Abstract
Membrane protein topology predictions can be markedly improved by the inclusion of even very limited experimental information. We have recently introduced an approach for the production of reliable topology models based on a combination of experimental determination of the location (cytoplasmic or periplasmic) of a protein’s C terminus and topology prediction. Here, we show that determination of the location of a protein’s C terminus, rather than some internal loop, is the best strategy for large-scale topology mapping studies. We further report experimentally based topology models for 31 Escherichia coli inner membrane proteins, using methodology suitable for genome-scale studies.
Keywords: membrane proteins, topology, prediction, bioinformatics, fusion protein, PhoA, green fluorescent protein, GFP
The topology of an integral membrane protein—that is, a specification of the membrane-spanning segments of the polypeptide chain and their in/out orientation relative to the membrane—is a basic characteristic that helps guide experimental studies in the absence of a high-resolution 3D structural model. A number of experimental approaches to topology mapping such as reporter fusions (Manoil 1991), Cys-labeling (Kimura et al. 1997), epitope mapping (Canfield and Levenson 1993), glycosylation mapping (Chang et al. 1994), and limited proteolysis (Wilkinson et al. 1996) have been developed over the years. In addition, theoretical topology prediction methods perform reasonably well, but still leave much room for improvement (Chen et al. 2002).
We have recently shown that the performance of topology prediction methods can be substantially improved by the inclusion of limited experimental information such as the location of the N- or C-terminal end of the protein relative to the membrane (Drew et al. 2002; Melén et al. 2003). In a typical experiment, C-terminal reporter fusions are made to the target protein, and the cytoplasmic/noncytoplasmic location of the C-terminal end of the protein is deduced from the experimental results. This information is then used as a constraint in the topology prediction step. Proof-of-principle studies have been carried out both in Escherichia coli (Drew et al. 2002) and Saccharomyces cerevisiae (Kim et al. 2003).
So far, studies of this kind have been limited to C-terminal reporter protein fusions, where the full-length target protein is fused to the reporters. However, it is not immediately apparent if full-length fusions are always the most informative; for instance, it could well be that more accurate predictions would be obtained if the location of an internal loop segment in a protein is mapped instead. Here, we have tested this possibility within the framework of the TMHMM topology prediction method (Krogh et al. 2001), and have tried various rules based on the output from an unconstrained TMHMM prediction to identify the best fusion point in a protein. Interestingly, we find that full-length C-terminal reporter fusions give better constraints for the TMHMM prediction than fusions to predicted loops. From an experimental point of view, fusions to full-length target proteins are also preferred over internal fusions because the risk of artifactual results is reduced.
Encouraged by this result, we have made a new set of 34 C-terminal reporter fusions to E. coli inner membrane proteins, and present topology models for 31 of these (including two control proteins with previously known topologies). From our data, we estimate that around 90% of the ~770 proteins with two or more transmembrane helices predicted to be present in the inner membrane proteome of E. coli (Krogh et al. 2001) should be amenable to this kind of analysis.
Results
Optimal placement of a topology reporter
We have based our analysis on an up-to-date data set of helix bundle-type membrane proteins with experimentally determined topologies (129 prokaryotic, 92 eukaryotic, 12 viral, and archaeal proteins; L. Käll, A. Krogh, and E.L.L. Sonnhammer, in prep.). When TMHMM was applied to this data set, the predicted topology was correct for 161 of the 233 proteins (69%; Table 1). Of the 72 incorrect predictions, 52 had an incorrect number of transmembrane helices (39 underpredictions and 13 overpredictions), 19 had the correct number of transmembrane helices but the wrong overall orientation relative to the membrane, and one had the correct number of transmembrane helices and the correct overall orientation but one of the predicted transmembrane helices did not overlap by at least five residues with the corresponding experimentally determined helix (which was our criterion for a correctly predicted helix). The most common error made by TMHMM is thus an incorrect number of transmembrane helices, with underpredictions being more frequent than overpredictions. There were no significant differences in the performance of TMHMM on the prokaryotic versus the eukaryotic proteins (data not shown).
Table 1.
TMHMM prediction performance for various choices of the residue to be constrained to its experimentally known location
| Reference predictions | Percent correctly predicted topologies |
| Unconstrained predictions | 69 |
| Maximum obtainable accuracy | 89 |
| Rules | |
| Always fix N terminus | 79 |
| Always fix C terminus | 81 |
| Fix terminal residue of the largest of the N- and C-terminal regions | 82 |
| Fix the terminal residue with lowest prediction probability | 82 |
| Fix the loop residue with lowest prediction probability (LPLR) | 70 |
The “maximum obtainable accuracy” is an upper bound, because the topology is never correctly predicted for 26 of the 233 test set proteins no matter which residue is fixed to its known location.
