium along with the somewhat decreased expression in dermal papillae. Runx2 expression in the bulb epithelium is most marked at stage 8 and in most instances is more pronounced on the side closest to the skin surface in these angulated follicles.
To analyze the expression of Runx2 in cycling follicles we induced synchronized hair cycles by plucking the dorsal hair during the second telogen phase (Muller-Rover et al., 2001; Paus et al., 1990). As depicted in Figure 2B, the pattern of Runx2 expression in cycling follicles is similar to that of developing follicles. The follicles of unplucked control animals are in uniform telogen with scant Runx2 expression in their small, compacted dermal papillae (Supplementary Figure). On day 1 post plucking, some follicles are in anagen I and display continued sparse expression confined to the dermal papilla. Anagen II follicles are seen on day 3 post plucking and show increased Runx2 expression in the dermal papillae and some expression in the connective tissue sheath at the base and sides of the developing bulb epithelium. At day 5, anagen IIIb follicles predominate and show comparatively robust Runx2 expression in the dermal papilla, in the connective tissue sheath at the base and sides of the bulb, as distally as the sebaceous gland with some staining in the epithelium. Anagen V follicles, as observed at day 8 post plucking, display prominent expression in the dermal papilla and connective tissue sheath, and show some expression in bulb epithelium. On day 12 post-plucking, the follicles are mainly in anagen VI and display reduced dermal papilla expression. However, at that stage fairly prominent staining is found in the epithelium of the bulb (Fig. 2B and Supplementary Figure) and along the connective tissue sheath, the latter extending as far distally as the sebaceous gland. Runx2 expression in the bulb epithelium is usually asymmetrically located most often on the anterior (superficial) side (Fig. 2B and Supplementary Figure). This is similar to the epithelial expression in developing follicles. On day 18 post plucking, early catagen follicles are found in which there is strong Runx2 expression in the epithelium undergoing apoptosis, dermal papilla and connective tissue sheath (Fig. 2B and Supplementary Figure). On day 22 post plucking, almost all follicles are again in telogen with sparse staining in the dermal papilla and subjacent connective tissue sheath.
The changing expression pattern of Runx2 during hair follicle morphogenesis and cycling suggests that it has a regulatory role in development.
Allografted Runx2 null skin undergoes full development and the follicles cycle
Runx2 null mice die at birth so that it is not certain that their skin is capable of growing a full coat of hair or that their hair follicles are capable of cycling. To address these questions, we compared allografts of Runx2 null skin at E18.5 with those of wild-type controls.
At 2 weeks post placement the hair was almost totally grown in the grafts with both under- and over-hair types evident. There were no apparent differences in the rate of hair growth or in the gross appearance of the hair coats of fully matured wild-type and Runx2 null grafts (Figs. 3A, D). During serial observations of up to 183 days after placement wild-type and Runx2 null grafts continued to be indistinguishable. The underlying skin of both genotypes was noted to change color from black to gray to pink and it became possible to pluck hair with the fingertips easily, indicative of club hair in telogen (Chase, 1954; Fiedler, 1994). Together these observations provided evidence of cycling follicles in both genotypes (Paus et al., 1990; Slominski et al., 1994; Stenn and Paus, 2001).
Fig. 3.
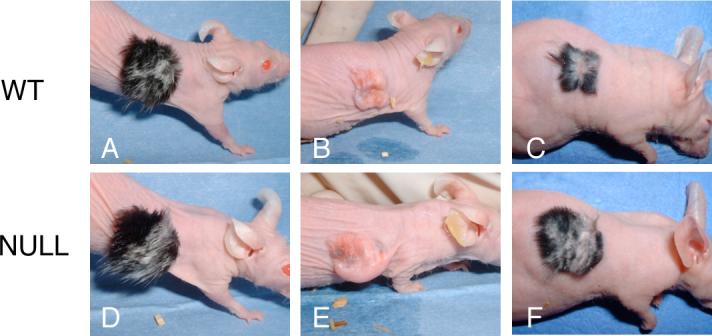
Hair growth in wild-type and Runx2 null skin allografts during development and cycling is grossly indistinguishable from that of wild-type. (A and D) There is no apparent difference in the robust growth of hair and hair types (see text) between wild-type and null grafts. The randomly distributed patches of white hair shown here were found in the majority of the grafts irrespective of genotype of the embryos (C57BL background). These patches persisted with cycling and after plucking (see below) and were presumed to result from a temporary ischemic insult inherent in grafting perhaps affecting the migration and/or maturation of melanocytes. (B and E) Skin grafts immediately after plucking. Note the focal pigmentation and the trauma to the grafted skin (see text). (C and F) Hair re-growth 13 days after plucking is similar in wild-type and null grafts.
We compared cycle progression in Runx2 null mice with that of wild-type animals by hair plucking to induce synchronized cycling (Chase et al., 1953; Paus et al., 1994; Paus et al., 1990). Unfortunately, a fully synchronized, prolonged telogen phase after the second postnatal cycle did not occur in the grafted skin as it does in intact skin (Chase et al., 1953; Paus et al., 1994; Slominski et al., 1991). Nevertheless, we plucked the hair of 7 wild-type and 6 Runx2 null grafts. There was evident focal trauma and isolated areas of pigmentation attributable to a lack of synchronization in telogen (Figs. 3B, E). Comparable hair re-growth was observed in null and wild-type grafts (Figs. 3C, F). Microscopic analysis of sections showed both telogen and anagen follicles in unplucked control grafts and, in grafts harvested at 6 and 13 days after plucking, that anagen was induced and progressed through all anagen stages in both genotypes (data not shown). No temporal or morphological differences in anagen progression between null and wild-type skin were evident, although such differences could well have been present but not detectable because of the lack of synchronization.
Microscopic study of the plucked hair from this experiment revealed the normal panoply of hair types (Sundberg, 1994) including both under-hair and over-hair (e.g. zigzag and awl) and no morphological differences between mutants and wild-type (data not shown).
It is clear from these experiments that Runx2 is not essential for the complete morphogenesis of hair follicles, hair growth and cycling capability.
Hair follicle development is delayed in Runx2 null mice
Despite the findings in the allografting experiments, examination of random microscopic sections of pre-term and newborn skin of Runx2 null mice suggested a delay in their hair follicle development. Quantitative morphometric staging (Paus et al., 1999) was employed to confirm or refute this impression.
