Abstract
Undecaprenyl pyrophosphate synthase (UPPs) catalyzes eight consecutive condensation reactions of farnesyl pyrophosphate (FPP) with isopentenyl pyrophosphate (IPP) to form a 55-carbon long-chain product. We previously reported the crystal structure of the apo-enzyme from Escherichia coli and the structure of UPPs in complex with sulfate ions (resembling pyrophosphate of substrate), Mg2+, and two Triton molecules (product-like). In the present study, FPP substrate was soaked into the UPPs crystals, and the complex structure was solved. Based on the crystal structure, the pyrophosphate head group of FPP is bound to the backbone NHs of Gly29 and Arg30 as well as the side chains of Asn28, Arg30, and Arg39 through hydrogen bonds. His43 is close to the C2 carbon of FPP and may stabilize the farnesyl cation intermediate during catalysis. The hydrocarbon moiety of FPP is bound with hydrophobic amino acids including Leu85, Leu88, and Phe89, located on the α3 helix. The binding mode of FPP in cis-type UPPs is apparently different from that of trans-type and many other prenyltransferases which utilize Asprich motifs for substrate binding via Mg2+. The new structure provides a plausible mechanism for the catalysis of UPPs.
Keywords: prenyltransferase, farnesyl pyrophosphate, isopentenyl pyrophosphate, crystal structure, substrate binding, metal ion
Undecaprenyl pyrophosphate synthase (UPPs) is a cis-prenyltransferase which catalyzes eight consecutive condensation reactions of isopentenyl pyrophosphate (IPP) with far-nesyl pyrophosphate (FPP) to generate C55 undecaprenyl pyrophosphate (UPP; Allen 1985; A.P.-C. Chen et al. 2002; Liang et al. 2002). Its biological function is to synthesize UPP as a lipid carrier for bacterial cell wall peptidoglycan assembly (Robyt-1998). UPPs along with other cis-prenyltransferases catalyze cis-double bond formation during IPP condensation but synthesize different chain-length products (Ogura and Koyama 1998). Dehydrodolichyl pyrophos-phate synthases for glycoprotein lipid carrier biosynthesis generate products with chain lengths ranging from C55 to C120 (Sato et al. 1999; Chang et al. 2001). Rubber prenyl-transferase synthesizes a huge polymer containing thousands of IPP units (Cornish 2001). In contrast, trans-prenyltransferases, which generate trans-double bonds in IPP condensation, synthesize shorter chain-length products, C15–C50 (Chen et al. 1994; Wang and Ohuma 2000).
The most striking difference between cis- and trans-prenyltransferases is that trans-type enzymes use two conserved aspartate-rich DDXXD motifs for binding with both FPP and IPP substrates via Mg2+ (Chen et al. 1994), whereas cis-type enzymes have no DDXXD motif in their amino acid sequences (Shimizu et al. 1998; Apfel et al. 1999). The crystal structures of both trans-type farnesyl pyrophosphate synthase (FPPs), which catalyzes the condensation between C10 geranyl pyrophosphate (GPP) and an IPP, and cis-type UPPs have been solved, providing the structural basis for comparison of the reaction mechanisms catalyzed by two groups of prenyltransferases (Tarshis et al. 1994; Fujihashi et al. 2001; Ko et al. 2001). In the FPPs crystal structure, the pyrophosphate head group of GPP is bound with Mg2+ chelated by the first DDXXD motif (Tarshis et al. 1994). The pyrophosphate of the other substrate IPP is bound with Mg2+ coordinated by the second DDXXD. Site-directed mutagenesis studies on FPPs demonstrated the essential roles of the Asp amino acids in the two DDXXD motifs for substrate binding and catalysis (Marrero et al. 1992; Joly and Edwards 1993; Koyama et al. 1994, 1995, 1996; Song and Poulter 1994). We recently solved the 3-D structure of longer-chain C40 octaprenyl pyrophosphate synthase of trans-type, and its FPP and IPP substrates were proposed to bind to the first and the second conserved DDXXD motif mediated by Mg2+, respectively (Guo et al. 2004).
The apo-UPPs shows a completely different active-site structure from those of the trans-prenyltransferases. A structural P-loop (G30—R33) of the UPPs from Micrococcus luteus BP-26, which appears in the pyrophosphate binding proteins such as nucleotide triphosphate hydrolase, phos-phofructokinase, c-AMP binding domain, and sugar phosphatase (Kinoshita et al. 1999) was proposed to be responsible for FPP binding (Fujihashi et al. 2001). The Asp29 (equivalent to Asp26 in E. coli UPPs) nearby was speculated to bind Mg2+ for the association of FPP with the enzyme, just like the Asp in DDXXD of trans-prenyltransferases (Fujihashi et al. 2001). However, in the structure of E. coli UPPs in complex with the sulfate ions S1 and S2 (pyrophosphate analog), Mg2+, and two Triton molecules (product-like), we found only a single Mg2+ coordinated by Glu213, His199, and waters in each subunit (Chang et al. 2003a). This Mg2+ is near the proposed IPP site (S2) rather than the putative FPP binding site (S1), and possibly is responsible for maintaining a proper protein conformation for IPP binding and the enzyme reaction. In addition, Arg194 and Arg200 likely provide the positively charged side chains for binding with IPP. On the other hand, FPP may be held in place by the enzyme through the side chains of Arg30 and Arg39. By solving the complex structure of UPPs with FPP, we can clearly see the amino acids which have direct contacts with FPP in the pyrophosphate head group and the hydrocarbon tail. No metal was required for FPP binding, consistent with previous observations (Y.H. Chen et al. 2002). Finally, we propose a plausible catalytic mechanism for E. coli UPPs based on the new structural information together with previous results.
