Abstract
The protease from type 1 human immunodeficiency virus (HIV-1) is a critical drug target against which many therapeutically useful inhibitors have been developed; however, the set of viral strains in the population has been shifting to become more drug-resistant. Because indirect effects are contributing to drug resistance, an examination of the dynamic structures of a wild-type and a mutant could be insightful. Consequently, this study examined structural properties sampled during 22 nsec, all atom molecular dynamics (MD) simulations (in explicit water) of both a wild-type and the drug-resistant V82F/I84V mutant of HIV-1 protease. The V82F/I84V mutation significantly decreases the binding affinity of all HIV-1 protease inhibitors currently used clinically. Simulations have shown that the curling of the tips of the active site flaps immediately results in flap opening. In the 22-nsec MD simulations presented here, more frequent and more rapid curling of the mutant’s active site flap tips was observed. The mutant protease’s flaps also opened farther than the wild-type’s flaps did and displayed more flexibility. This suggests that the effect of the mutations on the equilibrium between the semiopen and closed conformations could be one aspect of the mechanism of drug resistance for this mutant. In addition, correlated fluctuations in the active site and periphery were noted that point to a possible binding site for allosteric inhibitors.
Keywords: HIV-1 protease, V82F/I84V drug-resistant mutant, molecular dynamics, structure-based drug design, protein flexibility, ensemble of conformations, allosteric inhibitor, drug resistance
The development of resistance to the different drugs administered in the Highly Active Anti-Retroviral Therapy (HAART) cocktails is an ever-escalating problem. In a study of strains isolated from patients recently infected with HIV, the frequency of high-level resistance to one drug increased from 3.4% (1995–1998) to 12.4% (1999–2000), P = 0.002, and the frequency of resistance to multiple different drugs increased from 1.1% to 6.2% during those same few years, P = 0.01 (Little et al. 2002). Thus, the protease inhibitors currently available are becoming less effective, because the entire pool of viral strains within the United States is shifting to a more drug-resistant state.
Importance of flap dynamics
Examination of different ligand-bound conformations of HIV protease suggests that some mutations alter the equilibrium between the closed and open conformations of the protease (Rose et al. 1998). These differences, in turn, may alter the dissociation rates and affinities of drugs (Rose et al. 1998). Here, simulation data are presented that suggest that the V82F/I84V double mutation could be shifting that equilibrium between closed and semiopen forms in a manner that causes the mutant to favor the semiopen conformations more than the wild type prefers them.
There are published data indicating that certain active-site mutations affect the dynamics of conformational changes, causing the decreases in drug binding affinity. Kinetic experiments show that the decreased affinity of drugs for the L90M, G48V, and L90M/G48V mutants is caused by an increase in dissociation rates, which is due to increases in the flap opening rates, decreases in the flap closing rates, or both (Maschera et al. 1996). The association rates for the wild type and for those mutants were not significantly different (Maschera et al. 1996). MD simulations of a wild type and of an M46I mutant for 11 nsec performed by Rothlisberger’s group (Piana et al. 2002b) indicated that although M46I does not significantly affect the global, average structure, it does induce subtle differences in the dynamics; specifically, M46I stabilized the flaps in the closed conformation. Previous MD simulations from Erickson’s group (Collins et al. 1995) produced a similar conclusion: M46I increases the stability of the conformation with closed flaps, which decreases the flexibility of the flaps. Weber’s group, which crystallized several different mutants of HIV-1 protease with different peptide inhibitor analogs and also performed kinetic studies, reported that the K45I mutant has lower B-factors than the wild type, it has decreased mobility in its flaps (particularly residues 42–52) compared to the wild type, and it has increased activity (Mahalingam et al. 2001, 2002).
Further support for the importance of flap dynamics comes from NMR studies (Katoh et al. 2003), which suggested that the particular composition of the flap residues has little impact on substrate specificity, because most of the flap residues do not have unique interactions with the peptide/peptidomimetic substrates. This idea is supported by the fact that mutational studies involving the flap residues tend to produce mutants with near wild-type specificity (Katoh et al. 2003). If the particular sequence of most of the flap residues is not the major determinant of specificity, then this implies that flap dynamics could be a possible source of the specificity that surely does exist. NMR studies also suggest that association of substrates is controlled by a rare event, such as opening of the flaps (Katoh et al. 2003). Similarly, dissociation of products or release of the inhibitor would probably be significantly affected by the rate of flap opening as well. Thus, flap dynamics are likely involved in regulating both the association rates and the dissociation rates, both of which modulate the binding affinity of drugs.
Flap dynamics is also involved in the enzymatic mechanism itself. Ab initio quantum chemical calculations (i.e., ab initio MD) and classical MD simulations of the HIV-1 protease complex (Piana et al. 2002a) indicated that the activation free energy barrier of the enzymatic reaction is highly sensitive to the distance between the substrate and the catalytic aspartates, which is controlled by the flap dynamics. Similarly, conventional MD simulations (Piana et al. 2002b) indicated that the motion of the substrate towards the catalytic aspartates is tightly coupled to dynamics of the flap tips. Thus, the protein’s structural dynamics includes the initial opening of the flaps that allows the substrate/drug to access the active site, the closing of the flaps and the positioning of the substrate to make the enzyme catalytically competent, and the subsequent reopening of the flaps that allows the products or inhibitor to escape the active site. Mutations could perturb any of these critical events in the enzyme’s catalytic cycle.
Extent of flap motion possible on the nanosecond time scale
The simulations from Schiffer’s group showed complete flap opening (starting with the semiopen conformation and reaching the fully open form) after 3 nsec of conventional, solvated MD on the apo protease (Scott and Schiffer 2000). A previous simulation performed by Erickson’s group showed flap opening (from the closed to the semiopen conformation) within 140 psec, but in this case, the dynamics were perturbed by external forces that drove the protease core from a conformation seen in a closed structure to one seen in a semiopen structure (Collins et al. 1995).
NMR data from Torchia’s group indicate that the transition from the semiopen to the open conformation of the apo HIV-1 protease occurs on the 100-μsec time scale (Ishima et al. 1999). However, it was also observed that for the apo protease, the conformations with the semiopen flaps are the predominant form (Ishima et al. 1999), which is an observation supported by all NMR and crystallographic studies of apo HIV-1 protease. NMR and crystal structures of ligand-bound HIV-1 protease always produce the closed conformation. In addition, umbrella sampling MD calculations confirm that for apo protease the semiopen conformation is more stable than the closed conformation, due to differences in the entropy of the flap residues (Rick et al. 1998). The above data indicate that when HIV-1 protease is not bound by a ligand, it favors the semiopen conformation; thus, the transition from the semiopen form of apo protease to the fully open form of apo protease is at least a slightly unfavorable and rare occurrence that likely occurs in vitro on the microsecond time scale. By chance and/or because of features of the GROMOS force field, the flaps opened from the semiopen form to a fully open conformation during a published simulation of 10 nsec of conventional MD (Scott and Schiffer 2000). Torchia’s NMR studies indicated that fluctuations of the F53 ring are likely coupled to the motion of the entire flap backbone (Ishima et al. 1999). In the umbrella sampling MD calculations performed by Burt’s group, extensive flexibility was observed for the flap tip residues, and it was shown that the flap’s tips curl in (residues 48–52 undergo large changes in their φ and ψ torsion values and fold back onto themselves to give a bent L structure) before they open (Rick et al. 1998). The notion of the curling behavior of the flap tips is consistent with the NMR data from Torchia’s group (Freedberg et al. 2002), which indicated that significant motion of the flap tip residues 49–53 does occur on a subnanosecond time scale.