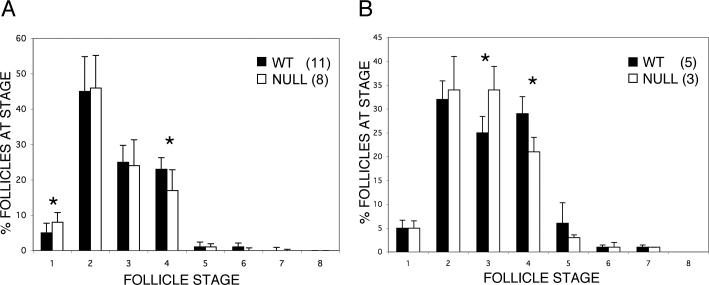
We compared the progression of hair follicle maturation in Runx2 null embryos with littermate wild-type controls from E14.5 through E19.0 using only litters that contained both genotypes. At E14.5 through E16.5, there are increasing numbers of follicles in embryos of both genotypes, but too few for any meaningful quantification. At E17.5, E18.5 and E19, a sufficient number of follicles have developed to allow staging of ∼200 follicles for each embryo. At E17.5 no significant differences are found (2 null and 2 wild-type embryos; data not shown). At E18.5 (Fig. 4A), there is a significantly increased percentage of stage 4 follicles in wild-type (23%) compared to Runx2 null animals (17%). Although there are also significantly fewer stage 1 follicles in wild-type than in null animals, this may well be a spurious result. In practical terms stage 1 follicle recognition is based solely on very faint sub-epithelial alkaline phosphatase staining so that the presence or absence of stain can be somewhat subjective. At E19.0 (Fig. 4B), the lag in progression from Stage 3 to Stage 4 in Runx2 null animals vs. wild-type persists (21% vs. 29%). In addition, there are significantly fewer follicles at stage 3 in wild-type (25%) than in Runx2_null mice (34%). The percentage of stage 5 follicles in wild-type compared to Runx2 null mice is also higher (6% vs. 3%) but this trend is not statistically significant.
Fig. 4.
Hair follicle development is delayed in Runx2 null embryos at E18.5 and E19.0. (A) Using quantitative morphometric staging there is a significant delay in Stage 4 follicle development at E18.5. Significantly fewer Stage 1 follicles are noted in WT than in KO animals (see text). (B) The delay in the development of Stage 4 follicles continues at E19.0 and, in addition, there are significantly fewer follicles at Stage 3. Note the different scales for the Y-axis in (A) and (B). * p=0.01 Error bars indicate ±1 standard deviation.
In hair phenotypes in which follicle development is delayed frequently the number of follicles is reduced as well. As counted on the sections used for measurements of skin thickness (see below) the mean number of hair follicles per millimeter skin length in wild-type animals is 11.7 ±2.6 and 10.2 ±3.8 in Runx2 null embryos (p=0.08). This non-significant trend is compatible with the modest delay shown in follicle development.
Runx 2 null skin is significantly thinner than that of wild-type mice
The skin of Runx2 null embryos or neonatal mice, when harvested for grafting or sectioning, appears to be markedly thinner than that of their wild-type or Runx2 heterozygous littermates. To confirm this gross observation, dorsal total skin and epidermal thickness were measured on sections of whole embryos at E 18.5.
The mean overall skin thickness of wild-type mice is 252±43 μm compared with 196±48 μm for the null animals (Fig. 5). The mean thickness of the epidermis for wild-type animals is 43±11 μm and 35±13 μm for Runx2 null littermates. The ratio of epidermal to overall skin thickness is almost identical in wild-type (17%) and null (18%) animals.
Fig. 5.
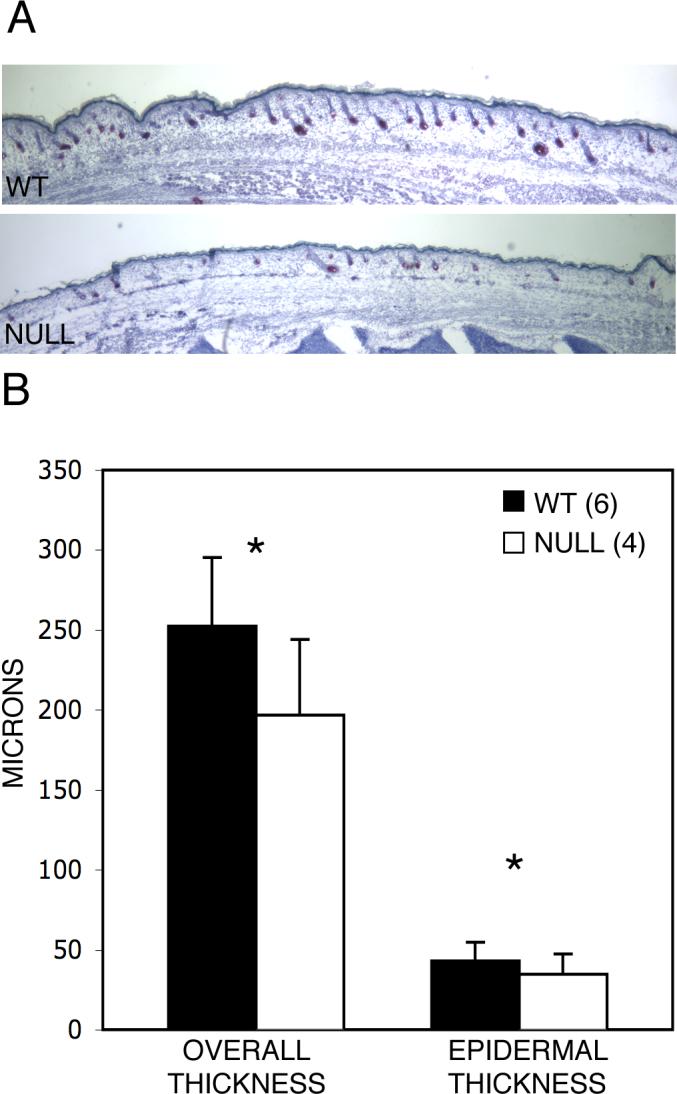
Runx2 null dorsal skin is thinner than wild-type skin. (A) Qualitative comparison of wild-type and Runx2 null embryos at E18.5. The differences are not always so obvious as shown in these littermates. There is considerable variation between animals of the same genotype within litters, between one section and another in the same animal and between areas in a given section. (B) Quantitative comparison shows that both overall skin and inter-follicular epidermal thickness of null embryos are significantly decreased compared with wild-type littermates. Mean values, variation and significance are based on measurements at 10 randomly determined points on each of 4 mid-sagittal sections such as shown here (total 40) for each animal (Materials and methods). *p<<0.0001. Error bars indicate ±1 standard deviation.
It is well established that skin thickness varies markedly with hair follicle development and the hair cycle (Chase et al., 1953; Hansen et al., 1984; Paus et al., 1990). The striking reduction in overall and epidermal skin thickness in the mutant mice, however, is out of proportion to the modest reduction in hair follicle development.