To obtain an upper bound on the possible improvement in prediction performance obtainable by experimental determination of the in/out location of one and only one loop or tail residue in the target protein, we ran TMHMM on each protein with each loop or tail residue in turn fixed to its experimentally annotated location. Fourteen of the 72 proteins for which the topology was incorrectly predicted by the unconstrained TMHMM method were correctly predicted no matter which residue was fixed (in all these cases, the unconstrained prediction had the correct number of transmembrane helices but an incorrect overall orientation, and fixing any single loop residue gave the correct orientation). Twenty-six proteins were never correctly predicted no matter which loop or tail residue was fixed. For the remaining 32 proteins, the correct topology prediction was obtained only when some specific residues were fixed to their known location. Thus, the maximum number of correct predictions obtainable by fixing one and only one loop or tail residue in each of the 233 proteins in the data set to its experimentally known location is 207 (89%; Table 1). This upper bound is slightly higher for the prokaryotic proteins (91%) compared to the eukaryotic ones (85%).
We next tried different rules for choosing the residue to be fixed when the only prior topology information available is the unconstrained TMHMM prediction. The first two rules were the same as described previously (Melén et al. 2003), that is, to fix either the N- or the C-terminal residue of the target protein. As shown in Table 1, there was only a slight difference between these two choices: When the N terminus was fixed, the topology prediction was correct for 79% of the 233 proteins in the data set, and when the C terminus was fixed, the prediction was correct for 81%.
The upper bound for the prediction performance obtainable is 85% if, for each protein, one could choose to determine the location of either the N or the C terminus, but we were unable to find a rule for choosing which terminus to fix based on the unconstrained TMHMM prediction that reached this level. By choosing to fix the terminus that belongs to the longest of the two predicted terminal nontransmembrane domains, or to fix the terminus with the lowest prediction probability, we reached a prediction performance of 82%, an insignificant increase over the results obtained when the C terminus was always chosen.
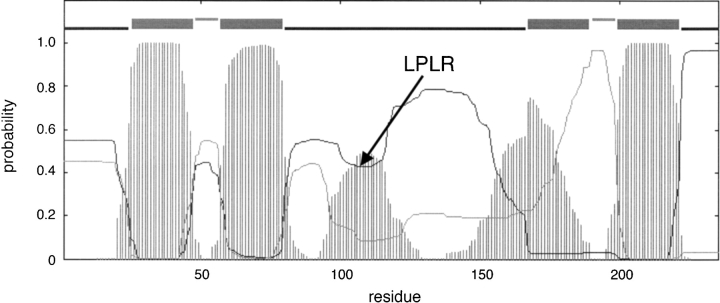
The TMHMM output includes the estimated posterior probabilities for each residue in the protein to be in an inside loop, an outside loop, or in a transmembrane segment (Krogh et al. 2001; Melén et al. 2003). We reasoned that the highest increase in prediction performance might be obtained by determining the location of the loop residue in the protein that has the lowest posterior probability for its predicted location (Fig. 1 ▶, point LPLR—lowest probability loop residue). For each protein we made an unconstrained prediction, identified the LPLR, and then fixed this residue to its known location. Other variations on this rule that were tested included fixing the first or last residue of the predicted loop containing the LPLR, or fixing a residue a given distance away from the ends of the predicted loop containing the LPLR. However, all rules based on the LPLR resulted in much poorer prediction performances than the N-and C-terminal rules discussed above: The best rule gave only 70% correct predictions (Table 1). The main reason for this poor performance is that the LPLR, which, while being predicted as belonging to a loop, in fact often belongs to a transmembrane segment (as exemplified in Fig. 1 ▶). Because all available experimental methods for topology mapping work only for loop regions, one cannot recover from this type of error.
Figure 1.
TMHMM topology prediction and probability profile for protein HISM_SALTY. The top line shows the predicted topology with the four predicted transmembrane helices. The thin black and gray curves show the posterior probabilities for inside and outside loop, respectively. The striped profile shows the probability for transmembrane helix. The lowest probability loop residue (LPLR) is indicated. The probability value for the LPLR (residue 108) is 0.43, reflecting the uncertainty in the topology predicted for this region. In the experimentally determined topology (Kerppola and Ames 1992), the LPLR is located in a transmembrane helix that is missed in the TMHMM prediction.