Although the transition from the semiopen form of the apo protease to the fully open form appears to be a somewhat unfavorable and rare event, the structural transition from the closed conformation of the apo protease to the semiopen conformation is quite favorable and should be expected to occur very rapidly. The simulations presented here began with the coordinates of the closed conformations of HIV-1 protease:inhibitor complexes. The inhibitors were deleted and then replaced with water before these MD simulations were initiated, and, as expected, large flap motions were observed. Most published simulations were performed on either the closed complex of HIV-1 protease still bound to a substrate/inhibitor (with very little flap motion during the simulations) or on the semiopen conformation of the apo protease (with substantial flap motion). One short simulation (400 psec, in pseudo vacuum) utilized an approach similar to the one presented here (Zoete et al. 2002). In that simulation (which was part of an exhaustive collection of many different MD simulations of HIV protease that were then used for normal mode analyses), the authors took the coordinates of an HIV-1 protease complex, deleted the inhibitor, and then performed MD on the closed conformation of the apo protease (Zoete et al. 2002). Their analyses (Zoete et al. 2002) indicated that the dynamic motion of the flaps was facilitated by deleting the ligand prior to initiating the MD simulation.
The significance of the curling of the flap tips
The early “activated” MD simulations by Burt’s group (in which the flaps opened from the closed conformation to the semiopen conformation; Rick et al. 1998) and the recent conventional MD simulations by Schiffer’s group (in which the flaps opened from the semiopen conformation to the fully open form; Scott and Schiffer 2000) both displayed curling behavior of the tips of the active site flaps (residues G48–G49–I50–G51–G52), and both studies indicated that the curling of the flap tips preceded flap opening. Scott and Schiffer (2000) stated that after 3 nsec of the MD simulation, the curling of the tips of the active site flaps quickly resulted in the flaps opening up to the fully open conformation (i.e., the curling behavior of the flap tips facilitated opening of the entire flaps from the semiopen form to a distance large enough to allow substrates to access the active site).
Such motions of the flap tip residues are consistent with NMR data. Torchia’s group observed rapid motion on a ≪ 10 nsec time scale for residues G49, I50, G51, and G52 (Ishima et al. 1999). In more recent NMR experiments, it was shown that the flap tip residues 49–53 experience significant subnanosecond time scale motion (Freedberg et al. 2002). Torchia’s group also noticed that fluctuations of F53 are likely coupled to motion of the entire flap backbone (Ishima et al. 1999). Thus, NMR studies indicate both that flap tip motion does actually occur in vitro on a subnanosecond time scale, and that the motion of the flap tip is likely coupled to motion of the entire flap backbone.
The importance of studying the V82F/I84V double mutant
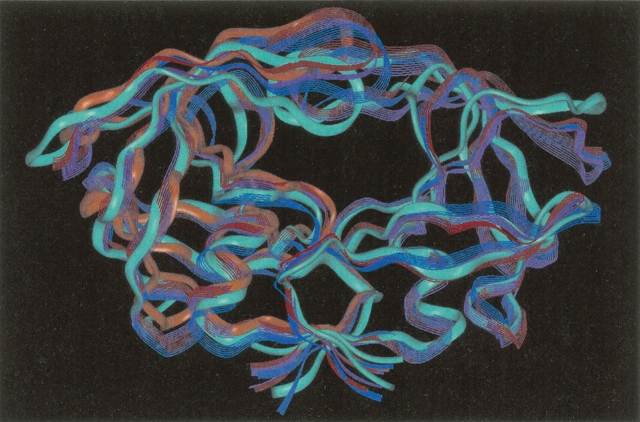
The V82F/I84V double mutation impedes the binding of all of the inhibitors currently used clinically (including Amprenavir) by making the inhibitor’s affinity for the mutant from 11-fold to 2000-fold worse than its affinity for the wild-type HIV-1 protease (Ala et al. 1997; Klabe et al. 1998; Ohtaka et al. 2002). For example, the V82F/I84V double mutant has a 700-fold increase in Ki for ritonavir, a 79-fold increase in Ki for indinavir, an 86-fold increase in Ki for nelfinavir, and a 1000-fold increase in Ki for DMP323 and DMP450 (Ala et al. 1997; Klabe et al. 1998). Even though the V82F/I84V mutation has a large effect on drug binding affinity, the structures of the backbones of the wild type (1KZK.pdb; Reiling et al. 2002) and of the mutant (1D4S.pdb; Thaisrivongs et al. 1996) are nearly identical (the RMSD between all backbone atoms of the crystal structures = 0.76 Å; see Fig. 1 ▶). Because the V82F/I84V double mutation deleteriously affects the affinities of all of the clinically used HIV-1 protease active site inhibitors (which includes inhibitors of several different classes), this suggests that the V82F/I84V mutant is likely displaying some of the indirect effects that are involved in drug resistance, which makes this a good system to study when trying to learn about those effects. (Note: If residue B has a hydrogen bond with a drug, that is an example of a direct effect on the drug; however, if residue X has interactions that affect the position of that residue B, then residue X has an indirect effect on the drug.)
Figure 1.
Topology of HIV protease: The mutant side chains 82F and 84V are displayed in red stick mode, while all other side chains are not displayed. The cyan ribbon depicts the backbone of the mutant 1D4S.pdb, while the purple ribbon displays the backbone of the wild-type 1KZK.pdb (backbone RMSD = 0.76 Å). A possible new convention for the terminology of the topology of HIV protease is proposed that involves the following: Flap (43–58), Ear (35–42), Cheek (Cheek Turn = 11–22 and Cheek Sheet = 59–75), Eye (23–30), and Nose (6–10). Unlike the current convention, this new terminology is independent of any particular hypothesis of the mechanism of flap motion.
The current convention for the terminology of the topology of HIV protease is heavily based upon one particular hypothesis of the mechanism governing flap motion. A possible new convention for the terminology of the topology of HIV protease that is independent of all mechanistic hypotheses is as follows: The Flap = residues 43–58 (the active site flaps will retain their name), the Ear or Ear Flap = 35–42 (currently called the Elbow of the Flap), the Cheek Turn = 11–22 (i.e., the Fulcrum), the Cheek Sheet = 59–75 (currently called the Cantilever), the Eye = 23–30 (it contains the catalytic Asp that “sees” the drug), and the Nose = 6–10 (which contains the R8 that blocks the front entrance to the active site). Similarly, the Whiskers (1–5 and 95–99) would refer to the termini involved in forming the dimerization interface, residues 86–90 form the helix, and 79–84 form the Wall Turn (the turn composing the active site’s wall). This new convention also seems much easier for people from many different disciplines to remember: Almost every depiction of HIV protease shows the same view, which looks like the head of a fat cat or a bulldog.
The ensembles of conformations generated for this V82F/ I84V mutant and for the wild type are currently being used for the implementation of the new Relaxed Complex method (Lin et al. 2002, 2003), which incorporates the flexibility of both the ligand and the target protein by using a docking algorithm that allows full ligand flexibility and by targeting a significant percentage of the entire ensemble of protein conformations in an automated fashion. When implementing the Relaxed Complex method, the drugs will be optimized by following the advice, which indicates the residues that should be targeted to evade resistance (Wang and Kollman 2001; Ohtaka et al. 2002), and the general strategies developed by the McCammon group (Hodge et al. 1997) will be utilized. Thus, this particular mutant was studied both for the basic information that it might provide about general features involved in drug resistance as well as for the direct applications involving the design of an inhibitor that is effective against that V82F/I84V mutant.
Results
Properties of the global structures
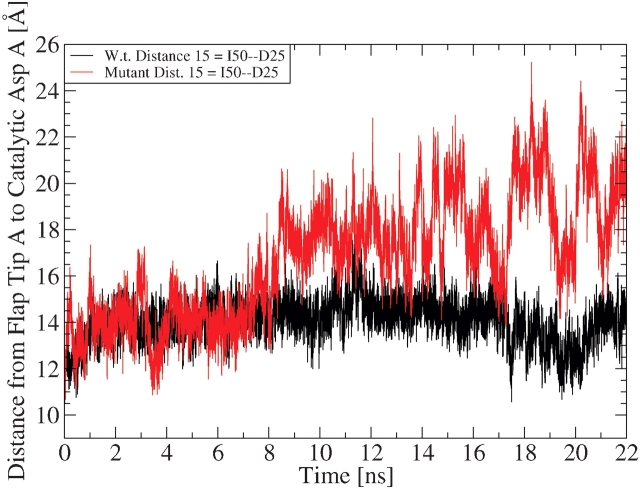
Conventional MD simulations were initiated using the closed conformations of the apo wild-type and the apo V82F/I84V mutant of HIV-1 protease, and the simulations were performed for 22 nsec on each system. Both systems began with nearly identical backbone structures (the RMSD of all backbone atoms between the two crystal structures = 0.76 Å; Fig. 1 ▶), and both systems departed from their crystal structures to a similar extent during most of the simulations (Fig. 2 ▶). However, the mutant did have a larger RMSD than the wild type during the last 8 nsec, which was likely due to the fact that the mutant’s flaps continued to open even further, while the wild-type’s flaps transiently closed and then began opening again during that interval (see Fig. 7 ▶).