Inter-follicular epidermal development is impaired in Runx2 mice
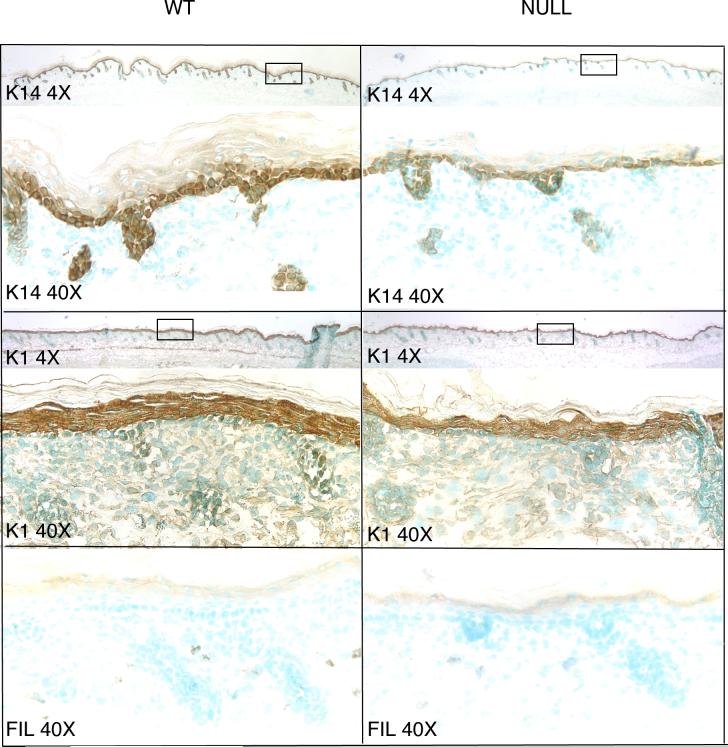
To further explore the thin-skin phenotype of Runx2 null mice, we studied the development of the epidermis using immuno-staining for keratin markers of differentiation (Yuspa et al., 1989). Keratin 14 expression is decreased in the basal layer of Runx2 null skin compared to wild-type skin in that staining is less intense and, although K14 in both genotypes is seen mainly in the skin basal layer, focal staining in two layers of cells is more likely to be found in wild-type than in wild-type than in null animals (Fig. 6). Keratin 14 staining is also decreased in the hair follicles of Runx2 null embryos. The suprabasal, differentiating layer of cells expressing K1 is thinner and the staining is less intense in the null than in wild-type mice (Fig. 6). No consistent differences are seen in filaggrin expression in terminally differentiating cells (Fig. 6).
Fig. 6.
Development is impaired in Runx2 null epidermis (E18.5). Immuno-staining for keratin markers of differentiation. K14 in Runx2 null embryos is less intense than in the wild-type embryos and has a greater tendency to be found focally in two cell layers in the wild-type embryos as compared to null animals. The layer of cells expressing K1 is thinner and the staining is less intense in the null mice than in the wild-type animals. Filaggrin staining was similar in wild-type and null embryos. The sections in this figure also illustrate that the null skin overall is thinner than that of wild-type, an observation that is compatible with the findings shown in Figure 5. Wild-type (WT), Keratin 14 (K14), Keratin 1 (K1), Filaggrin (Fil).
To quantify and verify the down-regulation of K14 and K1 in Runx2 null mice we employed real time PCR to compare the transcription of the two genes at E18.5. Keratin 14 and keratin 1 mRNA levels are both reduced by 62% in null embryos compared to the wild-type littermate controls (Fig. 7).
Fig. 7.
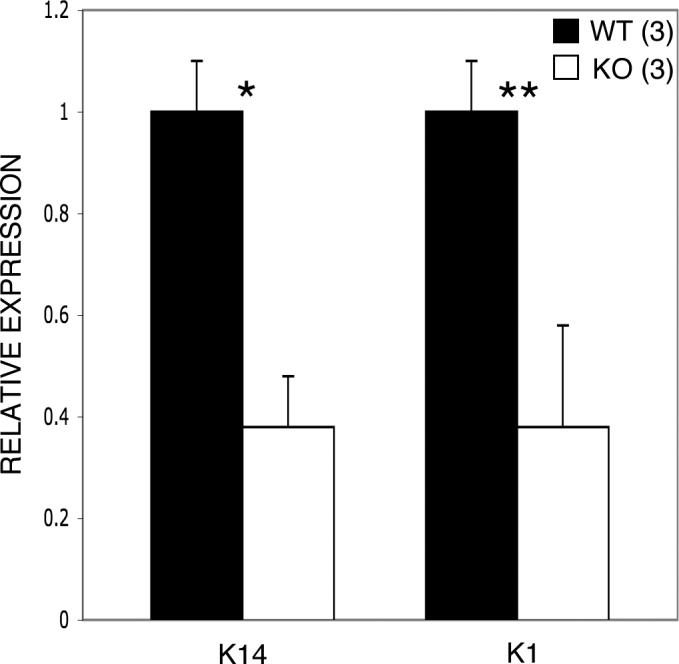
Keratin 14 and Keratin 1 mRNA levels are down-regulated in null embryos at E18.5. K14 and K1 expression levels in null mice are both decreased by 62% compared with their wild-type littermates. Note that the bars represent relative expression levels with wild-type levels set at 1. Wild-type (WT), Keratin 14 (K14), Keratin 1 (K1). *p=0.0002, **p<<0.0001. Error bars indicate ±1 standard deviation.
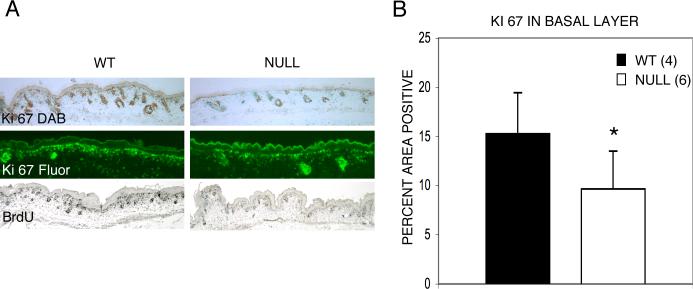
The decreased expression of K14 in the basal layer of inter-follicular epithelium together with the decreased epidermal thickness of Runx2 null animals suggested that basal cell proliferation might be decreased. Proliferation was first assessed using immunostaining for Ki 67. Ki 67 expression is decreased in the basal layer and also in the hair follicles in the null as compared to wild-type mice (Fig. 8A). For a quantitative comparison of Ki 67 antigen we used immuno-fluorescence staining combined with Metamorph® analysis. The results demonstrate that Ki 67 expression is reduced in the basal layer by 37% in Runx2 null compared to wild-type mice, thus confirming the qualitative assessment (Figs. 8A, B). Finally, as a completely different method of assessing mitotically active cells, we employed immunostaining for the incorporation of bromodeoxyuridine (BrdU) during DNA replication. Results similar to those using the Ki 67 antigen are found including the increased incorporation of BrdU in hair follicles (Fig. 8A).
Fig 8.
Proliferation is decreased in the basal layer of inter-follicular epidermis of Runx2 null mice. (A) Ki 67 antigen when detected with diaminobenzidine (DAB) is decreased in the basal layer of the skin and in hair follicles in null animals as compared to wild-type littermates (top panel). This assessment was confirmed by bromodeoxyuridine (BrdU) immuno-staining (bottom panel). (B) A quantitative comparison using immuno-fluorescence staining (see (A) middle panel) combined with Metamorph® analysis showed a 37% decrease in Ki 67 expression in null embryos compared with the wild-type controls. Note that in the quantitative analysis using Metamorph® indicates relative fluorescence in the basal layer of skin comparing wild-type and null animals. The numbers are based on the area occupied by basal cells and do not indicate the percent of cells expressing Ki 67 above an arbitrary baseline level of fluorescence. *p=0.03. Error bars indicate ±1 standard deviation.
These findings suggest that the thin epidermis of the Runx2 null state results from hypoplasia rather than, for example, an impaired differentiation program.