Experimental determination of C-terminal locations
Encouraged by the finding that results from C-terminal reporter fusions to full-length target proteins seem to yield the most consistent improvement in prediction performance, we have expanded our previous set of 12 E. coli inner membrane proteins analyzed in this way (Drew et al. 2002) to include an additional 34 proteins.
These new target proteins were selected as in the previous study, that is, by applying three criteria: (1) All target proteins must have two or more transmembrane helices predicted by TMHMM; (2) five different topology prediction methods (see Materials and Methods) must agree on the periplasmic or cytoplasmic location of the N terminus; and (3) the five prediction methods must collectively predict only one or two different topologies, and these must differ by no more than one predicted transmembrane segment. The first criterion is necessary to avoid problems with secretory proteins being mistakenly identified as integral membrane proteins; the second criterion ensures that the location of the N terminus is predicted with very high reliability, which, together with the third criterion, means that a reliable topology model can be indicated once the location of the C terminus has been experimentally determined, either by a “consensus” approach (Nilsson et al. 2000; Drew et al. 2002), or by applying TMHMM alone (Kim et al. 2003; Melén et al. 2003).
For two of the 34 target proteins (BtuC and SdhC), an experimentally determined 3D structure was available (Locher et al. 2002; Yankovskaya et al. 2003). These proteins were included in the study as internal controls for the reliability of our approach.
As before, we used two topology reporters: alkaline phosphatase (PhoA) and green fluorescent protein (GFP). PhoA can fold into an enzymatically active conformation only when located in the periplasm (Manoil and Beckwith 1986). In contrast, GFP becomes fluorescent only when located in the cytoplasm and not in the periplasmic space (Feilmeier et al. 2000; Drew et al. 2002). The location of a protein’s C terminus can thus be determined by making both a PhoA and a GFP fusion to the C terminus: A high PhoA and low GFP activity indicates a periplasmic location, while a low PhoA and high GFP activity indicates a cytoplasmic location. If both or neither of the fusions show significant activity, no conclusion can be drawn.
For the PhoA fusions, we constructed two vectors that differ in the restriction sites available for cloning. Similarly, for the GFP fusions, we used two vectors with different combinations of restriction sites. One of the GFP vectors also has a TEV-protease site in the linker between the target protein and GFP, together with a C-terminal His8-tag. The restriction sites in the different vectors were chosen based on the gene sequences of the 770 E. coli inner membrane proteins for which TMHMM predicts a minimum of two transmembrane helices (Krogh et al. 2001); the vectors will ultimately allow PhoA and GFP fusions to be made to around 95% of these proteins. As another crucial step towards large-scale topology mapping projects, we adapted the PhoA and GFP activity measurements to a microtiter plate format for rapid analysis (see Materials and Methods).