Results
Overall structure
The refined structure of UPPs complexed with FPP shown in Figure 1A,B ▶ can be well superimposed with the closed and open forms of previously solved structure of UPPs with sulfates, Mg2+ and Tritons (Chang et al. 2003a), respectively, except that residues 83–86 in the α3 helix of monomer A extended the α3 helix by more than one turn at the N terminus. Like the reported structures (Ko et al. 2001; Chang et al. 2003a), the present complex structure reveals a parallel α/β topology for the UPPs catalytic domain which contains a total of six β-strands and seven α-helices. The six β-strands are arranged in a βC-βD-βB-βA-βE-βF topology, and all helices are mostly straight except α3, which is kinked.
Figure 1.
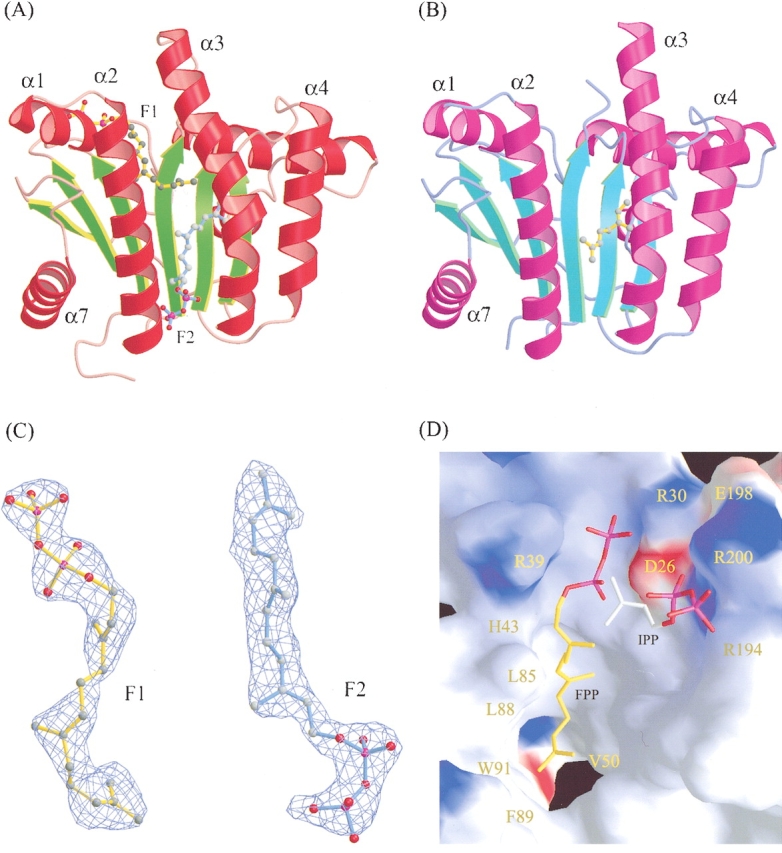
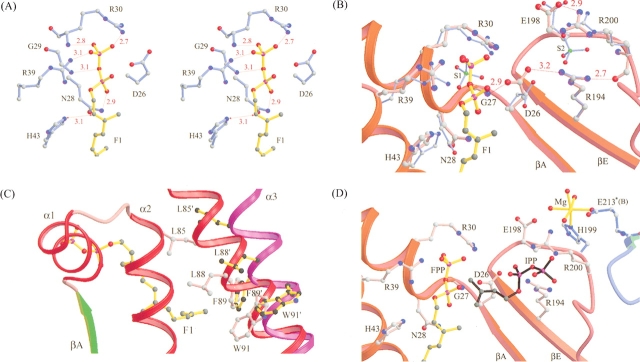
Overall structure of UPPs complexed with FPP, and the molecular structure of FPP (F1 and F2) and the amino acids surrounding FPP (F1) and IPP (proposed) in the active site. Monomer A is colored red and green for α-helices and β-strands, respectively (A); monomer B is magenta and cyan (B). Two FPP (F1 and F2) molecules (yellow and cyan) are bound in monomer A (closed form), but only the geranyl part of F2 (yellow) was detected in monomer B (open form). The α3 helix in monomer A with F1 bound is kinked toward α2 to form a narrower active site (closed form) for better enzyme-substrate interaction. (C) The molecular structure of the two FPP molecules in the UPPs active-site tunnel of monomer A, F1 on the top and F2 on the bottom. The model is superimposed on the final 2FO-FC map contoured at the 1.0 σ level. Densities in the initial Fourier maps were comparable to the clear densities shown here, especially for the carbons of hydrocarbon tail. (D) The overall active-site structure represented by solid surface-charged potential. The surface is colored from red to blue according to charge potential from -15 to 15 kBT, where kB is the Boltzmann constant (1.381 × 10-23 J•k-1) and T is Kelvin temperature. The FPP molecule (F1) and a hypothetical IPP binding model in the active site are denoted by a stick in yellow and white, respectively. Amino acids surrounding the active-site pocket and hydrophobic tunnel are labeled.