Expression of Sonic hedgehog and its targets is reduced in Runx2 null mice
As noted above, Runx2 expression in the bulb epithelium of late stage developing and cycling follicles is found predominately on the side closest to the skin surface. A strikingly similar asymmetric and restricted pattern is characteristic of Sonic hedgehog expression in hair follicles (Gat et al., 1998; Mill et al., 2003; Millar, 1997; Millar, 2002; St-Jacques et al., 1998). These observations raised the question of a potentially important functional relationship between Runx2 and hedgehog signaling in the skin since, in the absence of Sonic hedgehog, hair follicle development is arrested at stage 2 (Chiang et al., 1999; St-Jacques et al., 1998; Wang et al., 2000). If abnormal hedgehog signaling were present in the mutant mice, this could contribute to the hair and skin phenotypes of Runx2 deficiency. Therefore, we compared the expression of Sonic hedgehog and its mediators in Runx2 null and wild-type littermates at E18.5.
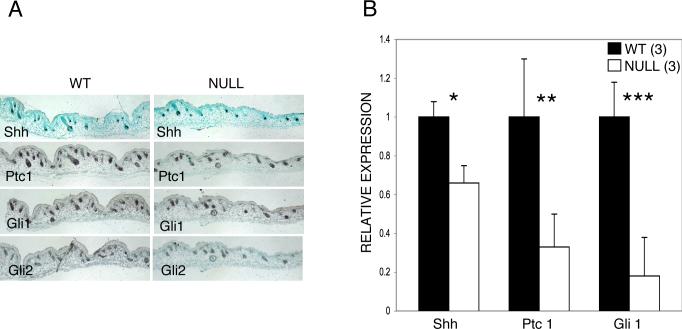
The expression patterns of Sonic hedgehog, Patched1, Gli1 and Gli2 using in situ hybridization are identical to those previously reported (Gat et al., 1998; Mill et al., 2003; St-Jacques et al., 1998) and no detectable differences are found between the wild-type and null animals (Fig. 9A).
Fig. 9.
Hedgehog signaling is significantly reduced in Runx2 null skin at E18.5. (A) By in situ hybridization the patterns of expression of Sonic hedgehog and downstream mediators are similar to those previously reported. There are no differences in apparent expression levels between wild-type and null animals. Note that the expression of Sonic hedgehog is restricted to the proximal (proliferating end) of the more mature follicles and tends to predominate on the superficial side. This asymmetrical pattern is strikingly similar to the epithelial expression of Runx2 (see text and Fig.1D). (B) Sonic hedgehog mRNA is reduced by 34%, Patched1 by 67% and Gli1 by 82% utilizing real-time PCR. Note that the bars represent relative expression levels with wild-type values set at 1. Wild-type (WT), Sonic hedgehog (Shh), Patched1 (Ptc1). *p=0.06, **p=0.04, ***p=0.001. Error bars indicate ±1 standard deviation.
Since in situ hybridization is not strictly a quantitative technique, we used real time quantitative PCR to quantify the relative transcript levels of Sonic hedgehog and its mediators in mutant and wild-type animals. The levels of expression of Sonic hedgehog, Patched1 and Gli1 in Runx2 null animals are reduced by 34%, 67% and 82%, respectively, when compared to wild-type mice (Fig. 9B).
Patched 1 is expressed in the basal layer of the inter-follicular epidermis
Although the molecular control of skin thickness has attracted little direct attention, most of the available evidence would suggest no role for hedgehog signaling in the growth or differentiation of the inter-follicular epidermis (McMahon et al., 2003). Since this prevailing view is largely based on data extracted from studies not directed specifically to skin development, we addressed the hypothesis that hedgehog signaling does play such a role. In fact, supporting this supposition, both Patched1 protein and transcription have been demonstrated in the basal layer of the skin (Adolphe et al., 2006; Nieuwenhuis et al., 2006). This observation stands in contrast to the results of in situ hybridization in the present (Fig. 8) and most other studies (Mill et al., 2003; St-Jacques et al., 1998). We sought additional evidence that this immediate target of Sonic hedgehog is normally expressed in the epidermis.
We studied the expression of Patched1 in the dorsal skin at E18.5 utilizing a mouse strain in which one Patched1 allele is inactivated and in which the Lac Z gene is present in the targeted allele (Goodrich et al., 1997). Thus, the detection of Lac Z expression in tissue of these mice provides a read-out of Patched1 expression (Goodrich et al., 1997). Using an immunohistologic method to detect Lac Z we demonstrate that Patched1 is expressed in the basal layer of skin as well as in the hair follicles (Fig. 10).
Fig. 10.
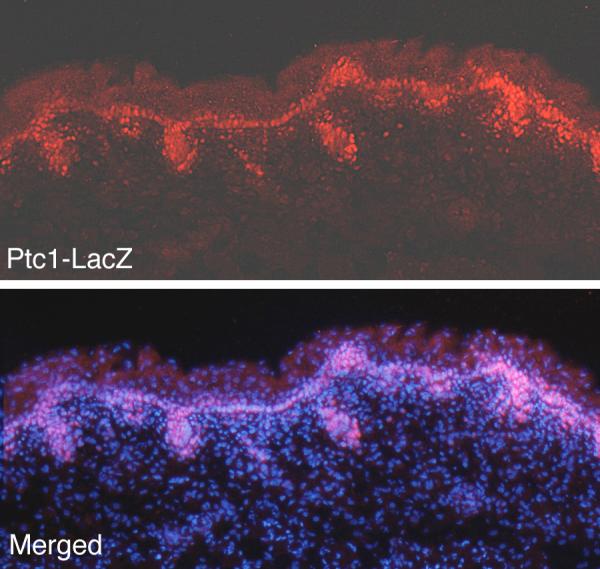
Patched1 is expressed in the basal layer of dorsal skin. Using a mouse strain that gives an accurate read-out of Patched1 expression (Goodrich et al., 1997), Patched1 is seen to be expressed in the basal cells of the inter-follicular epidermis as well as in hair follicles where it is detected by in situ hybridization in this and other studies.
The presence of Patched1 transcription and protein expression in the inter-follicular epithelium suggests a mechanism whereby Sonic hedgehog signaling could affect epidermal development. We sought more direct support for this hypothesis.
The skin of heterozygous Sonic hedgehog embryos is thinner than that of wild-type littermates
Although it has been reported that loss of both Sonic hedgehog alleles does not affect skin development (Chiang et al., 1999; St-Jacques et al., 1998), the thickness of the skin in the intact mutant animal has not been systematically studied. We compared skin thickness in heterozygous Sonic hedgehog mice and their wild-type littermates at E18.5 using the same method employed for assessing skin thickness in Runx2 null embryos and their wild-type controls . The results show that heterozygous Sonic hedgehog mice have a mean reduction of 10% in overall skin thickness and a 4% reduction in epidermal thickness when compared with wild-type littermate controls (Fig. 11).
Fig. 11.