Figure 2.
Root-mean-square deviation of the backbone atoms (i.e., N, α C, and carbonyl C) with respect to time. For qualitative comparisons, an RMSD of 1.75 Å was observed between certain semiopen and closed conformations in one published study.
Figure 7.
Comparing the motion of monomer A’s flap: The black curve maps the trajectory of the wild-type’s distance from I50 C α to D25 C β, while the red curve shows the mutant’s flap tip-to-catalytic Asp distance trajectory. Any snapshot with an I50–D25 distance greater than or equal to 15.8 Å is defined as a snapshot in the semiopen conformation (according to the 1HHP.pdb crystal structure). To observe the motion of monomer B’s flap for these systems, see the Supplemental Material, Figure 3 ▶.
As can be seen in Figure 2 ▶, the wild-type and the mutant HIV-1 protease systems departed from their crystallographic structures by an RMSD of approximately 1.75 to 2.25 Å. For a qualitative comparison, note that in the simulations from the Karplus group (Zoete et al. 2002) an RMSD of 1.75 Å (for all backbone atoms) was observed between the minimized forms of the closed and certain semiopen conformations. Thus, a qualitative examination of the RMSD trajectories from these simulations of the wild-type and mutant systems suggests that both protease molecules might have undergone enough flap opening to enable reaching the semiopen conformations. More direct measurements will be shown later to prove that opening of the flaps to the semiopen form did indeed occur in these simulations (for at least the mutant system).
Curling behavior of the flap tips
According to published simulations, the curling of the tips of the active site flaps triggers opening of the entire flaps (see the introduction). To investigate this flap tip behavior, the variability in certain “TriCa Angles” (i.e., the angle between three adjacent α carbons) was measured. Although it is certainly possible that other ways of measuring the curling behavior of the flap tips could be devised, any curling of the flap tips that does occur should involve changes in the TriCa Angles of the residues at or near the tips. The flaps and Ears were scanned using the TriCa Angle analysis in a sliding window approach that involved residues 33–57 and 133–157 (e.g., scans were done of residues 33–34–35, 34–35–36, 35–36–37, etc.). This determined that for both wild-type and mutant systems, the exact same TriCa Angles for the tips of the active site flaps (angles G48–G49–I50 and G148–G149–I150) and for the tips of the Ear Flaps (angles L38–P39–G40, G40–K41–W42, L138–P139–G140, and G140–K141–W142) displayed the most variation among all the residues scanned (as judged by having much larger values of both the standard deviations and the ranges). An observation was made that a few of the TriCa Angles for those flap tip and Ear tip residues also displayed a distinct, bimodal distribution (angles G48–G49–I50 and G148–G149–I150 had the most well-separated bimodal distributions, but angles L38–P39–G40, G40–K41–W42, L138–P139–G140, G140–K141–W142, I47–G48–G49, and I147–G148–G149 also had bimodal distributions, although they were not as distinct). The particular TriCa angle for the flap tip residues G48–G49–I50 displayed the greatest differences between wild-type and mutant simulations, and its trajectory is shown in Figure 3 ▶.
Figure 3.
Comparing the curling of the flap tips: The black line displays the curling behavior of wild-type monomer A’s flap tip, while the red line depicts the curling of the mutant’s flap tip. When a flap tip curls, its backbone geometry must change. To demonstrate curling behavior, the variability in the angle between the three α C’s of G48–G49–I50 is shown.
In Figure 3 ▶ it is apparent that the mutant switched between those two subsets of conformational space many more times than the wild type switched, especially during 0–8 nsec. The curled-in and curled-out states are displayed in Figure 4 ▶. The qualitative residence times spent within each curled-in (~115°) or curled-out (~145°) state indicate that the mutant’s flap tip was actually curling faster than the wild-type’s flap tip (see Fig. 3 ▶). Thus, the V82F/I84V mutant’s flap tip curled more rapidly and more frequently than the wild-type’s flap tip. The large flap displacements that were observed in the wild-type and mutant simulations frequently involved substantial curling behavior of the flap tips (see Fig. 5 ▶). Because curling of the flap tip likely produces flap opening, the more rapid and more frequent curling behavior displayed by the V82F/I84V mutant in these simulations suggests that more flap opening should have been observed in the mutant system. This was indeed the case.
Figure 4.

Curled-in vs. curled-out flap tips: Ribbon representations are shown of the two extrema from the largest flap opening event that occurred in the wild-type simulation. The snapshot from 17,505 psec is colored red, it has a I50 C α–D25 C β distance of 10.6 Å, and it has a value of 149° for TriCa Angle 6 (G48–G49–I50, which is colored white). Thus, the red ribbon is displaying the curled out state. The snapshot from 17,666 psec is colored violet, it has a I50 C α–D25 C β distance of 16.0 Å, and it has a value of 108° for TriCa Angle 6. Thus, the violet ribbon has its flap tip curled in.
Figure 5.

Rapid curling behavior was displayed: The above seven ribbon diagrams depict a fast flap-opening event (3.0 Å displacement in 20 psec) that involved rapid curling behavior of the mutant’s flap tip. The snapshot from 4210 psec is colored orange, it has a I50 C α–D25 C β distance of 13.8 Å, and it has a value of 147° for TriCa Angle 6 (G48–G49–I50, which is colored white). The time points, flap–Asp distances, and TriCa Angle 6 values for the other images are as follows: 4214 psec in cyan, 14.9 Å, 120°; 4217 psec in light blue, 14.9 Å, 138°; 4225 psec in purple, 15.5 Å, 118°; 4226 psec in violet, 16.5 Å, 131°; 4227 psec in magenta, 15.5 Å, 114°; and 4228 psec in white, 14.9 Å, 135°. The displacement of flap A proceeded from 13.5 Å at 4206 psec to 16.5 Å at 4226 psec (see also Supplemental Material, Fig. 2 ▶).
Extent of flap opening observed in these simulations
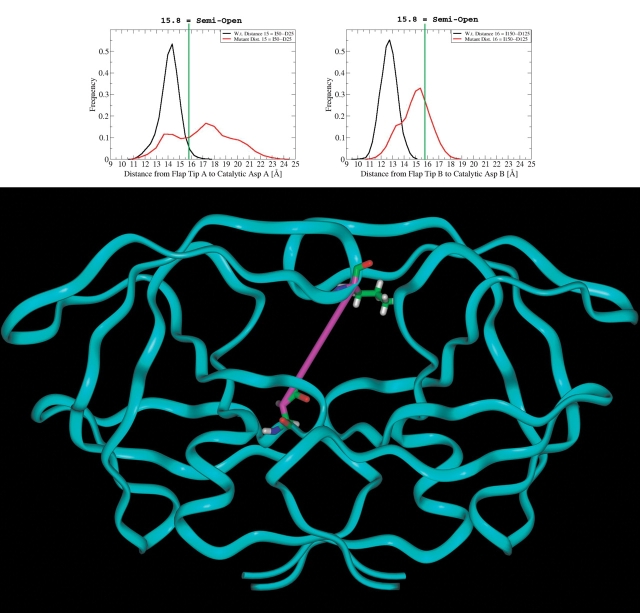
By several different ways of measuring flap opening, this drug-resistant mutant’s active site flaps opened more than the wild-type’s flaps during these simulations. The mutant’s flaps opened farther (i.e., they sampled much larger distance values), and the mutant’s flap–catalytic Asp distance values fluctuated much more erratically than the wild-type’s values. Although observing the same average dynamic properties for both of the flaps is expected (provided that the simulations were of sufficient duration), observing the same motions in both flaps at any one point in time should not be expected (see the Discussion). Because it is now known that the nature of the motions of the flaps at any given point in time is asymmetric, and because of the recent discovery of the importance of the curling behavior at the tips of the flaps, nontraditional distance measurements were selected to monitor flap motion. Instead of monitoring the I50–I150 distance, which could be affected by both flap tip curling and by flap asymmetry, different distances between the tip of one flap and one of the catalytic Asp’s (and the corresponding distances in the other monomer) were measured (see Fig. 6 ▶).