Two protein conformations, named closed and open forms, appear in the two monomers (Figure 1A,B ▶, respectively, and the α3 is more kinked in the closed conformer (Fig. 1A ▶). The monomer A containing two molecules of FPP (F1 on the top and F2 on the bottom of the tunnel) adopts a closed conformation, whereas the other monomer (monomer B) with a single FPP (F2) bound on the bottom (only the electron density of the geranyl fragment of the FPP could be detected) adopts an open conformation. Unlike the two FPP molecules, two larger Triton molecules together in the active site make the tunnel crowded, thereby forcing it into open form (Chang et al. 2003a). In contrast, a single Triton occupying at the position similar to that of F1 renders UPPs to form a narrower active site (closed conformation) for better UPPs-ligand interactions, in which α3 moves closer to α2. Moreover, pyrophosphate of F1 is located coincident with the active-site sulfate ion (S1) in the UPPs structure (Chang et al. 2003a). This FPP (F1) can be thus regarded as the reactive substrate. The additional FPP (F2) bound to the lower portion of the tunnel is through nonspecific hydrophobic interaction, and its pyrophosphate head group is located outside of the tunnel, unlikely to react with IPP. In this paper, we focus only on the interactions of FPP (F1) with the nearby amino acids.
Structures of bound FPP and UPPs
The electron densities assigned to the two FPP molecules in monomer A clearly indicate that the double bonds of both FPP molecules are in trans configuration ( Fig. 1C ▶). As shown in Figure 1D ▶, the FPP (F1) sits in an active-site pocket with Arg30 and Arg39 near its pyrophosphate head group, His43 is close to C2, and several hydrophobic residues, Leu85, Leu88, Phe89, and Val50 surround its hydrocarbon moiety. Based on the position of the sulfate ion (S2) in the active site (Chang et al. 2003a), we modeled in an IPP beside FPP, which is bound with Arg200 and Arg194 (Fig. 1D ▶). A closer look into the FPP site reveals that the oxyanions on the pyrophosphate of the FPP molecule form hydrogen bonds with the backbone NH atoms of Gly29 and Arg30 as well as the side chains of Asn28, Arg30, and Arg39 (Fig. 2A ▶). The interactions between the side chains and FPP lie in the range of 3.2–4.2 Å, as shown in the Supplemental Material (Table S1).
Figure 2.
Interactions between FPP pyrophosphate head group and the nearby amino acids in the active side. In A, the pyrophosphate of FPP (F1) is hydrogen-bound to the backbone NH and side chain of R30, and backbone NH of G29 as well as the side chains of R39 and N28. Oxygen and nitrogen atoms are shown as red and blue dots, respectively. All of the distances of possible hydrogen bonds are indicated in Å, shown with red dotted lines. In B, side chain of D26 forms hydrogen bonds with the backbone NH of Gly27 and the side chain of R194 with the distances indicated in Å. Moreover, R194 interacts with R200, which is hydrogen-bound to E198. The segments of UPPs containing residues 23–43 and 192–205 are represented by the red ribbon. (C) Superimposition of the βA strand and three α-helices (α1, α2, and α3) in the active-site area from the closed and open conformations of UPPs. The α2 helix is shown in red in the closed conformer and purple in the open conformer. Several amino acids including L85, L88, F89, and W91 on this helix become closer to the bound FPP compared to their positions (L85′, L88′, F89′, and W91′) in the open form. In D, the Mg2+ (shown in yellow) near IPP is coordinated with H199 from A subunit, E213 from B subunit, and four waters. The hydrocarbon parts of FPP and hypothetical IPP are represented by ball-and-stick in yellow and black, respectively. The oxygen and phosphate atoms in the pyrophosphate moiety are shown in red and purple, respectively. The segments of A subunit of UPPs 23–43 and 192–205 are represented by red ribbon, and segments of B subunit of apo-UPPs 210–215 by cyan ribbon.
The catalytic roles of Asp26, Arg194, and Arg200
It was first suggested that Asp29 in M. luteus UPPs (equivalent to Asp26 in E. coli UPPs) in the neighborhood of FPP may coordinate with Mg2+ for binding with the pyrophosphate group of FPP (Fujihashi et al. 2001). However, based on our study of E. coli UPPs, Asp26 may be responsible for UPPs catalysis but not for FPP binding, because no metal was observed to be associated with it (Chang et al. 2003a). Moreover, the D26A mutation of E. coli UPPs did not change the FPP Km value but resulted in a 103-fold smaller kcat value (Pan et al. 2000a). The proposed model for E. coli UPPs catalysis is that Asp26 likely acts as a general base to remove a proton from IPP for its nucleophilic attack on FPP (Chang et al. 2003a). In the present structure of UPP in complex with FPP, Asp26 is positioned correctly in the active site by several hydrogen bonds contributed from Gly27 and Arg194 (Fig. 2B ▶) with its carboxylate group pointing to the S2 site (IPP site), whereas FPP binds at the S1 site. The side-chain NH of Arg194 forms a hydrogen bond with the backbone oxygen atom of Arg200, which in turn forms a 2.9 Å hydrogen bond with the side-chain oxygen OE2 atom of Glu198. In light of the structure (Fig. 2B ▶), Arg200 and Glu198 are in close proximity of IPP (S2) and likely contribute to its binding.