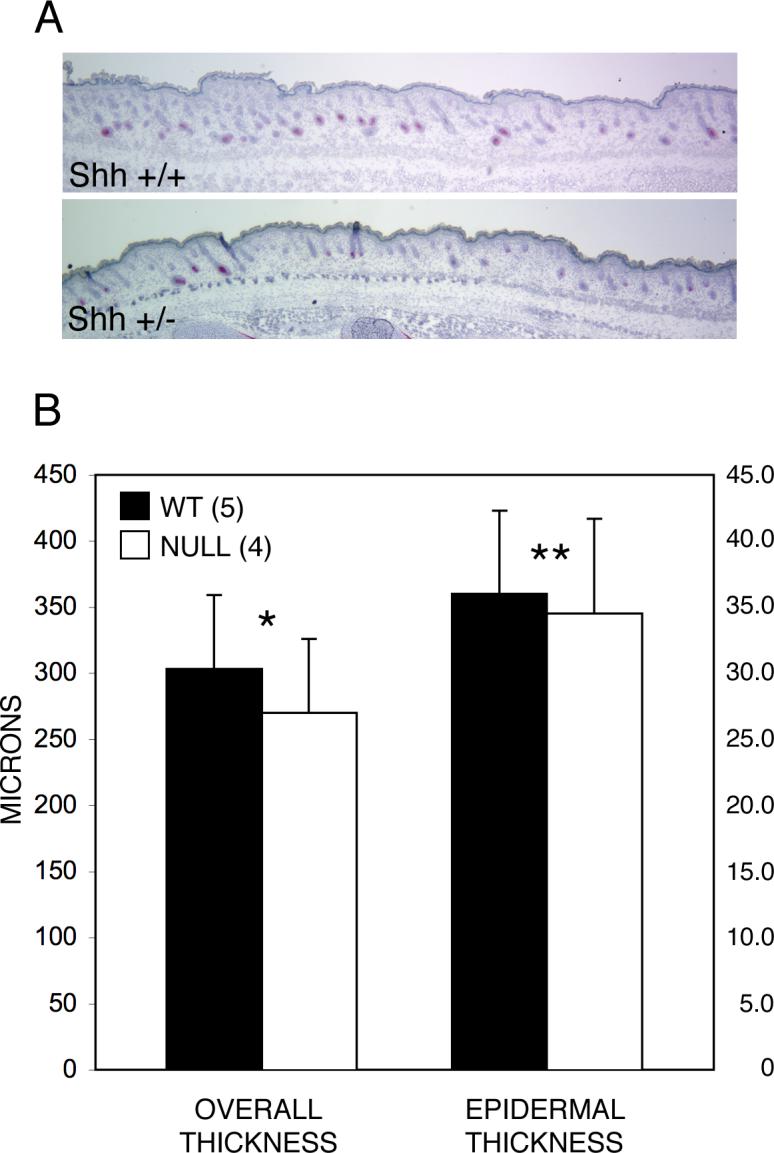
Skin thickness is reduced in the dorsal skin of heterozygous null Sonic hedgehog mice at E18.5. (A) Qualitative comparison of dorsal skin of wild-type and Sonic hedgehog +/− littermates. The difference in skin thickness between the WT and their Sonic hedgehog +/− littermates illustrated here is small but obvious. However, there is variation between animals of the same genotype and between levels of section in the same animal. As in the measurements of skin thickness in the Runx2 mutants it was therefore necessary to make a large number of measurements for each animal to determine if a real difference existed between wild-type and the Sonic hedgehog mutants. (B) Quantitative comparison shows that overall skin thickness in Sonic hedgehog +/− embryos is reduced by 10% while epidermal thickness is reduced by 4%. Note that the scales for overall and epidermal thickness differ by a factor of 10. The mean values, variation and significance are based on measurements at 11 randomly determined points on each of 4 mid-sagittal sections (total 44) for each animal. p<<0.0001, **p=0.02. Error bars indicate ±1 standard deviation.
DISCUSSION
In this study we show that Runx2 is expressed in the hair follicles of the skin in a dynamic pattern pari passu with development and cycling. Hair follicle development is delayed in the absence of Runx2. Although Runx2 expression is restricted to hair follicles, we find that the skin of null embryos is abnormally thin and that the inter-follicular epithelium is hypoplastic. It is well recognized that skin thickness dramatically increases as hair follicles develop, but the molecular mechanism for this phenomenon is not understood. The results of this study indicate that Runx2 is a component of a thickness-controlling pathway. We also show that lack of Runx2 results in impaired hedgehog signaling in the skin and demonstrate that Patched1, the immediate target of Sonic hedgehog, is transcribed in the epidermis. These findings together with our additional observation that the skin of heterozygous Sonic hedgehog deficient mice is thinner than in wild-type controls lend support to the hypothesis that Runx2 affects skin thickness in part through a hedgehog-mediated pathway.
Expression of Runx genes in the skin
Runx2 expression in the skin is restricted to the hair follicles in a dynamic pattern, progressively being expressed in the dermal papilla, the connective tissue sheath and the epithelium as follicles mature. Two other Runx genes, Runx1 and Runx3, are expressed in the skin (Raveh et al., 2005; Raveh et al., 2006). Runx3 is expressed primarily in dermal papilla cells and in the connective tissue sheath (Raveh et al., 2005), similar to our findings in the case of Runx2. In postnatal mice, isolated Runx3 expression is found in cells of the inter-follicular epidermis and in the distal outer root sheath. Postnatal mice also show staining of some cells in the bulb matrix that were identified as bulb melanocytes rather than epithelial cells. Runx1 is also expressed in dermal papillae and in connective tissue sheaths (Raveh et al., 2006). However, in contrast to Runx2 and Runx3, Runx1 expression is especially strong in the epithelial elements of hair follicles, including the upper matrix, the differentiating hair shaft and the infundibulum (Raveh et al., 2006).
In summary, all three genes are expressed in the dermal papilla and connective tissue sheaths. Runx1 expression seems to be especially strong in epithelium whereas the Runx3 expression pattern appears more like that of Runx2. However, unlike Runx3, Runx2 is expressed in the epithelium of later stage follicles as well as in the dermal papilla and connective tissue sheath.
Runx2 null skin phenotype
The grossly normal coat of hair found in the Runx2 null skin allografts and the plucking experiments confirm that Runx2 is not crucial for hair growth, the cycling of mature hair follicles or the ultimate development of normal appearing skin. However, Runx2 null mice have a significant delay in hair follicle development and a striking decrease in epidermal and overall skin thickness compared to wild-type controls. Thus, the impact of Runx2 deficiency on the skin phenotype is more severe than in the case of either Runx1 or Runx3 deficiency. In the studies of Runx3 and Runx1 deficient mouse skin changes in hair types or structure were described (Raveh et al., 2005; Raveh et al., 2006) but no impairment of hair follicle or skin development was reported.
Since there is co-expression of the three Runx genes in the skin, redundancy of their function conceivably could account for the relatively mild skin and hair phenotype of Runx2 deficiency. This could be especially relevant in the case of Runx3 that, like Runx2, is principally expressed in the dermal papilla and the connective tissue sheath. In fact, Runx3 was shown to be up-regulated in the upper but not the lower molar teeth of Runx2 null mice (Aberg et al., 2004a). Thus, it is possible that double null mice would show more extreme phenotypes than we observed in Runx2 mutants. However, the molar tooth phenotype exhibited by Runx2 null mice did not worsen in Runx2−3 double null animals (Wang et al., 2005). Moreover, the down-regulation of Sonic hedgehog and its mediators in the lower molar teeth of Runx2 null mice was not further reduced in the double null animals. In the end, the question of whether or not a more severe skin phenotype would be present in Runx double null mice must wait until this experiment has been performed.