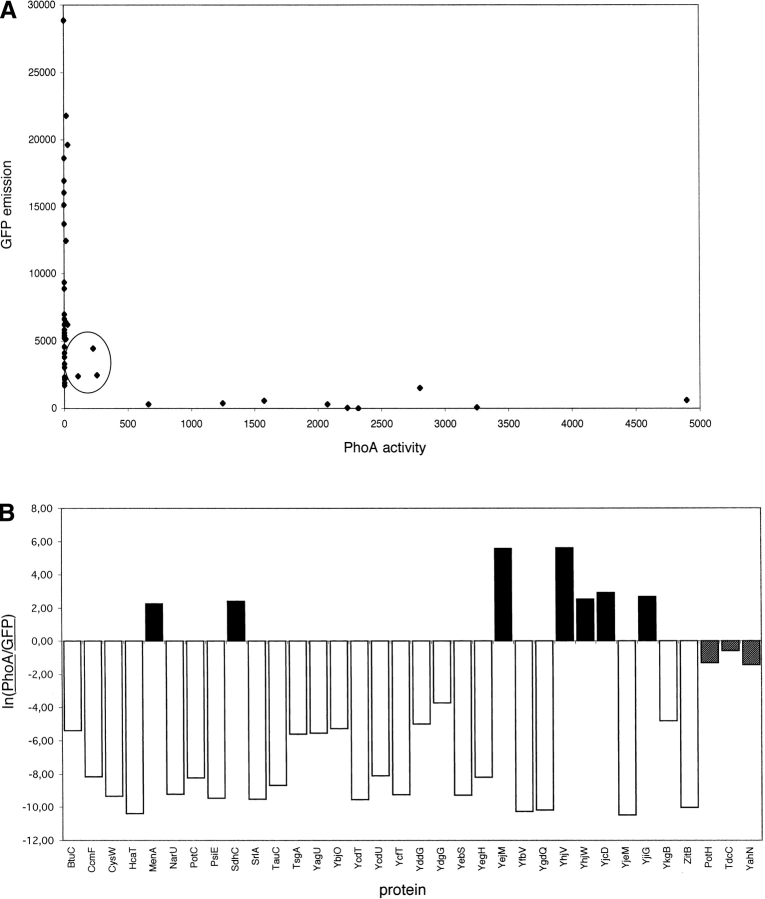
The PhoA activity and the GFP fluorescence emission for each of the 34 proteins are shown in Figure 2A ▶. To allow a quantitative comparison between the PhoA and GFP activities, we further normalized all PhoA measurements to the mean PhoA activity measured for the “active” fusions (those with PhoA activity higher than 500 units, not counting the three encircled fusions in Fig. 2A ▶), and likewise for the “active” GFP fusions (those with a PhoA activity lower than 100 and a GFP emission intensity higher than 600 units). We then calculated the quotient between the normalized PhoA and GFP activities, and finally plotted the natural logarithm of this quotient for each protein (Fig. 2B ▶). From this plot, the location of the C terminus can be read off for 31 of the 34 proteins, while for three proteins (PotH, TdcC, and YahN; encircled in Fig. 2A ▶, gray bars in Fig. 2B ▶) the data do not allow an unambiguous assignment. As found already in our previous studies (Drew et al. 2002; Kim et al. 2003), and as predicted by TMHMM (Krogh et al. 2001), the majority of the proteins (24 of 31) have the C terminus in the cytoplasm.
Figure 2.
PhoA activities and GFP fluorescence intensities for the 34 inner membrane proteins. (A) Measured PhoA vs. GFP activities. The three encircled proteins (PotH, TdcC, YahN) have comparable PhoA and GFP activities and their C-terminal locations cannot be determined from the current data. (B) ln(PhoA/GFP) where PhoA is the measured PhoA activity normalized by the mean PhoA activity of all “active” PhoA fusions and GFP is the measured GFP activity normalized by the mean GFP activity of all “active” GFP fusions (see Results section). Because two different vectors were used for the GFP fusions (pWaldo and pGFPe), the GFP values were normalized by the mean fluorescence intensities of all “active” constructs for which the corresponding vector was used (the mean intensity for the pWaldo constructs is about threefold higher than for the pGFPe constructs). Black bars indicate proteins with a periplasmic C terminus; white bars, proteins with a cytoplasmic C terminus. The three gray bars correspond to the three encircled proteins in A for which the C-terminal location cannot be determined from the data.
The expression of each fusion protein was monitored by Western blotting with antibodies against either PhoA or GFP on samples collected at the time of harvest for the PhoA and GFP activity assays. All proteins were detected, but for some only a PhoA- or GFP-sized fragment was visible (Table 2), indicating proteolysis of the fusion joint. Although not shown in the table, for many fusion proteins for which a full-length product was visible, a PhoA and/or GFP fragment was also visible.
Table 2.