FPP binding does not require Mg2+
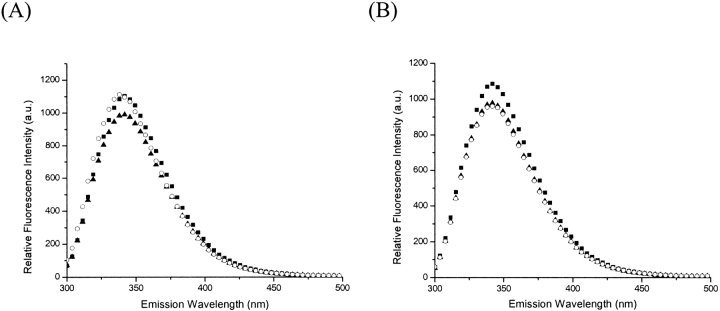
To confirm that FPP binding does not require Mg2+, we added excess EDTA (1 mM) into E. coli UPP solution (1 μM) to remove the metal from the enzyme by dialysis. After dialysis, only 1% residual activity of UPPs was detected. By adding 15 μM FPP into the enzyme solution, the intrinsic fluorescence was quenched in the presence (Fig. 3A ▶) and the absence of 5 mM Mg2+ (Fig. 3B ▶), indicating that FPP binding does not require Mg2+. This quenching of intrinsic fluorescence is due to the reduction of Trp91 fluorescence, which is located in α3 helix and moves during the conformational change induced by FPP binding (Y.H. Chen et al. 2002). Unlike the result obtained in the presence of Mg2+ (Fig. 3A ▶), the addition of IPP into the UPPs-FPP mixture without Mg2+ failed to change the fluorescence intensity (Fig. 3B ▶), suggesting that IPP binding absolutely requires Mg2+. This fluorescence experiment was performed in a manner similar to that used in the previous study (Y.H. Chen et al. 2002), except that the UPPs used here was dialyzed into the EDTA-containing buffer to remove any tightly bound metal ion (as indicated by the 1% residual activity) prior to the measurements of fluorescence spectra.
Figure 3.
The fluorescence spectra of EDTA-treated and nontreated UPPs, the sample with FPP, and with further addition of IPP in the presence and absence of Mg2+. The UPPs solution containing 1 μM enzyme was added with 1 mM EDTA and dialyzed twice overnight to remove the metal from UPPs. The activity of EDTA-treated was reduced to 1% after dialysis. The fluorescence emission spectrum of the EDTA-treated (B) and nontreated UPPs (A) was measured (▪). The samples were then added with 15 μM FPP and 5 mM MgCl2 (A) and only FPP (B), and the fluorescence spectra were collected (▴). The mixtures were further added with 40 μM IPP (A,B), and the spectra were taken (○). The addition of FPP changed the fluorescence of UPPs in the presence and absence of Mg2+, whereas the further addition of IPP failed to alter the intrinsic fluorescence of the EDTA-treated UPPs without Mg2+. These results indicate that the binding of FPP does not require Mg2+, but IPP binding needs the metal ion.
Amino acids surrounding the hydrocarbon moiety of FPP
Interactions between the 15-carbon hydrocarbon tail of the FPP (F1) molecule and the active-site residues of UPPs are mostly hydrophobic. The side chains of Leu85, Met25, Leu88, Ala47, Val50, Ile141, Ala69, Ala92, and Phe89 of UPPs are involved in the hydrophobic interactions, as summarized in Table S1. Three of these residues, Leu85, Leu88, and Phe89, are located in the α3 helix that reorients during protein conformational change. This helix represents a key determinant in regulating UPPs activity (Chang et al. 2003b). It moves toward the α2 helix to form the closed conformation when FPP is bound to the active site, and this closed form is active (Y.H. Chen et al. 2002; Chang et al. 2003b). In this active closed form, the side chains of Leu85, Leu88, and Phe89 on the α3 helix (shown as a red ribbon in Fig. 2C ▶) face and are closer to the bound FPP. In contrast, these residues (L85′, L88′, F89′, and W91′) are away and more distant from the position of FPP in the open conformer (α3 at the position of the purple ribbon in Fig. 2C ▶), and the open form has a much lower activity (Chang et al. 2003b). This suggests that Leu85, Leu88, and Phe89 directly participate in the catalysis by interacting with FPP.
Discussion
FPP serves as a common substrate for many prenyltransferases. In nature, there are several classes of prenyltransferases, including: (1) prenyltransferases such as UPPs (cis-type) and FPPs (trans-type) which add IPP molecules into allylic substrate, (2) prenyltransferases catalyzing the cyclization of FPP itself, (3) protein farnesyltransferases (FTase) that catalyze the transfer of FPP to a specific Cys residue of a protein substrate through a thioether linkage, and (4) aromatic prenyltransferases catalyzing the formation of a carbon-carbon bond between a prenyl group and an aromatic nucleus, which are involved in the biosynthesis of ubiquinones, menaquinones, tocopherols, and plastoquinones (Liang et al. 2002). As demonstrated in the present study, the backbone NHs and the side chains of the Asn28, Gly29, Arg30, and Arg39 of UPPs provide hydrogen bonding and electrostatic interactions to the pyrophosphate of FPP, making it a good leaving group. In contrast, the negatively charged FPP pyrophosphate is bound to the DDXXD-coordinated Mg2+ in the trans-prenyltransferases (Marrero et al. 1992; Song and Poulter 1994). In pentalenene synthase (an FPP cyclase), the top of the active-site pocket is surrounded by several hydrophilic amino acids, particularly Asp80 and Asp84, which provide carboxylate groups to complex with Mg2+ for FPP binding (Lesburg et al. 1997). Moreover, all of the aromatic prenyltransferases include the prenyl diphosphate binding site (N/D)DXXD similar to that of the trans-prenyltransferases (Suvarna et al. 1998; Collakova and DellaPenna 2001; Meganathan 2001; Schledz et al. 2001). The X-ray structure of rat FTase illustrates that the diphosphate moiety of the FPP molecule is hydrogen bonded with His248β, Arg291β, and Tyr300β (Park et al. 1997; Long et al. 1998). The catalytic zinc ion which activates the thiolate anion as a nucleophile to attack the peptide substrate is coordinated by Asp297b, Cys299b, and His362b.