The most prominent feature of the Runx2 null skin phenotype is the marked reduction in skin thickness. It is well known that rather spectacular changes in skin thickness take place pari passu with maturation and regression of hair follicles (Chase et al., 1953; Hansen et al., 1984; Paus et al., 1990). While the delay in hair follicle development in the Runx2 null mouse is significant, it is quantitatively small and, therefore, on a purely anatomic basis cannot account for the decrease in skin thickness. The decreased proliferation in the epithelial basal layer of the Runx2 null mice we show here may explain the thin inter-follicular epithelium: We postulate that fewer daughter cells are available to move up along the differentiation pathway and add to the thickness of the epithelium. Although a number of genes have been identified that affect epithelial proliferation and differentiation (Fuchs and Raghavan, 2002; Millar, 1997) the molecular signals coordinating the changes in skin thickness with follicle growth and regression are as yet undefined.
As pointed out by Hansen et al. (1984) data on skin thickness can be invalid if evaluated on routine, random microscopic sections because the skin is elastic, resulting in shrinkage when it is excised. These investigators pinned out excised specimens to their original size on a template before fixation in order to eliminate contracture or overstretching. In the present study, skin thickness was measured on whole embryo sections so that the skin was still attached, thus eliminating shrinkage or overstretching artifact. Moreover, measurements were made at a large number of randomly determined points to mitigate the effect of the undulations created by panniculus carnosus contracture during imbedding.
Decreased hedgehog signaling and its postulated impact
The similar restricted and asymmetrical expression patterns of Runx2 and Sonic hedgehog in the bulb epithelium of mature hair follicles led us to ask whether there is a functional link between the two. We were further encouraged to pursue this hypothesis because Runx2 is known to be a regulator of Indian hedgehog in bone (Kim et al., 1999; Wang et al., 2002; Yoshida et al., 2004) and of Sonic hedgehog in the developing tooth (Wang et al., 2005). Although we demonstrate a substantial reduction in hedgehog signaling in the skin of Runx2 null mice, the question must be raised as to the relevance of this finding to the observed hair and skin phenotypes.
Hedgehog signaling is essential for development of hair follicles: In the absence of Sonic hedgehog or of Gli2, hair follicle development, although initiated, is arrested at the peg stage (Chiang et al., 1999; Mill et al., 2003; St-Jacques et al., 1998; Wang et al., 2000). Since Sonic hedgehog is such a major driver of hair follicle maturation we believe that decreased hedgehog signaling may account for the modest delay in hair follicle development in Runx2 null mice. Although heterozygous Sonic hedgehog deficient mice show no obvious hair phenotype (Levy et al., 2005), only a systematic, careful study could reveal the modest, albeit significant, delay in hair follicle development shown in this study.
Could reduced hedgehog signaling also play a role in the genesis of the thin, hypoplastic skin phenotype of Runx2 null mice? It is well established that hedgehog signaling can be utilized not only in cell fate determination and patterning, but also in driving cell proliferation during development (Ingham and McMahon, 2001; McMahon et al., 2003). Sonic hedgehog induces proliferation in keratinocytes in cell culture (Fan and Khavari, 1999)(#51). Ectopic expression of Sonic hedgehog in the basal layer of the skin can result in epithelial hyperplasia, increased skin thickness and neoplasia (Ellis et al., 2003; Fan and Khavari, 1999; Fan et al., 1997; Oro et al., 1997). Mice lacking the repressor function of Patched1 in the skin basal layer also develop hyperplasia and neoplasia (Adolphe et al., 2006; Nieuwenhuis et al., 2007). In humans constitutive hedgehog signaling caused by inactivating mutations in Patched1 have been demonstrated in both hereditary and sporadic basal cell tumors (Johnson et al., 1996; Levanat et al., 1996). Activating Smoothened mutations have been found in sporadic human basal cell cancers (Xie et al., 1998).
Despite these natural and experimental models involving unleashed hedgehog signaling in skin and keratinocytes, most of the published evidence suggests no role for hedgehog signaling in the normal development of the inter-follicular epidermis despite its key role in hair follicle morphogenesis (McMahon et al., 2003). This prevailing opinion is based on the lack of expression of Sonic hedgehog and its mediators in the inter-follicular epithelium (Gat et al., 1998; Mill et al., 2003; Millar, 1997; Millar, 2002; St-Jacques et al., 1998); (Levy et al., 2005) and on observations of normal skin development when hedgehog signaling is ablated (Chiang et al., 1999; St-Jacques et al., 1998; Wang et al., 2000). Moreover, allografts of Sonic hedgehog null embryonic skin develop a thickened, hyperplastic epidermis that manifests increased basal cell proliferation, effects attributed to either juxtaposed abnormal hair follicles or to up-regulated Smoothened expression in the grafted skin (Chiang et al., 1999; St-Jacques et al., 1998). On the other hand K14-Cre-mediated ablation of Smoothened also results in thickened, hyperplastic inter-follicular epithelium (Gritli-Linde et al., 2007). Whether or not these somewhat contradictory findings elucidate normal development is conjectural. However, arguing against our hypothesis that lack of hedgehog signaling results in epidermal hypoplasia (and in apparent contrast to the implications of misexpression studies cited earlier) exposure of banked human epidermis to a hedgehog agonist (Paladini et al., 2005) or exposure of mouse skin to Sonic hedgehog protein via a viral vector (Sato et al., 1999) did not result in increased mitotic activity in the basal layer of the epidermis.
Our findings are not necessarily incompatible with much of the data just cited which parenthetically are culled from studies focused on hair follicle development. Although we and most other investigators do not find transcripts of Sonic hedgehog and its mediators in the inter-follicular epithelium by in situ hybridization, we do demonstrate Patched1 transcription therein using immuno-fluorescence and an anti-ß-galactosidase antibody (Fig. 9). This observation is concordant with the reported strong Patched1 protein expression in the basal layer of mouse skin using immuno-staining with a very specific antibody (Adolphe et al., 2006) and Patched1 transcription therein by in situ hybridization (Nieuwenhuis et al., 2006). In Patched2 null mice there is up-regulation of Patched1 and focal hyperproliferation in the skin basal cell layer of male mice implying that Patched2 may normally repress basal cell Patched1 expression therein (Nieuwenhuis et al., 2006). Ablation of Patched1 from the basal layer (Adolphe et al., 2006) or homozygosity for a C-terminal truncation of this gene (Nieuwenhuis et al., 2007) results in basal cell hyperplasia. Thus, there is abundant evidence that Patched 1 is normally expressed in basal cells and has an important function therein. We suggest that in addition to serving as a gatekeeper in epidermal basal cells Patched1 could also be a target of Sonic hedgehog protein.