Summary of the predicted and experimental topology data for the 34 inner membrane proteins included in the study (protein names are from the Colibri database at http://genolist.pasteur.fr/Colibri (Rudd 2000)
| Protein | Length | TMHMM | HMMTOP | MEMSAT | PHD | TOPPRED | Consensus prediction | S3 Score | Accuracy | Expression | Experimentally determined C terminus | TMHMM C term fix | S3 Score C-term fix | Accuracy C-term fix |
| SdhC | 129 | 3P | 3P | 2C | 3P | 3P | P(4:1) | 0.94 | 0.95 | P | 3P | 1.00 | 1.00 | |
| CysW | 291 | 6C | 6C | 5P | 6C | 5P | C(3:2) | 0.91 | 0.93 | C | 6C | 0.98 | 0.98 | |
| YhjW | 563 | 5P | 5P | 6C | 5P | 5P | P(4:1) | 0.27 | 0.42 | P | 5P | 0.94 | 0.96 | |
| YjiG | 153 | 3P | 4C | 4C | 4C | 3P | C(3:2) | 0.70 | 0.76 | P | 3P | 0.91 | 0.93 | |
| PotC | 264 | 6C | 6C | 6C | 7P | 6C | C(4:1) | 0.88 | 0.90 | F | C | 6C | 0.90 | 0.93 |
| YfbV | 151 | 2C | 2C | 2C | 1P | 2C | C(4:1) | 0.67 | 0.74 | C | 2C | 0.88 | 0.92 | |
| TauC | 275 | 6C | 6C | 7P | 7P | 6C | C(3:2) | 0.85 | 0.88 | F | C | 6C | 0.87 | 0.91 |
| YagU | 204 | 3P | 3P | 3P | 4C | 3P | P(4:1) | 0.39 | 0.51 | C | 4C | 0.86 | 0.90 | |
| YcdT | 452 | 8C | 7P | 8C | 8C | 8C | C(4:1) | 0.81 | 0.85 | C | 8C | 0.85 | 0.90 | |
| CcmF | 647 | 15C | 15C | 15C | 14P | 15C | C(4:1) | 0.81 | 0.85 | G | C | 15C | 0.81 | 0.87 |
| YhjV | 423 | 11P | 11P | 11P | 12C | 11P | P(4:1) | 0.78 | 0.82 | G | P | 11P | 0.78 | 0.85 |
| YbjO | 162 | 4C | 4C | 3P | 3P | 4C | C(3:2) | 0.60 | 0.68 | C | 4C | 0.78 | 0.85 | |
| YcdU | 328 | 8C | 8C | 8C | 8C | 8C | C(5:0) | 0.72 | 0.77 | C | 8C | 0.72 | 0.80 | |
| YejM | 586 | 5P | 5P | 5P | 5P | 6C | P(4:1) | 0.70 | 0.76 | P | 5P | 0.70 | 0.79 | |
| YcfT | 357 | 8C | 9P | 8C | 9P | 8C | C(3:2) | 0.59 | 0.67 | C | 8C | 0.69 | 0.78 | |
| PsiE | 136 | 4C | 4C | 4C | 4C | 3P | C(4:1) | 0.49 | 0.59 | C | 4C | 0.49 | 0.64 | |
| SrlA | 187 | 3C | 3C | 3C | 4P | 3C | C(4:1) | 0.44 | 0.55 | C | 3C | 0.48 | 0.64 | |
| YjeM | 500 | 12C | 12C | 12C | 13P | 12C | C(4:1) | 0.47 | 0.58 | G,F | C | 12C | 0.48 | 0.63 |
| YddG | 293 | 10C | 10C | 10C | 9P | 9P | C(3:2) | 0.42 | 0.54 | F | C | 10C | 0.43 | 0.60 |
| MenA | 308 | 9P | 9P | 9P | 8C | 8C | P(3:2) | 0.27 | 0.42 | P | 9P | 0.42 | 0.60 | |
| YgdQ | 237 | 7C | 7C | 7C | 7C | 7C | C(5:0) | 0.39 | 0.51 | C | 7C | 0.41 | 0.59 | |
| YdgG | 344 | 8C | 8C | 8C | 8C | 8C | C(5:0) | 0.23 | 0.39 | C | 8C | 0.38 | 0.57 | |
| NarU | 462 | 12C | 12C | 12C | 13P | 12C | C(4:1) | 0.38 | 0.50 | C | 12C | 0.38 | 0.56 | |
| YegH | 527 | 7C | 7C | 7C | 7C | 7C | C(5:0) | 0.27 | 0.42 | C | 7C | 0.36 | 0.55 | |
| TsgA | 393 | 12C | 12C | 12C | 11P | 12C | C(4:1) | 0.27 | 0.42 | F | C | 12C | 0.27 | 0.49 |
| YebS | 427 | 8C | 8C | 7P | 8C | 8C | C(4:1) | 0.19 | 0.35 | F | C | 8C | 0.23 | 0.46 |
| ZitB* | 313 | 5P | 5P | 6C | 6C | 5P | P(3:2) | 0.37 | 0.49 | C | 6C | 0.19 | 0.43 | |
| HcaT | 379 | 12C | 12C | 12C | 11P | 11P | C(3:2) | 0.14 | 0.31 | G | C | 12C | 0.17 | 0.42 |
| YkgB | 197 | 3P | 4C | 3P | 4C | 4C | C(3:2) | 0.24 | 0.39 | C | 3C | 0.11 | 0.38 | |
| BtuC | 326 | 9P | 9P | 9P | 9P | 9P | P(5:0) | 0.13 | 0.30 | C | 8C | 0.08 | 0.35 | |
| YjcD | 449 | 13P | 13P | 13P | 13P | 13P | P(5:0) | 0.04 | 0.23 | P | 13P | 0.05 | 0.33 | |
| PotH | 317 | 6C | 6C | 6C | 7P | 6C | C(4:1) | 0.85 | 0.88 | ? | ||||
| YahN | 223 | 6P | 6P | 6P | 6P | 5C | P(4:1) | 0.56 | 0.65 | F | ? | |||
| TdcC | 443 | 11P | 11P | 10C | 11P | 11P | P(4:1) | 0.27 | 0.41 | G,F | ? |
The topologies predicted by five commonly used prediction programs are summarized by indicating the predicted cytoplasmic (C) or periplasmic (P) location of the C terminus together with the predicted number of transmembrane helices (columns 3–7). The consensus topology prediction is summarized by the location of the C terminus followed by the majority level (5 : 0 means that all five methods give the same prediction, 4 : 1, that four methods agree, etc.; column 8). The reliability score (S3 score) and the estimated prediction accuracy calculated by TMHMM with no inclusion of information on the location of the C-terminus are given in columns 9 and 10. Fusions for which only a fragment corresponding in size to the PhoA or GFP reporter moieties, but no full-length protein was visible on Western blots are indicated by F and G, respectively (column 11). The experimentally determined location of the C terminus is given in column 12 (c.f., Fig. 2B ▶). The topology, S3 score, and estimated accuracy predicted by TMHMM with information on the experimentally determined C-terminal location included are given in columns 13–15.