In light of the findings of the present study, UPPs represents a unique enzyme, because all of the above-mentioned prenyltransferases require Mg2+ for binding with the pyrophosphate of FPP to increase its ability as a leaving group or facilitate cyclization of farnesyl cation (Lesburg et al. 1997; Park et al. 1997; Long et al. 1998). Even for Ras FTase, it has been shown that a Zn2+ is catalytically important to activate the thiolate anion but a Mg2+ chelating with pyrophosphate of FPP increases the FPP transfer activity by 100-fold (Saderholm et al. 2000). In contrast, no metal was detected in the FPP site of the crystal structure of UPPs in complex with FPP. Indeed, UPPs lacks the conserved DDXXD motifs as seen in FPPs and other trans-type pren-yltransferases for binding with substrates via Mg2+. Although an Asp26 is in the active site, it is not likely to bind the pyrophosphate of FPP based on the crystal structure. Moreover, in the presence of EDTA, the FPP can still quench the UPPs intrinsic fluorescence of Trp91, indicating that the binding of FPP does not require Mg2+. On the other hand, IPP binding as well as the subsequent condensation reaction require Mg2+ (Y.H. Chen et al. 2002).
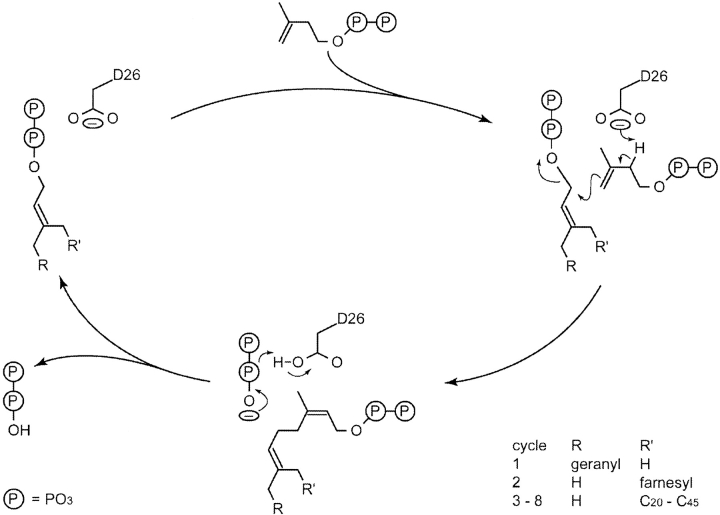
The amino acids essential for substrate binding and the enzyme reaction are summarized in Figure 2D ▶. A Mg2+ ion coordinated by His199 (subunit A) and Glu213 (subunit B) is near but not in direct contact with IPP in its putative binding site (S2), which may play a structural rather than a catalytic role. The plausible reaction mechanism of UPPs deduced from the present crystallographic studies is outlined in Figure 4 ▶. Asp26 may serve as a general base that removes a proton from IPP and later transfers the proton to the pyrophosphate group of FPP. The importance of Asp26 in catalysis was shown by the 1000-times reduction in the kcat value of D26A mutant (Pan et al. 2000a). In this model (Fig. 1D ▶), Asp26 is near the proton at the C2 carbon of IPP and ready to remove it. The remaining electrons after de-protonation shift to form a cis-double bond, and the carban-ion at C4 of IPP attacks the C1 carbocation of FPP to form a condensation product.
Figure 4.
The proposed reaction mechanism of UPPs. Based on the present crystal structure, Asp26 serves as a general base to subtract a proton from IPP. This essential active-site amino acid is near the proton at C2 carbon of IPP and ready to remove it. The remaining electrons after deprotonation shift to form a cis-double bond, and the carbanion intermediate attacks the C1 carbocation of FPP to form a condensation product. Eight cycles total of IPP condensation generate the UPP product.
On the other hand, His43 is closed to C2-H of the substrate FPP (Fig. 2 ▶). This amino acid probably offers a lone pair of electrons to interact with the C2-H, and thus partially stabilizes the carbocation formed after elimination of the pyrophosphate group. This partially positively charged character on C2 of the allylic substrate was also found in FPPs, as the fluoro-substituted GPP analog was shown to be a poor substrate for FPPs due to fluorine’s strong electro-negativity to destabilize the intermediate (Poulter et al. 1977). The mutation of H43A causes the decrease of kcat value by 1000-fold, suggesting its importance in catalysis (Chang et al. 2003a).