The fact that heterozygous Sonic hedgehog deficient mice have thinner skin than their wild-type littermates adds strength to the argument that reduced hedgehog signaling is involved in the thin-skin phenotype of Runx2 null mice. At first glance this observation itself would appear to be at odds with previous observations that the skin of Sonic hedgehog null mice is not abnormal and, unlike hair follicles, inter-follicular epithelium does not require hedgehog signaling to undergo a complete differentiation program (Chiang et al., 1999; Mill et al., 2003; St-Jacques et al., 1998; Wang et al., 2000); (Mill et al., 2003). Runx2 null skin, although thin and hypoplastic, also undergoes a full differentiation program as is the case with heterozygous Sonic hedgehog skin. With regard to the thickness of Sonic hedgehog null skin, in view of the arrested hair follicle development in the absence of hedgehog signaling, it seems unlikely that the skin would be as thick as normal controls at a time point when the follicles are nearing full development, e.g. at E18.5 as in the current study. Moreover, actual measurements of epithelial or overall skin thickness were not reported in the studies of the skin phenotype of Sonic hedgehog null mice. Thus, the finding of normal skin development so far as thickness is concerned appears to be based on observations of random sections and thus subject to artifact as discussed earlier.
The abnormally thin skin of Runx2 null mice involves the dermis as well as the epidermis. At least one component of the dermis has been shown to be capable of responding to hedgehog signaling: Mill et al. (2003) showed that dermal fibroblasts harvested from wild-type skin respond to Sonic hedgehog signaling whereas those derived from Gli2 null mice (thus lacking the activating function of Gli2 in this pathway) did not. This implies that dermal fibroblasts may be a target of hedgehog signaling in vivo. There could be additional dermal components that respond to hedgehog signaling or to other cytokines that are regulated by Runx2.
In the current study there is a difference in the degrees of thinning observed in Runx2 null mice (22%) and in heterozygous Sonic hedgehog deficient animals (∼10%). Moreover, the Runx2 skin was found to have a 34% decrease in Sonic hedgehog mRNA whereas heterozygous Sonic hedgehog deficient animals would be presumed to have a 50% reduction in Sonic transcription. Therefore, it could be argued that the Sonic hedgehog heterozygously deficient skin should be even thinner than Runx2 null skin. This discrepancy in reduction of skin thickness could be explained in at least two ways. One possibility is that the actual reduction of Sonic hedgehog mRNA or protein in Sonic hedgehog heterozygous animals is less than 50%. The second and the more likely possibility is that the absence of Runx2 might well adversely impact the transcription of genes other than Sonic hedgehog that affect skin thickness. Further studies must take into account that both Runx2 (a transcription factor) and Sonic hedgehog (a soluble cytokine) are expressed only in the hair follicles and not in the skin per se.
In summary, the dynamic expression of Runx2 in hair follicles modulates their development and appears to play an important role in determining skin thickness during morphogenesis. Since Runx2, directly or indirectly, is a regulator of Sonic hedgehog in the skin this signaling pathway may be one link in the coupling of skin thickness to follicle development.
ACKNOWLEGEMENTS
We thank the following for their generous help during the course of this work: V. Botcharev, E. McGlinn, C. Tabin, K. Hu, Y. Li, B. Morgan, J. Zhang, S. Lyle, C. Adolphe, N. Fukai and members of the Olsen laboratory. We are also grateful to Yulia Pittel and Sofiya Plotkina for their expert secretarial and technical assistance respectively. The Nikon Imaging Center at Harvard Medical School and the help of Jennifer Waters and Lara Petrak are gratefully acknowledged. This work was supported by NIH grants AR36819 and AR 36820 (to B.R.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Appearance of Lac Z (Runx2 expression) in developing and cycling follicles. Note that Lac Z stain is not found in every follicle at a given stage and, when found, all the features characteristic of a stage are not necessarily found in a given follicle in which expression is found (see text). No Runx2 expression is found at early stage 2 but is seen at later stage 2. Increasing stain is found in the dermal papilla as the follicles mature and is seen progressively in the connective tissue sheath at the base of the follicle and along the sides as distally as the sebaceous gland. As the follicle matures dermal papilla expression is decreased. Epithelial expression is seen in the stage 6 and 8 developing follicles as well as in the fully mature cycling follicle shown here. As follicles mature the stain dots tend to be rather small as seen in the dermal papillae (solid arrow shafts) and the epithelium (dashed arrows) of the stage 8 and the anagen VI follicles shown. Strong Runx2 expression is seen in catagen. Note that the magnification varies between some of the follicles illustrated here. Arabic numerals refer to the stages of developing follicles. Telogen (Tel), Anagen (An), Catagen (Cat).
REFERENCES
- Aberg T, et al. Phenotypic changes in dentition of Runx2 homozygote-null mutant mice. J Histochem Cytochem. 2004a;52:131–9. doi: 10.1177/002215540405200113. [DOI] [PubMed] [Google Scholar]
- Aberg T, et al. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004b;270:76–93. doi: 10.1016/j.ydbio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Adolphe C, et al. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–8. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- Chase HB. Growth of the hair. Physiol Rev. 1954;34:113–26. doi: 10.1152/physrev.1954.34.1.113. [DOI] [PubMed] [Google Scholar]
- Chase HB, et al. Changes in the skin in relation to the hair growth cycle. Anat Rec. 1953;116:75–81. doi: 10.1002/ar.1091160107. [DOI] [PubMed] [Google Scholar]
- Chase HB, et al. Critical stages of hair development and pigmentation in the mouse. Physiol Zool. 1951;24:1–8. doi: 10.1086/physzool.24.1.30152098. [DOI] [PubMed] [Google Scholar]
- Chiang C, et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev Biol. 1999;205:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- D'Souza RN, et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–20. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- Ellis T, et al. Overexpression of Sonic Hedgehog suppresses embryonic hair follicle morphogenesis. Dev Biol. 2003;263:203–15. doi: 10.1016/s0012-1606(03)00394-4. [DOI] [PubMed] [Google Scholar]
- Fan H, Khavari PA. Sonic hedgehog opposes epithelial cell cycle arrest. J Cell Biol. 1999;147:71–6. doi: 10.1083/jcb.147.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, et al. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic Hedgehog. Nat Med. 1997;3:788–92. doi: 10.1038/nm0797-788. [DOI] [PubMed] [Google Scholar]
- Farago AF, et al. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–18. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Fiedler V. C. a. H. A. Diffuse Alopecia: Telogen Hair Loss. In: Olsen EA, editor. Disorders of Hair Growth: Diagnosis and Treatment. McGraw-Hill, Inc; New York: 1994. pp. 241–255. [Google Scholar]
- Fuchs E, et al. At the roots of a never-ending cycle. Dev Cell. 2001;1:13–25. doi: 10.1016/s1534-5807(01)00022-3. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nature reviews. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- Gat U, et al. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–14. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, et al. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, et al. Abnormal hair development and apparent follicular transformation to mammary gland in the absence of hedgehog signaling. Dev Cell. 2007;12:99–112. doi: 10.1016/j.devcel.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handjiski BK, et al. Alkaline phosphatase activity and localization during the murine hair cycle. Br J Dermatol. 1994;131:303–10. doi: 10.1111/j.1365-2133.1994.tb08515.x. [DOI] [PubMed] [Google Scholar]