Figure 6.
Flap tip to catalytic asp distances: The black histograms represent the wild-type frequencies, while the red histograms display the drug-resistant V82F/I84V mutant’s frequencies. The graph on the left indicates the values that were sampled by the I50 C α–D25 C β distance during nsec 2–22, while the graph on the right signifies the values sampled for the I150 C α–D125 C β distance. The green line marks the I50–D25 distance value from the apo [semiopen] 1HHP.pdb structure; thus, any snapshot with an I50–D25 (or I150–D125) distance greater than or equal to 15.8 Å is defined as a snapshot in the semiopen conformation. For comparison, those distances in the closed crystallographic complexes were as follows: I50–D25 in 1KZK (w.t.) = 12.4 Å and in 1D4S = 12.1 Å; I150–D125 in 1KZK = 13.0 Å and in 1D4S = 12.1 Å. The ribbon diagram displays one of the flap tip-to-Asp distance measurements as a magenta stick.
The distance values from the simulations were compared to those values measured from the 1HHP.pdb crystal structure, which is a structure of apo HIV-1 protease in the semiopen form (Spinelli et al. 1991). Thus, any snapshot with a distance value greater than or equal to the value of that distance in the 1HHP.pdb structure will be defined as a snapshot in the semiopen conformation. Choosing a particular definition of the “semiopen” conformations is a somewhat subjective process.
Figure 7 ▶ displays the trajectories of the data that are shown as histograms in Figure 6 ▶. From these trajectories it is obvious that the mutant’s flap opened much more than the wild-type’s flap (i.e., the mutant’s flap sampled much larger flap–Asp distance values, and the mutant’s flap–Asp distance values fluctuated much more erratically than the wild-type’s values). Compare the flap-opening trajectory in Figure 7 ▶ to the flap tip curling behavior shown in Figure 3 ▶. When that flap tip is curling frequently in both systems during 0–8 nsec, that flap is opening in both the wild-type and the mutant systems (i.e., the distances between the flaps and the catalytic aspartates are generally increasing). During 8–16 nsec, little to no flap tip curling is observed for the wild type, and its flap-to-Asp distance remains fairly consistent. Around the 17th nsec the flap tips curl frequently again in both systems, and that curling behavior is followed by further flap opening for the mutant and by transient flap closing and then flap opening for the wild type.
For visual depictions of the maximum flap–Asp distances sampled in these simulations, see Figures 8 ▶–11 ▶ ▶ ▶. The global maxima sampled for the distance between I50 and D25 in each simulation are compared to a space-filling view of the closed conformation of protease in Figure 8 ▶. From Figure 8 ▶ it is readily apparent that the mutant simulation sampled much larger flap–Asp distance values than the wild-type simulation. To indicate the relative size of the active site in the mutant snapshot that had the largest I50–D25 distance, that maximum mutant snapshot is shown with a natural substrate in the space-filling representation (see Fig. 9 ▶). That image was generated by using the backbone atoms of residues 6–30 and 106–130 to superimpose the complex of the D25N mutant of HIV-1 protease with its natural substrate KARVLAEAM (1F7A.pdb = Prabu-Jeyabakin et al. 2000) onto the snapshot from 18,282 psec of the mutant simulation. The protease molecule from the 1F7A crystal structure was then hidden, leaving the natural substrate at a reasonable position within that mutant snapshot’s active site. Figure 10 ▶ displays the global maxima sampled for the I50–D25 distance from each simulation superimposed (using the backbone atoms of residues 6–30 and 106–130) onto a closed and onto a semiopen crystal structure. In Figure 10 ▶ it appears that only the wild-type monomer on the right reached the semiopen form; however, both monomers of the mutant clearly sampled flap–Asp distance values greater than those observed in the semiopen crystal structure. Figure 11 ▶ indicates that the mutant simulation produced many different snapshots with flaps that are at least semiopen.
Figure 8.
Global maxima for flap tip A-to-catalytic Asp 25 distances compared to a closed crystal structure: The cyan space-filling model depicts the mutant’s closed crystal structure (1D4S.pdb, with the ligand not displayed), the red space-filling image shows the mutant snapshot that sampled the largest distance from I50 C α to D25 C β (i.e., snapshot 18,282 psec, distance = 25.2 Å), the small ligand colored by atom type is the substrate KARVLAEAM from the 1F7A.pdb crystal structure of the D25N mutant of HIV-1 protease bound to that peptide, and the green CPK model displays the wild-type snapshot that sampled the largest flap–Asp distance (i.e., snapshot 11,353 psec, distance = 18.1 Å).
Figure 9.
The most open snapshot from the mutant’s simulation: The red space-filling model displays snapshot 18,282 psec from the mutant’s simulation, which is the mutant snapshot that displayed the largest value for the distance between I50 and D25. The green space-filling model represents the substrate KARVLAEAM from the 1F7A.pdb crystal structure of the D25N mutant of HIV-1 protease bound to that peptide. The backbone atoms of residues 6–30 and 106–130 were used to superimpose the 1F7A complex onto the snapshot from 18,282 psec, and then the protease molecule from the 1F7A complex was hidden. This mutant snapshot has more than enough space to easily accommodate its natural substrate.
Figure 10.
Global maxima for flap tip A-to-catalytic Asp 25 distances compared to the crystal structures of the closed and semiopen conformations: The closed conformation of the wild-type crystal structure (1KZK) is shown as the violet ribbon, and the semiopen conformation of the 1HHP crystal structure is displayed as the monomer on the right in white line-ribbon mode. The mutant snapshot that sampled the largest distance from I50 C α to D25 C β (i.e., snapshot 18,282 psec, distance = 25.2 Å) is depicted as the red ribbon, and the green ribbon displays the wild-type snapshot that sampled the largest flap–Asp distance (i.e., snapshot 11,353 ps, distance = 18.1 Å). For the distance values displayed by the crystal structures for this dimension, see the caption for Figure 6 ▶.
Figure 11.
The most open snapshots from the mutant’s simulation compared to the closed and semiopen crystallographic conformations: The solid ribbon in cyan represents the mutant’s crystal structure of the closed complex (1D4S), while the monomer on the right with a solid orange ribbon shows the semiopen conformation of the 1HHP crystal structure of the apo protease. The ribbons shown in line mode depict the local maxima for the distance between I50 and D25 in the mutant’s simulation. These maxima were the peaks from different flap opening events. The following snapshots are displayed: 18,282 = red (I50–D25 distance was 25.2 Å), 20,210 = orange (24.4 Å), 18,821 = green (23.6 Å), 15,296 = light blue (23.0 Å), 12,066 = purple (22.9 Å), and 17,620 = magenta (22.9 Å). For the maxima from the wild-type simulation along this dimension, see Fig. 16 ▶.
Although it is best to measure one particular flap at a time when investigating the flap’s flexibility and the extent of flap motion sampled, assessing the behavior of both flap tips with respect to the catalytic Asp’s can provide insight about the accessibility of the active site to drugs/substrates. Measurements involving the motion of the center of mass of the tips of both flaps relative to a catalytic Asp were thus collected (see Figs. 12 ▶ and 13 ▶). No matter what distance was used to monitor flap motion, the conclusion was always the same. This drug-resistant mutant’s flaps opened more than the wild-type’s flaps during the same time period (i.e., the mutant’s flaps opened up to larger distances, and the mutant’s flap–Asp distances fluctuated more erratically).
Figure 12.