Leu85, Leu88, and Phe89 in the active site of UPPs ensure the correct position and orientation of the allylic substrate FPP for an efficient nucleophilic attack by IPP. It is notable that these three amino acids located on the α3 helix interact with the substrate only when FPP is bound and the closed conformation is formed. It was shown that the flexible loop (amino acids 72–83) preceding the α3 helix may pull the helix closer to the bound FPP substrate to form a closed conformation. By inserting extra Ala residues to freeze the UPPs structure in the open conformation, we demonstrated that the closed form of UPPs is active and the open form is 104-fold less active (Chang et al. 2003b). As shown in the present study, the amino acids such as Leu85, Leu88, and Phe89 in the α3 helix are near the bound FPP in the closed conformation.
Materials and methods
Materials
The following materials were obtained from the respective suppliers: Radiolabeled [14C]IPP (55 mCi/mmole), Amersham Pharmacia Biotech; FPP, Sigma; reversed-phase thin layer chromatography (TLC) plates, Merck; Taq DNA polymerase, Life Technologies; the plasmid mini-prep kit, DNA gel extraction kit, and NiNTA resin, QIAGEN; potato acid phosphatase (2 units/mg), Roche Molecular Biochemicals; FXa and the protein expression kit (including the pET32Xa/LIC vector and competent JM109 and BL21 cells), Novagen. All commercial buffers and reagents were of the highest grade.
Crystallization of UPPs with FPP
We first grew crystals of the E. coli UPPs and later soaked them with FPP. For purification of UPPs, our previously reported protocol of using NiNTA column chromatography was followed (Pan et al. 2000b). The purified UPPs was crystallized using the hanging drop set-up purchased from Hampton Research. The crystallization conditions we used previously were under the high concentration of sulfate or phosphate ion in the mother liquid which prevented FPP and/or IPP from binding to UPPs. Moreover, high concentrations (>1%) of Triton X-100 caused the detergent to be bound in the active-site tunnel, which also interferes with the binding of FPP to UPPs. We therefore used a negatively charged peptide (an UPPs inhibitor, S.Y. Chang, Y.S. Sun, H.M. Yu, A.H.-J. Wang, and P.H. Liang, unpubl.) to facilitate the crystallization of UPPs under both low salt and low Triton X-100. In the end, 2 μL mother liquid containing 25% ethylene glycol at pH 7.5 was mixed with 2 μL protein solution of 10 mg/mL UPPs, 1 mM peptide, 0.05% Triton X-100, and 0.5 mM MgCl2. The mixture was equilibrated against 200 μL of the mother liquid at 25°C. Crystals started to appear within 5 d. Crystals of UPPs bound with FPP bound were obtained by soaking the crystals of apo-enzyme in solution containing the substrate. FPP was soaked into the crystal by incremental addition to a final concentration of 230 μM in the mother liquid. This crystallization condition was different from that previously used (Chang et al. 2003a), but the crystals turned out to be isomorphous.
Crystallographic data collection and structural analysis
Diffraction experiments of the UPPs crystal was carried out at −150°C using an MSC MicroMax-007 equipped with an R-AXIS IV++ image-plate detector at Academia Sinica (Taipei, Taiwan). Data were processed and scaled by employing the HKL package (Otwinowski and Minor 1997). For computational refinement, manual modification, and analysis of the crystal structure, the programs CNS (Brunger and Warren 1998), O (Jones et al. 1991), and CCP4 (Collaborative Computational Project No. 4 1994) were used. The parameters for ideal protein geometry (Engh and Huber 1991) were used for the refinements, and the stereochemical quality of the refined structure was checked with the program PRO-CHECK (Laskowski et al. 1993).
Using 2.4 Å resolution data and the Triton-bound UPPs structure that we previously solved as a template (Chang et al. 2003a), without all solvent and cofactor molecules, an initial R-value of 0.53 was calculated, and it was reduced immediately to 0.43 after rigid-body refinement. Electron density maps in the active-site tunnel of one subunit (monomer A) clearly showed two FPP molecules, and those in the other subunit (monomer B) contained one partial FPP (only the geranyl portion was detected). Subsequently more water and FPP molecules were added to the models, and the amino acid side chains were also modified according to the Fourier maps.
Both R- and Rfree-values were used to monitor the progress of refinement (Brunger 1992). The final R- and Rfree-values and refinement statistics are given in Table 1. Figures were generated using MOLSCRIPT (Kraulis 1991), RASTER3D (Merritt and Murphy 1994), and BobScript (Esnouf 1997). The refined model contains amino acid residues 13–239 in one subunit (monomer A) and 17–73, 78–240 in the other (monomer B).
Table 1.
Data collection and refinement statistics
| UPPs + FPP | |||
| Data collection | |||
| Space group | P212121 | ||
| Unit cell (a, b, c Å) | 63.03 | 69.20 | 108.26 |
| Resolution (Å) | 10–2.35 (2.43–2.35) | ||
| No. of observations | 95659 (9202) | ||
| Unique reflections | 19508 (1882) | ||
| Completeness (%) | 97.1 (95.1) | ||
| Average I/σ (I) | 11 (4.1) | ||
| Rmerge (%) | 12.9 (36.3) | ||
| Refinement | |||
| No. of reflections | 19026 (1786) | ||
| (F > 0 σ (F)) | |||
| Rworking | 0.173 (0.215) | ||
| Rfree (%) for 5% data | 0.254 (0.332) | ||
| R.m.s. deviation from ideal bond lengths (Å) | 0.021 | ||
| R.m.s. deviation from ideal bond angles (°) | 1.87 | ||
| Residues in most favored regions of Ramachandran plot (%) | 92.8 | ||
| Residues in additional allowed regions (%) | 7.2 | ||
| Average B-value (Å2) for 1,788 back-bone atoms | 30.7 | ||
| 1,769 side-chain atoms | 34.2 | ||
| 456 water molecules | 50.5 | ||
| 58 FPP atoms (2 FPP + 1 geranyl) | 56.3 | ||
All refinement and calculation of R-factor were done by CNS using all reflections. Numbers in parentheses are for the outmost resolution shell.