- Hansen LS, et al. The influence of the hair cycle on the thickness of mouse skin. Anat Rec. 1984;210:569–73. doi: 10.1002/ar.1092100404. [DOI] [PubMed] [Google Scholar]
- Hanson J. The histogenesis of the epidermis in the rat and mouse. Journal of anatomy. 1947;81:174–97. [PMC free article] [PubMed] [Google Scholar]
- Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–28. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Huang LF, et al. Mouse clavicular development: analysis of wild-type and cleidocranial dysplasia mutant mice. Dev Dyn. 1997;210:33–40. doi: 10.1002/(SICI)1097-0177(199709)210:1<33::AID-AJA4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Inada M, et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–90. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Kim IS, et al. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159–70. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Laird PW, et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker RM, et al. Hair follicle stem cells: their location, role in hair cycle, and involvement in skin tumor formation. J Invest Dermatol. 1993;101:16S–26S. doi: 10.1111/1523-1747.ep12362556. [DOI] [PubMed] [Google Scholar]
- Levanat S, et al. A two-hit model for developmental defects in Gorlin syndrome. Nat Genet. 1996;12:85–7. doi: 10.1038/ng0196-85. [DOI] [PubMed] [Google Scholar]
- Levy V, et al. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell. 2005;9:855–61. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Link RE, et al. Epithelial growth by rat vibrissae follicles in vitro requires mesenchymal contact via native extracellular matrix. J Invest Dermatol. 1990;95:202–7. doi: 10.1111/1523-1747.ep12478002. [DOI] [PubMed] [Google Scholar]
- McMahon AP, et al. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Mill P, et al. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–94. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar S. The Role of Patterning Genes in Epidermal Differentiation. In: P. a. K. Cowin MW, editor. Cytoskeletal-Membrane Interactions and Signal Transduction. Landes Bioscience; Austin, Texas: 1997. [Google Scholar]
- Millar SE. Molecular mechanisms regulating hair follicle development. J Invest Dermatol. 2002;118:216–25. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–9. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, et al. Epidermal hyperplasia and expansion of the interfollicular stem cell compartment in mutant mice with a C-terminal truncation of Patched1. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, et al. Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Molecular and cellular biology. 2006;26:6609–22. doi: 10.1128/MCB.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro AE, et al. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276:817–21. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Paladini RD, et al. Modulation of hair growth with small molecule agonists of the hedgehog signaling pathway. J Invest Dermatol. 2005;125:638–46. doi: 10.1111/j.0022-202X.2005.23867.x. [DOI] [PubMed] [Google Scholar]
- Panteleyev AA, et al. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am J Pathol. 1999;155:159–71. doi: 10.1016/S0002-9440(10)65110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Cotsarelis G. The biology of hair follicles. The New England journal of medicine. 1999;341:491–7. doi: 10.1056/NEJM199908123410706. [DOI] [PubMed] [Google Scholar]
- Paus R, et al. Expression of classical and non-classical MHC class I antigens in murine hair follicles. Br J Dermatol. 1994;131:177–83. doi: 10.1111/j.1365-2133.1994.tb08488.x. [DOI] [PubMed] [Google Scholar]
- Paus R, et al. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J Invest Dermatol. 1999;113:523–32. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- Paus R, et al. Telogen skin contains an inhibitor of hair growth. Br J Dermatol. 1990;122:777–84. doi: 10.1111/j.1365-2133.1990.tb06266.x. [DOI] [PubMed] [Google Scholar]
- Raveh E, et al. Runx3 is involved in hair shape determination. Dev Dyn. 2005;233:1478–87. doi: 10.1002/dvdy.20453. [DOI] [PubMed] [Google Scholar]
- Raveh E, et al. Dynamic expression of Runx1 in skin affects hair structure. Mech Dev. 2006 doi: 10.1016/j.mod.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Sato N, et al. Induction of the hair growth phase in postnatal mice by localized transient expression of Sonic hedgehog. J Clin Invest. 1999;104:855–64. doi: 10.1172/JCI7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, et al. Differential expression and activity of melanogenesis-related proteins during induced hair growth in mice. J Invest Dermatol. 1991;96:172–9. doi: 10.1111/1523-1747.ep12460956. [DOI] [PubMed] [Google Scholar]
- Slominski A, et al. Melanogenesis during the anagen-catagen-telogen transformation of the murine hair cycle. J Invest Dermatol. 1994;102:862–9. doi: 10.1111/1523-1747.ep12382606. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–68. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Stenn KS, Paus R. Controls of hair follicle cycling. Physiol Rev. 2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- Sundberg J. P. a. H., M. E. Hair Types and Subtypes in the Laboratory Mouse. In: Sundberg JP, editor. Handbook of Mouse Mutations with Skin and Hair Abnormalities: Animal Models and Biomedical Tools. CRC Press; Ann Arbor: 1994. pp. 57–68. [Google Scholar]
- Van Exan RJ, Hardy MH. The differentiation of the dermis in the laboratory mouse. The American journal of anatomy. 1984;169:149–64. doi: 10.1002/aja.1001690204. [DOI] [PubMed] [Google Scholar]
- Wang LC, et al. Conditional disruption of hedgehog signaling pathway defines its critical role in hair development and regeneration. J Invest Dermatol. 2000;114:901–8. doi: 10.1046/j.1523-1747.2000.00951.x. [DOI] [PubMed] [Google Scholar]
- Wang WF, et al. Growth defect in Grg5 null mice is associated with reduced Ihh signaling in growth plates. Dev Dyn. 2002;224:79–89. doi: 10.1002/dvdy.10089. [DOI] [PubMed] [Google Scholar]
- Wang XP, et al. Runx2 (Cbfa1) inhibits Shh signaling in the lower but not upper molars of mouse embryos and prevents the budding of putative successional teeth. J Dent Res. 2005;84:138–43. doi: 10.1177/154405910508400206. [DOI] [PubMed] [Google Scholar]
- Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90– 2. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuspa SH, et al. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J Cell Biol. 1989;109:1207–17. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, et al. Tissue specific regulation of VEGF expression during bone development requires Cbfa1/Runx2. Mech Dev. 2001;106:97–106. doi: 10.1016/s0925-4773(01)00428-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appearance of Lac Z (Runx2 expression) in developing and cycling follicles. Note that Lac Z stain is not found in every follicle at a given stage and, when found, all the features characteristic of a stage are not necessarily found in a given follicle in which expression is found (see text). No Runx2 expression is found at early stage 2 but is seen at later stage 2. Increasing stain is found in the dermal papilla as the follicles mature and is seen progressively in the connective tissue sheath at the base of the follicle and along the sides as distally as the sebaceous gland. As the follicle matures dermal papilla expression is decreased. Epithelial expression is seen in the stage 6 and 8 developing follicles as well as in the fully mature cycling follicle shown here. As follicles mature the stain dots tend to be rather small as seen in the dermal papillae (solid arrow shafts) and the epithelium (dashed arrows) of the stage 8 and the anagen VI follicles shown. Strong Runx2 expression is seen in catagen. Note that the magnification varies between some of the follicles illustrated here. Arabic numerals refer to the stages of developing follicles. Telogen (Tel), Anagen (An), Catagen (Cat).