Distances between the center of mass of the top of both flap tips to a catalytic Asp: The distances between the center of mass of G51 and G151 to either D25 C β or D125 C β were measured. The black and red curves symbolize the distance between the center of mass of two residues at the tops of the tips of both flaps to the catalytic Asp of monomer B (i.e., distance L) for wild-type and for mutant, respectively. The blue and magenta curves depict the distance from the C.O.M. of the tips of both flaps to the catalytic Asp of monomer A for wild-type and for mutant (i.e., dist. L2), respectively. For comparison, that distance had a value of 19.7 Å in the 1HHP.pdb [semiopen] structure, while its values in the closed crystallographic complexes for distances L and L2 were 15.0 Å and 14.9 Å in 1KZK and 14.6 Å and 14.1 Å in 1D4S. The ribbon diagram displays the location of distance L2; the red stick connects the two Gly residues whose center of mass was utilized, while the magenta stick connects that center of mass to the respective catalytic Asp.
Figure 13.
Distances between the center of mass of the bottom of both flap tips to a catalytic Asp: The distances between the center of mass of G49 and G149 to either D25 C β or D125 C β were measured. The black and red curves symbolize the distance between the center of mass of two residues at the bottom of the tips of both flaps to the catalytic Asp of monomer A for wild-type and for mutant, respectively. The blue and magenta curves depict the distance from the C.O.M. of the tips of both flaps to the catalytic Asp of monomer B for wild-type and for mutant, respectively. For this dimension, 14.6 Å = semiopen, and the values of distances M and M2 in the closed crystallographic complexes were as follows: 11.0 Å and 11.5 Å in 1KZK (w.t.) and 10.4 Å and 10.5 Å in 1D4S. The ribbon diagram displays the location of distance M2; the red stick connects the two Gly residues whose center of mass was utilized, while the magenta stick connects that center of mass to the respective catalytic Asp.
Possible relationship connecting motion of the flaps and the ears
The Ear Flaps (see Fig. 1 ▶) are connected to the active site flaps in sequence, and likely in dynamic structure—half of the Ear Flap is, in fact, the opposite end of one of the β-strands composing half of the active site flap. Because the same β-strand forms half of the Ear Flap and half of the active site flap, it makes sense that their motions would be connected in some way. The mutant ensemble displayed larger distances between the flaps and the catalytic Asp’s (i.e., the mutant’s flaps opened farther than the wild-type’s flaps; see Figs. 6 ▶, 7 ▶, 8 ▶, 10 ▶, 12 ▶, and 13 ▶); conversely, the mutant ensemble sampled smaller distances between the Ear Flaps and the Cheek region.
From Figure 14 ▶ it is apparent that the wild-type ensemble sampled larger Ear-to-Cheek distance values than the mutant. The ensemble that sampled larger Ear-to-Cheek values also displayed smaller flap-to-Asp values, and vice versa. The same relationship was observed for both monomers (i.e., the wild-type ensemble also sampled larger values for the P139–G116 and the G140–Q161 distances; see Supplemental Material, Fig. 4 ▶). The opposite relationship that was observed in both ensembles between Ear–Cheek distances and flap–Asp distances prompted an investigation of these distance values with respect to time (and to each other).
Figure 14.
Distances between the ear and the cheek: The histograms on the left depict the distance from the Ear Flap tip (P39 C α) to the Cheek Turn (G16 C α), while the histograms on the right display the distance from the Ear Flap Tip (G40 C α) to the Cheek Sheet (Q61 C α). As in all figures, red = mutant (i.e., 1D4S), and black = wild type (i.e., 1KZK). The values for these distances in the crystal structures were as follows: P39–G16 in 1KZK = 7.9 Å, in 1D4S = 9.0 Å, and in 1HHP = 8.4 Å; G40–Q61 in 1KZK = 9.6 Å, in 1D4S = 9.7 Å, and in 1HHP = 9.4 Å. The ribbon diagram below each graph displays the particular distance value measured as a magenta stick.
A qualitative analysis of the two trajectories in Figure 15 ▶ indicates that in the wild-type’s simulation, the motion of the flap–Asp distance was anticorrelated with the motion of the Ear–Cheek distance (i.e., when the Ear-to-Cheek distance was generally decreasing, the flap to Asp distance was increasing). The same anticorrelated relationship was observed in the mutant’s simulation; however, the trend was not quite as clean due to the increased flexibility of the mutant’s flaps (see Supplemental Material, Figs. 5 ▶ and 6 ▶).
Figure 15.
Qualitative anticorrelation between flap–Asp distances and Ear–Cheek distances: Both lines represent distance trajectories from the wild-type ensemble, but the same trend was observed in the mutant’s trajectories. The black line above displays the trajectory of the flaps-to-Asp distance (C.O.M. of G51 + G151 to D125 C β), while the blue curve below depicts the Ear-to-Cheek distance (G40 C α–Q61 C α). The two trajectories have a pseudomirror symmetry plane between them, which indicates that they are displaying anticorrelated motion.
Discussion
To examine the fundamental structural underpinnings affecting drug-resistance against HIV-1 protease inhibitors, 22 nsec of molecular dynamics (MD) were simulated on both a wild-type HIV-1 protease (1KZK.pdb) and on the drug-resistant V82F/I84V mutant of HIV-1 protease (1D4S.pdb) in all-atom mode with thousands of explicit waters present. The mutant structure 1D4S.pdb is the Tipranavir-bound conformation, and it contains the V82F and I84V active site mutations that significantly decrease the affinities of all HIV-1 protease inhibitors currently used clinically. The wild-type structure 1KZK.pdb is the JE-2147–bound conformation of protease. These studies began with crystal structures of the protease : inhibitor complexes, but the inhibitors were deleted prior to initiating these MD simulations. Deletion of the inhibitors before performing these MD studies on the closed forms of the apo protease molecules facilitated the large flap motions that were observed, because the apo protease thermodynamically favors the semiopen form. Large displacements of the active site flaps were observed, which included the structural transition wherein the flaps opened from the closed conformation to a semiopen form.
Previous simulations in which flap opening was assessed often used the same definition to monitor flap behavior: the distance between the α Carbons of I50 and I150. However, the tip of each flap can curl in an independent and asymmetric fashion, and the full flaps also move asymmetrically. Even though the MD simulations performed by Schiffer’s group on the apo HIV-1 protease began with a perfectly symmetrical structure (the asymmetric unit of the semiopen crystal structure used was, in fact, a monomer rather than the dimer), the flaps behaved asymmetrically at any one point in time (Scott and Schiffer 2000). Similarly, in crystal structures of HIV-1 protease complexed with perfectly symmetrical inhibitors, differences of 0.2 to 0.6 Å were observed between the two monomers for flap–drug and for drug–catalytic Asp distances (Ala et al. 1997). Calculations (Kurt et al. 2003) using the Gaussian Network Model (GNM) to study the dynamics of HIV-1 protease also indicated asymmetric flap motion occurs: In calculations of apo HIV-1 protease, either one flap or the other had more mobility at different snapshots. It should be mentioned that those GNM calculations were performed using structures of snapshots from Schiffer’s conventional MD run (Kurt et al. 2003). Because the I50–I150 distance value could be affected by both the asymmetric motion of the flaps and by the curling behavior of the flap tips, different distance measurements were used to analyze these trajectories (see Figs. 6 ▶, 7 ▶, 12 ▶ , and 13 ▶).