Fluorescence experiments
The fluorescence emission spectra of 1 μM UPPs, the UPPs solution with the addition of 15 μM FPP, and the mixture with a further addition of 40 μM IPP in the presence or absence of Mg2+ were monitored using an F-4500 fluorescence spectrophotometer (Hitachi). The buffer solution was 100 mM HEPES (pH 7.5), 50 mM KCl, with or without 5 mM MgCl2. The UPPs used in this study was dialyzed into a buffer containing 1 mM EDTA prior to the fluorescence measurements to remove any tightly bound metal in the UPPs. The samples were excited at 285 nm, and the emission spectra from 300 to 450 nm were recorded.
Acknowledgments
We thank Mr. Kai-Fa Huang and Dr. Chun-Hua Hsu for technical assistance. This work was supported by grants from Academia Sinica and the National Science Council to P.-H.L. and A.H.-J.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
UPPs, undecaprenyl pyrophosphate synthase
IPP, iso-pentenyl pyrophosphate
FPP, farnesyl pyrophosphate
UPP, undecaprenyl pyrophosphate
FPPs, farnesyl pyrophosphate synthase
GPP, geranyl pyrophosphate
TLC, thin-layer chromatography
NiNTA, nickel nitrilotriacetic acid
HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
EDTA, ethylenediaminetetraacetic acid
FTase, farnesyltransferase
Supplemental material: see www.proteinscience.org
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03519904.
References
- Allen, C.M. 1985. Purification and characterization of undecaprenylpyrophosphate synthetase. Methods Enzymol. 110 281–299. [DOI] [PubMed] [Google Scholar]
- Apfel, C.M., Takacs, B., Fountoulakis, M., Stieger, M., and Keck, W. 1999. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: Cloning, expression, and characterization of the essential uppS gene. J. Bacteriol. 181 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355 472–475. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. and Warren, G.L. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Chang, S.Y., Tsai, P.C., Tseng, C.S., and Liang, P.H. 2001. Refolding and characterization of a yeast dehydrodolichyl diphosphate synthase overex-pressed in Escherichia coli. Protein Exp. Purif. 23 432–439. [DOI] [PubMed] [Google Scholar]
- Chang, S.Y., Ko, T.P., Liang, P.H., and Wang, A.H.-J. 2003a. Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton. J. Biol. Chem. 278 29298–29307. [DOI] [PubMed] [Google Scholar]
- Chang, S.Y., Chen, Y.K., Wang, A.H.-J., and Liang, P.H. 2003b. Identification of the active conformation and the importance of length of the flexible loop 72–83 in regulating the conformational change of undecaprenyl pyrophosphate synthase. Biochemistry 42 14452–14459. [DOI] [PubMed] [Google Scholar]
- Chen, A., Kroon, P.A., and Poulter, C.D. 1994. Isoprenyl diphosphate synthases: Protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 3 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A.P.-C., Chen, Y.H., Liu, H.P., Li, Y.C., Chen, C.T., and Liang, P.H. 2002. Synthesis and application of a fluorescent substrate analogue to study ligand interactions for undecaprenyl pyrophosphate synthase. J. Am. Chem. Soc. 124 15217–15224. [DOI] [PubMed] [Google Scholar]
- Chen, Y.H., Chen, A.P.-C., Chen, C.T., Wang, A.H.-J., and Liang, P.H. 2002. Probing the conformational change of Escherichia coli undecaprenyl pyrophosphate synthase during catalysis using an inhibitor and tryptophan mutants. J. Biol. Chem. 277 7369–7376. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4. 1994. The CCP 4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- Collakova, E. and DellaPenna, D. 2001. Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127 1113–1124. [PMC free article] [PubMed] [Google Scholar]
- Cornish, K. 2001. Biochemistry of natural rubber, a vital raw material, emphasizing biosynthetic rate, molecular weight and compartmentalization, in evolutionarily divergent plant species. Nat. Prod. Rep. 18 182–189. [DOI] [PubMed] [Google Scholar]
- Engh, R.A. and Huber, R. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47 392–400. [Google Scholar]
- Esnouf, R.M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graphics 15 132–134. [DOI] [PubMed] [Google Scholar]
- Fujihashi, M., Zhang, Y.-W., Higuchi, Y., Li, X.-Y., Koyama, T., and Miki, K. 2001. Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. 98 4337–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R.T., Kuo, C.J., Chou, C.C., Ko, T.P., Shr, H.L., Liang, P.H., and Wang, A.H.-J. 2004. Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J. Biol. Chem. 279 4903–4912. [DOI] [PubMed] [Google Scholar]
- Joly, A. and Edwards, P.A. 1993. Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J. Biol. Chem. 268 26983–26989. [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 392–400. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Sadanami, K., Kidera, A., and Go, N. 1999. Structural motif of phosphate-binding site common to various protein superfamilies: All-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 12 11–14. [DOI] [PubMed] [Google Scholar]
- Ko, T.P., Chen, Y.K., Robinson, H., Tsai, P.C., Gao, Y.-G., Chen, A.P.-C., Wang, A.H.-J., and Liang, P.H. 2001. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undeca-prenyl-pyrophosphate synthase catalysis. J. Biol. Chem. 276 47474–47482. [DOI] [PubMed] [Google Scholar]
- Koyama, T., Obata, S., Saito, K., Takeshita-Koike, A., and Ogura, K. 1994. Structural and functional roles of the cysteine residues of Bacillus stearothermophilus farnesyl diphosphate synthase. Biochemistry 33 12644–12648. [DOI] [PubMed] [Google Scholar]
- Koyama, T., Tajima, M., Nishino, T., and Ogura, K. 1995. Significance of Phe-220 and Gln-221 in the catalytic mechanism of farnesyl diphosphate synthase of Bacillus stearothermophilus. Biochem. Biophys. Res. Commun. 212 681–686. [DOI] [PubMed] [Google Scholar]
- Koyama, T., Tajima, M., Sano, H., Doi, T., Koike-Takeshita, A., Obata, S., Nishino, T., and Ogura, K. 1996. Identification of significant residues in the substrate binding site of Bacillus stearothermophilus farnesyl diphosphate synthase. Biochemistry 35 9533–9538. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Laskowski, R.A., Moss, D.S., and Thornton, J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231 1049–1067. [DOI] [PubMed] [Google Scholar]
- Lesburg, C.A., Zhai, G., Cane, D.E., and Christianson, D.W. 1997. Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science 275 1820–18224. [DOI] [PubMed] [Google Scholar]
- Liang, P.H., Ko, T.P., and Wang, A.H.-J. 2002. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269 3339–3354. [DOI] [PubMed] [Google Scholar]
- Long, S.B., Casey, P.J., and Beese, L.S. 1998. Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Biochemistry 37 9612–9618. [DOI] [PubMed] [Google Scholar]
- Marrero, P.F., Poulter, C.D., and Edwards, P.A. 1992. Effects of site-directed mutagenesis of the highly conserved aspartate residues in domain II of farnesyl diphosphate synthase activity. J. Biol. Chem. 267 21873–21878. [PubMed] [Google Scholar]
- Meganathan, R. 2001. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203 131–139. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D. 50 869–873. [DOI] [PubMed] [Google Scholar]
- Ogura, K. and Koyama, T. 1998. Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98 1263–1276. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pan, J.J., Yang, L.W., and Liang, P.H. 2000a. Effect of site-directed mutagenesis of the conserved aspartate and glutamate on E. coli undecaprenyl pyrophosphate synthase catalysis. Biochemistry 39 13856–13861. [DOI] [PubMed] [Google Scholar]
- Pan, J.J., Chiou, S.T., and Liang, P.H. 2000b. Product distribution and pre-steady-state kinetic analysis of Escherichia coli undecaprenyl pyrophosphate synthase reaction. Biochemistry 39 10936–10942. [DOI] [PubMed] [Google Scholar]
- Park, H.W., Boduluri, S.R., Moomaw, J.F., Casey, P.J., and Beese, L.S. 1997. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science 275 1800–1804. [DOI] [PubMed] [Google Scholar]
- Poulter, C.D., Argyle, J.C., and Mash, E.A. 1977. Letter: Prenyltransferase. New evidence for an ionization-condensation-elimination mechanism with 2-fluorogeranyl pyrophosphate. J. Am. Chem. Soc. 99 957–959. [DOI] [PubMed] [Google Scholar]
- Saderholm, M.J., Hightower, K.E., and Fierke, C.A. 2000. Role of metals in the reaction catalyzed by protein farnesyltransferase. Biochemistry 39 12398–12405. [DOI] [PubMed] [Google Scholar]
- Sato, M., Sato, K., Nishikawa, S.-I., Hirata, A., Kato, J.-I., and Nakano, A. 1999. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in doli-chol synthesis. Mol. Cell Biol. 19 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledz, M., Seidler, A., Beyer, P., and Neuhaus, G. 2001. A novel phytyl-transferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett. 499 15–20. [DOI] [PubMed] [Google Scholar]
- Shimizu, N., Koyama, T., and Ogura, K. 1998. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase. No sequence similarity between E- and Z-prenyl diphosphate synthases. J. Biol. Chem. 273 19476–19481. [DOI] [PubMed] [Google Scholar]
- Song, L. and Poulter, C.D. 1994. Yeast farnesyl-diphosphate synthase: Site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc. Natl. Acad. Sci. 91 3044–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarna, K., Stevenson, D., Meganathan, R., and Hudspeth, M.E. 1998. J. Bacteriol. 180 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarshis, L.C., Yan, M., Poulter, C.D., and Sacchettini, J.C. 1994. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-Å resolution. Biochemistry 33 10871–10877. [DOI] [PubMed] [Google Scholar]
- Varki, A., Cummings, R., Esko, J., Freeze, H., Hart, G., and Marth, J. 1999. Bacterial polysaccharides. In Essentials of glycobiology, pp. 322–325. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wang, K.C. and Ohnuma, S.-I. 2000. Isoprenyl diphosphate synthases. Biochim. Biophys. Acta 1529 33–48. [DOI] [PubMed] [Google Scholar]