As is apparent from Figures 6 ▶, 7 ▶, 8 ▶, 10 ▶, 12 ▶ , and 13 ▶, the mutant’s flap–Asp values always sampled significantly larger distances relative to the wild-type’s simulation. Thus, no matter how the motion of the flaps was monitored, all measurements indicated that this drug-resistant mutant’s flaps opened several Angstroms farther than the wild-type’s flaps during the same time period. However, by analyzing certain distance values, both the wild type and the mutant could be classified as having reached the semiopen conformations (see the graph on the left in Fig. 6 ▶), while the use of other distance values to define the semiopen form caused the conclusion that only the mutant reached the semiopen conformations (see Figs. 12 ▶ and 13 ▶, and the graph on the right in Fig. 6 ▶). One possible explanation for that observation could be that perhaps the wild-type’s sampling behavior reflects transient fluctuations involved in trying to overcome whatever energetic or kinetic barriers impede reaching the semiopen conformation’s local energetic minimum, while the values of the mutant’s histogram peaks indicate that it did actually reach and begin sampling a local minimum encompassing some of the semiopen conformations. That is, the wild-type’s snapshots with large flap–Asp distance values could mainly correspond to extreme fluctuations within the local minima describing the closed conformations and perhaps the first transition structure along the way to reaching the semiopen form, whereas the mutant’s simulation produced a significant number of snapshots that were clearly sampling the semiopen conformations. A simpler explanation for that observation (which is likely related to the aforementioned explanation) is that one of the wild-type’s flaps briefly sampled values corresponding to the semiopen conformations, while both of the mutant’s flaps extensively sampled the semiopen conformations (see Fig. 10 ▶). Thus, the definitions of the semiopen form that involved only that one particular flap caused the conclusion that both the wild-type and mutant systems sampled semiopen conformations (see Fig. 6 ▶), but the definitions that involved the other flap or that involved the center of mass of both flap tips caused the conclusion that the wild type did not reach the semiopen form (see Figs. 6 ▶, 12 ▶ , and 13 ▶), because one of the wild-type’s flaps did not quite open enough to sample semiopen distance values. The most significant point is that regardless of the particular distance that was used to define the semiopen form, this drug-resistant mutant ensemble always had significantly more snapshots in the semiopen conformations than the wild-type ensemble had.
The specific details of individual dynamic relationships were quite insightful, such as the differences observed in the curling behavior of the flap tips of the mutant compared to the wild type. In these simulations the mutant protease displayed more frequent and more rapid curling of its flap tips (see Fig. 3 ▶). NMR studies by Torchia’s group (Ishima et al. 1999; Freedberg et al. 2002) showed that significant subnanosecond motion does indeed occur at the flap tips, and that fluctuations of F53 (which is at the flap tip) are likely coupled to motion of the entire flap backbone. Simulations from Schiffer’s group (Scott and Schiffer 2000) observed that the curling of the flap tips produces flap opening. Because this mutant’s flap tips experienced more rapid and more frequent curling behavior, it was expected that this mutant’s active site flaps would open more than the wild-type’s flaps. By all forms of measurement attempted, this mutant’s flaps did open significantly farther than the wild-type’s flaps, and the mutant’s flap displacements were much more erratic. This suggests that the fact that more flap opening was displayed in the mutant’s simulation than in the wild type’s might correspond to differences in equilibrium preferences.
Considering that the unfavorable enthalpic cost involved in ligand binding is due to closing and rearranging the flaps (Luque et al. 1998), if this mutant does actually prefer the semiopen conformations more than the wild type favors them, then that could be a general feature contributing to its drug resistance properties, because all of the drugs would have to pay a larger enthalpic cost to close the mutant’s flaps.
By comparing different active site distances to different peripheral distances in one ensemble or the other, an observation was made that compression of the Ear to Cheek region was correlated with opening of the flaps (see Fig. 15 ▶). Rigorous quantitative analyses performed by other groups on the results of several different MD simulations provide support for the notion that the Ear–Cheek and flap–Asp displacements can display an anticorrelated relationship. From the Gaussian Network Model calculations performed on the apo HIV-1 protease, it appears that in the third and fourth slowest principal modes of motion, each active site flap was anticorrelated with the motion of the Ear Flap of the same monomer (Kurt et al. 2003). The normal mode analyses performed by the Karplus group on the plethora of MD simulations that were ran on different complexes of HIV-1 protease and on the closed form of the apo HIV-1 protease also support the notion of this anticorrelated relationship (Zoete et al. 2002). In Figure 17 of that paper, one normal mode calculated from the simulation of the closed form of apo HIV-1 protease shows that each Ear Flap displayed anticorrelated motion with respect to the active site flap to which it is connected (Zoete et al. 2002). In the raw data in Figures 19B, 20A and B, and 21A of that paper, the Dynamic Cross Correlation Maps (DCCMs) also indicate that the motion of each Ear Flap tip can be anticorrelated with respect to the motion of its adjacent active site flap tip (Zoete et al. 2002). In the simulations by Piana et al. (2002a), when harmonic constraints were applied to the Cheek–Ear region that pushed apart the α C’s of residues 68–39, 168–139, 69–40, and 169–140 for a 0.35-nsec MD run, this caused a motion of the substrate towards the catalytic aspartates. When the Ear–Cheek region was forced to expand, the flaps were pushed down towards the catalytic aspartates (Piana et al. 2002a). In Figure 16 ▶ it shows that a comparison of the extrema for that flap–Asp distance from throughout the wild-type simulation supports that anticorrelated relationship as well. The wild-type snapshot with the smallest flap–Asp distance shown also had the largest Ear–Cheek distance. Similarly, Figure 17 ▶ indicates that the local extrema involved in that particular opening event also display this anticorrelated trend. In the opening event that reached the wild-type’s global maximum, the snapshot with the largest flap–Asp distance had the most compressed Ear–Cheek region. Thus, the rigorous quantitative analyses published by other groups and the qualitative analyses of the results of these simulations all support the notion that the motion of the tips of the Ear Flaps can be anticorrelated with respect to the motion of the active site flap tips.
Figure 16.
A comparison of the most open snapshots from the wild-type’s simulation to the most closed snapshot supports that anticorrelated relationship: The solid ribbon in cyan represents the wild-type’s snapshot that displayed the second smallest value for the I50–D25 distance (cyan = 19,471 psec, value = 10.7 Å; the absolute minimum was snapshot 17,505, with a value of 10.6 Å), while the monomer on the left with a solid orange ribbon shows the semiopen conformation of the 1HHP crystal structure of the apo protease. The ribbons shown in line mode depict the local maxima for the distance between I50 and D25 from the wild-type’s simulation. These maxima were the peaks from different flap opening events. The following snapshots are displayed: 11,353 = red (I50–D25 distance was 18.1 Å = the global max.), 11,574 = magenta (17.3 Å), 8647 = blue (16.6 Å). For the corresponding maxima from the mutant’s simulation, see Fig. 11 ▶. The snapshot with the smallest distance between its flap and Asp 25 (i.e., the cyan ribbon) has the largest distance between its Ear and Cheek regions.
Figure 17.
A comparison of the local extrema sampled during the opening event that reached the wild-type’s global maximum supports that anticorrelated relationship: The ribbon diagrams show snapshots of the local extrema involved in the flap opening event that proceeded from an I50–D25 value of 13.4 Å at 11,227 psec (the red ribbon) to a value of 18.1 Å at 11,353 psec (the purple ribbon). The other snapshots are as follows: 11,273 = orange (17.6 Å) and 11,284 = green (14.5 Å). Examine the left side of the figure. Note that the conformation with the largest flap–Asp distance (the purple ribbon) also has the most compressed Ear–Cheek region.
The presence of that anticorrelated relationship suggests a new site on the surface of HIV-1 protease that potentially could be used to design a new class of drugs to help treat HIV infections. The apparent anticorrelation of the flap and Ear displacements from the HIV protease core points to the Ear–Cheek interface region as a potential new target against which allosteric inhibitors could be designed. Although others have proposed the idea of designing allosteric inhibitors to regulate HIV protease dynamics, the idea of targeting the Ear–Cheek region has apparently not been raised. If compression of the Ear–Cheek region does correlate with flap opening in vivo, then an inhibitor that binds to the Ear–Cheek region could stabilize either the pinched or the expanded Ear–Cheek conformations. If a new class of inhibitors (e.g., class AFO = Allosteric-Flap-Openers) can be developed that binds to and stabilizes the subset of the ensemble with pinched Ear–Cheek distances, then those inhibitors could help keep the active site flaps in the semiopen to fully open conformations, which would help prevent the protease enzyme from ever becoming catalytically competent.
Alternatively, if a second class of allosteric inhibitors (e.g., class AFC = Allosteric-Flap-Closers) can be developed that binds to and stabilizes the conformations with expanded Ear–Cheek distances, then those inhibitors could help close the flaps and/or help keep the flaps closed. Because the massive enthalpic penalty paid by the active site protease inhibitors involves the cost of closing the active site flaps (Luque et al. 1998), then allosteric inhibitors that help close the flaps could help pay that enthalpic cost, which could improve the binding affinities of all active site protease inhibitors currently used clinically. That is, double protease cocktails involving two entirely different targets on HIV protease (i.e., the active site and the Ear–Cheek interface) could perhaps have synergistic effects. Similarly, if AFC inhibitors could be developed that help keep the flaps closed, then the release of products from the active site would be impeded, which would help prevent that protease molecule from undergoing another catalytic cycle.
Any inhibitor that binds to the Ear–Cheek interface region and disturbs or exploits the normal dynamic relationships involved in correlating the motions of the active site flaps to the rest of the protease enzyme would likely be a useful inhibitor. This potential new site is proposed publicly to guide drug design studies in the future towards the goal of developing allosteric inhibitors to regulate motion of the active site flaps. If such allosteric inhibitors can be created, that should aid in the evasion of the drug resistance that continually develops.
Materials and methods
Starting structures/protein extraction
The coordinates of the two complexes of HIV-1 protease (1KZK.pdb = the wild-type HIV-1 protease:JE-2147 complex and 1D4S.pdb = the V82F/I84V double mutant HIV-1 protease:Tipranavir complex) were downloaded from the Protein Data Bank (Thaisrivongs et al. 1996; Berman et al. 2000; Reiling et al. 2002). Although the two systems are referred to as “wild type” and “double mutant,” there are actually eight differences in their amino acid sequence/monomer. The mutant has only two drug-resistance mutations—V82F and I84V—but it also has the N37S substitution, which is a natural polymorphism. The “wild-type” system has the Q7K substitution that is used to decrease autoproteolysis (Q7K is present in many to most constructs that are used for structural, kinetic, and binding affinity studies), and the wild type also has the natural polymorphisms K14R, R41K, and I64V. The “wild-type” system also has the minor mutation L63P, which has little effect on binding affinity, and which is present in 56% of the isolates whose sequences are deposited in Stanford’s HIV DataBase (http://hivdb.stanford.edu/), which contains nearly all published sequences, including those in GenBank, as well as isolates from people participating in clinical trials (Rhee et al. 2003). Thus, the only significant differences between the primary structures of the two systems studied here are expected to be the presence of the V82F and I84V drug-resistance mutations in the mutant system. The locations of those mutations and a possible new terminology for the topology of HIV-1 protease are displayed in Figure 1 ▶.
Protein preparation and simulation details
The preparation of both of the protease systems was identical, and it followed a protocol established previously (Lin et al. 2003). The crystal structures 1KZK and 1D4S were obtained from the PDB, the inhibitors were deleted from the active sites, and hydrogens were added using the WHATIF program (Nielsen et al. 1999) of the PDB2PQR service (Hooft et al. 1996). The crystallographic water molecules and the chloride counterions from each crystal structure were maintained throughout the preparation process, and solvation of the proteins with additional TIP3P waters (Jorgensen et al. 1983) was then done using LEaP (Schafmeister et al. 1995). The buffer distance chosen was 10 Å for both systems, which means that there is at least a 10 Å-thick layer of water between each protein and the edges of its cubic box. To achieve electro-neutrality of the system, a few of the bulk waters were manually changed into Cl− counterions. Thus, the wild-type protease dimer was surrounded by 8464 water molecules (223 were crystallographic) and 6 Cl− ions (which makes the total size of the system 28,526 atoms) during the simulations, while the drug-resistant mutant protease dimer was encased in 9614 water molecules (133 were crystallographic) and 4 Cl− ions (for a total system size of 31,976 atoms). The difference in the number of water molecules added by LEaP to each system was due to differences in the orientation of each protein within its cubic box.
A restrained energy minimization was performed on each of those two protease systems using the SANDER module of AMBER7 (Case et al. 2002; note: The version of SANDER used was modified and recompiled by Jung-Hsin Lin to improve its utilization of the NMR-type restraints). For both the minimizations and the subsequent MD runs, the long-range Coulombic interactions were handled by the particle mesh Ewald method (Essmann et al. 1995) with cubic spline approximation (the direct sum tolerance was set to 0.00001). The minimizations were performed for 500 iterations of steepest descent, and a force constant of 30 kcal/mole/Å2 was applied to the restrained atoms. During those minimizations, the heavy atoms of the proteins and the oxygen atoms of the crystallographic waters were restrained, which allowed for the optimization of both the hydrogens (which were added both to the proteins and to the crystallographic waters) as well as the optimization of the thousands of waters added by LEaP.
After the restrained minimizations, Equilibration Molecular Dynamics (EqMD) runs were performed to gently heat the systems and to relax them to their proper densities. These EqMD runs used the exact same restraints that were used in the minimizations. Because the SHAKE algorithm (Ryckaert et al. 1977) was used in the EqMD runs and in the subsequent production quality MD runs, the hydrogen atoms had their bond stretching degrees of freedom prohibited, which allowed for the use of a 2-fsec time step. The minimized structures were used as the reference structures for the Equilibration MD runs, which caused the proteins to maintain their original structures while the solvent environments were allowed to reach their proper densities.
The heating and restraint procedure for EqMD was as follows: During 0–10 psec, the temperature was increased linearly from 50 to 100 K while a force constant of 30 kcal/mole/Å2 was applied to all of the restrained atoms. During 10–20 psec, the temperature was increased from 100 to 200 K (the same force constant was still being applied). For the 20–30-psec period, the temperature was increased from 200 to 300 K, while that same force constant was still being applied to the restrained atoms. The temperature was maintained at 300 K for the rest of the EqMD runs and for the entire production phase of the MD runs. That same force constant was still applied during 30–40 psec, and then the force constant linearly decreased to zero during 40–60 psec. The last 40 psec of EqMD (i.e., 60–100 psec) were performed without any restraints.
At the end of the EqMD runs, the density of the wild-type system was 1.016 g/cc, while the density of the mutant system was 1.015 g/cc. The restart files from those EqMD runs were then used as the inputs for all of the subsequent production quality MD runs. These long-scale MD runs (i.e., 22 nsec each for both systems) were performed at 300 K using the constant NPT conditions (isothermal-isobaric) on two 2-GHz Xeon processors, which produced ~70 psec of MD/day. The reference pressure was set equal to 1 Bar, and the Berendsen barostat (Berendsen et al. 1984) was used with the pressure-coupling constant of 0.1 psec. The temperature was maintained at 300 K using the Berendsen thermostat (Berendsen et al. 1984) with the coupling constant of 0.2 psec. The center of mass motion was removed at the end of each picosecond, and solvent and solute atoms were coupled to a single thermostat.
Acknowledgments
We thank Dr. Gennady Verkhivker of Pfizer Global Research and Development (La Jolla site) for his discussions concerning the biological details of the HIV protease system. We thank Dr. David A. Case and the late Dr. Peter A. Kollman for providing early access to AMBER 7. We thank Dr. Chung F. Wong, Dr. Steve Bond, and Dr. Robert Konecny for their assistance. We thank the Howard Hughes Medical Institute and the W.M. Keck Foundation for their continued and generous financial support. We thank Accelrys, Inc. for generously providing their InsightII software, which was used to make some of the figures. Additional support has been provided, in part, by grants to JAM from NIH, NSF, NPACI/SDSC, NBCR, and UCSD’s NSF Center for Theoretical Biological Physics. We thank Dr. Donald Hamelberg and Dr. Valentina Tozzini for proofreading assistance. We thank Dr. Tongye Shen and Dr. Elizabeth Komives for useful discussions. A.L.P. is an HHMI Predoctoral Fellow in the Biomedical Sciences Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material: See www.proteinscience.org
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03468904.
References
- Ala, P.J., Huston, E.E., Klabe, R.M., McCabe, D.D., Duke, J.L., Rizzo, C.J., Korant, B.D., DeLoskey, R.J., Lam, P.Y.S., Hodge, C.N., et al. 1997. Molecular basis of HIV-1 protease drug resistance: Structural analysis of mutant proteases complexed with cyclic urea inhibitors. Biochemistry 36 1573–1580. [DOI] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem.Phys. 81 3684–3690. [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D.A., Pearlman, D.A., Caldwell, J.W., Cheatham III, T.E., Wang J., Ross, W.S., Simmerling, C., Darden, T., Merz, K.M., Stanton, R.V., et al. 2002. AMBER 7. University of California, San Francisco.
- Collins, J.R., Burt, S.K., and Erickson, J.W. 1995. Flap opening in HIV-1 protease simulated by “activated” molecular dynamics. Nat. Struct. Biol. 2 334–338. [DOI] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M.L., and Darden, T. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103 8577–8593. [Google Scholar]
- Freedberg, D.I., Ishima, R., Jacob, J., Wang, Y.-X., Kustanovich, I., Louis, J.M., and Torchia, D.A. 2002. Rapid structural fluctuations of the free HIV protease flaps in solution: Relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 11 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, C.N., Straatsma, T.P., and McCammon, J.A. 1997. Rational design of HIV protease inhibitors. In Structural biology of viruses (eds. W. Chiu et al.), pp. 451–473. Oxford University Press, Oxford, UK.
- Hooft, R.W., Sander, C., and Vriend, G. 1996. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins Struct. Funct. Genet. 26 363–376. [DOI] [PubMed] [Google Scholar]
- Ishima, R., Freedberg, D.I., Wang, Y.-X., Louis, J.M., and Torchia, D.A. 1999. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Struct. Fold. Des. 7 1047–1055. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar, J., and Madura, J.D. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79 926–935. [Google Scholar]
- Katoh, E., Louis, J.M., Yamazaki, T., Gronenborn, A.M., Torchia, D.A., and Ishima, R. 2003. A solution NMR study of the binding kinetics and the internal dynamics of an HIV-1 protease–substrate complex. Protein Sci. 12 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabe, R.M., Bacheler, L.T., Ala, P.J., Erickson-Viitanen, S., and Meek, J.L. 1998. Resistance to HIV protease inhibitors: A comparison of enzyme inhibition and antiviral potency. Biochemistry 37 8735–8742. [DOI] [PubMed] [Google Scholar]
- Kurt, N., Scott, W.R.P., Schiffer, C.A., and Haliloglu, T. 2003. Cooperative fluctuations of unliganded and substrate-bound HIV-1 protease: A structure-based analysis on a variety of conformations from crystallography and molecular dynamics simulations. Proteins Struct. Funct. Genet. 51 409–422. [DOI] [PubMed] [Google Scholar]
- Lin, J.-H., Perryman, A.L., Schames, J.R., and McCammon, J.A. 2002. Computational drug design accommodating receptor flexibility: The Relaxed Complex scheme. JACS 124 5632–5633. [DOI] [PubMed] [Google Scholar]
- ———. 2003. The Relaxed Complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers 68 47–62. [DOI] [PubMed] [Google Scholar]
- Little, S.J., Holte, S., Routy, J.P., Daar, E.S., Markowitz, M., Collier, A.C., Koup, R.A., Mellors, J.W., Connick, E., Conway, B., et al. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347 385–394. [DOI] [PubMed] [Google Scholar]
- Luque, I., Todd, M.J., Gomez, J., Semo, N., and Freire, E. 1998. Molecular basis of resistance to HIV-1 protease inhibition: A plausible hypothesis. Biochemistry 37 5791–5797. [DOI] [PubMed] [Google Scholar]
- Mahalingam, B., Louis, J.M., Hung, J., Harrison, R.W., and Weber, I.T. 2001. Structural implications of drug-resistant mutants of HIV-1 protease: High-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins Struct. Funct. Genet. 43 455–464. [DOI] [PubMed] [Google Scholar]
- Mahalingam, B., Boross, P., Wang, Y.-F., Louis, J.M., Fischer, C.C., Tozser, J., Harrison, R.W., and Weber, I.T. 2002. Combining mutations in HIV-1 protease to understand mechanisms of resistance. Proteins Struct. Funct. Genet. 48 107–116. [DOI] [PubMed] [Google Scholar]
- Maschera, B., Darby, G., Palu, G., Wright, L.L., Tisdale, M., Myers, R., Blair, E.D., and Fufine, E.S. 1996. Human immunodeficiency virus: Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease–saquinavir complex. J. Biol. Chem. 271 33231–33235. [DOI] [PubMed] [Google Scholar]
- Nielsen, J.E., Andersen, K.V., Honig, B., Hooft, R.W., Klebe, G., Vriend, G., and Wade, R.C. 1999. Improving macromolecular electrostatics calculations. Protein Eng. 12 657–662. [DOI] [PubMed] [Google Scholar]
- Ohtaka, H., Velazquez-Campoy, A., Xie, D., and Freire, E. 2002. Overcoming drug resistance in HIV-1 chemotherapy: The binding thermodynamics of Amprenavir and TMC-126 to wild-type and drug-resistant mutants of HIV-1 protease. Protein Sci. 11 1908–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana, S., Carloni, P., and Parrinello, M. 2002a. Role of conformational fluctuations in the enzymatic reaction of HIV-1 protease. J. Mol. Biol. 319 567–583. [DOI] [PubMed] [Google Scholar]
- Piana, S., Carloni, P., and Rothlisberger, U. 2002b. Drug resistance in HIV-1 protease: Flexibility-assisted mechanism of compensatory mutations. Protein Sci. 11 2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabu-Jeyabalan, M., Nalivaika, E., and Schiffer, C.A. 2000. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 301 1207–1220. [DOI] [PubMed] [Google Scholar]
- Reiling, K.K., Endres, N.F., Dauber, D.S., Craik, C.S., and Stroud, R.M. 2002. Anisotropic dynamics of the JE-2147-HIV protease complex: Drug resistance and thermodynamic binding mode examined in a 1.09 A structure. Biochemistry 41 4582–4594. [DOI] [PubMed] [Google Scholar]
- Rhee, S.-Y., Gonzales, M.J., Kantor, R., Betts, B.J., Ravela, J., and Shafer, R.W. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, S.W., Erickson, J.W., and Burt, S.K. 1998. Reaction path and free energy calculations of the transition between alternate conformations of HIV-1 protease. Proteins 32 7–16. [DOI] [PubMed] [Google Scholar]
- Rose, R.B., Craik, C.S., and Stroud, R.M. 1998. Domain flexibility in retroviral proteases: Structural implications for drug resistance mutations. Biochemistry 37 2607–2621. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.-P., Ciccotti, G., and Berendsen, H.J.C. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23 327–341. [Google Scholar]
- Schafmeister, C.E.A.F., Ross, W.S, and Romanovski, V. 1995. LEaP. University of California, San Francisco, CA.
- Scott, W.R. and Schiffer, C.A. 2000. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Struct. Fold. Des. 8 1259–1265. [DOI] [PubMed] [Google Scholar]
- Spinelli, S., Liu, Q.Z., Alzari, P.M., Hirel, P.H., and Poljak, R.J. 1991. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochemie 73 1391–1396. [DOI] [PubMed] [Google Scholar]
- Thaisrivongs, S., Skulnick, H.I., Turner, S.R., Strohback, J.W., Tommasi, R.A., Johnson, P.D., Aristoff, P.A., Judge, T.M., Gammill, R.B., Morris, J.K., et al. 1996. Structure-based design of HIV protease inhibitors: Sulfonamide-containing 5,6-dihydro-4-hydroxy-2-pyrones as non-peptidic inhibitors. J. Med. Chem. 39 4349–4353. [DOI] [PubMed] [Google Scholar]
- Wang, W. and Kollman, P.A. 2001. Computational study of protein specificity: The molecular basis of HIV-1 protease drug resistance. Proc. Natl. Acad. Sci. 98 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Michielin, O., and Karplus, M. 2002. Relation between sequence and structure of HIV-1 protease inhibitor complexes: A model system for the analysis of protein flexibility. J. Mol. Biol. 315 21–52. [DOI] [PubMed] [Google Scholar]