Topology models
The experimentally determined location of the C terminus for the 31 proteins with clear-cut PhoA/GFP results were used as input to the latest version of TMHMM (Melén et al. 2003) to produce new, experimentally based topology models. For each prediction, a reliability score (S3 score) as well as an estimate of the probability that the prediction is correct (the estimated accuracy) was calculated both before and after inclusion of the experimental information (Table 2; the detailed topology models are available as Supplemental Material). Although only three proteins have accuracy values above 0.9 in the unconstrained predictions, nine proteins fall in this category when the information on the C-terminal location is included. Further, the accuracy values increase for 29 of the 31 proteins when the C terminus is fixed to the experimentally determined location. It is interesting to note that of the two proteins with a known 3D structure, the model predicted for SdhC has an estimated accuracy of 1.0 and is correct (Yankovskaya et al. 2003), whereas the model predicted for BtuC has an estimated accuracy of only 0.35 and is, in fact, incorrect (the orientation is correct but two transmembrane helices are missed; Locher et al. 2002).
Discussion
Incorporation of limited experimental information can substantially improve the performance of membrane protein topology prediction methods (Tusnady and Simon 2001; Melén et al. 2003). Most current experimental approaches allow the determination of the location of extramembraneous loops, but do not work (or give artifactual results) for residues in transmembrane segments. For bacterial proteins, the most popular approach is to make reporter fusions to the target protein; because the reporter has to be at the C-terminal end of the fusion protein, these methods cannot be used to determine the location of the N terminus.
We have previously indicated that an efficient approach to the generation of more reliable topology models on a genome-wide scale is to map the location of the C terminus of all predicted membrane proteins encoded in a given genome by making C-terminal reporter fusions to each protein (Drew et al. 2002; Kim et al. 2003). C-terminal fusions to full-length proteins should be minimally disruptive to the topology and structure of the target protein, and thus minimize the risk of getting artifactual results. However, from a theoretical point of view, one should consider the possibility that the experimental information might be more valuable if the choice of the fusion point is not a priori restricted to the very C terminus of the target protein.
Here, we have compared the increase in performance of the TMHMM predictor obtained for different choices of which site in a protein to fix by an experimental effort. The choice between the N- and C-terminal residue makes little difference, and in both cases the prediction performance increases from ~70% (unconstrained) to ~80% correctly predicted topologies (over a data set of 233 membrane proteins with known topologies).
Somewhat unexpectedly, the prediction performance actually gets worse when the residue to be fixed is not restricted to one of the terminal residues but is chosen based on the “lowest probability loop residue” in the probability profiles from the unconstrained TMHMM predictions. The main reason for this is that loop regions predicted with low probability, in fact, correspond to true transmembrane regions sufficiently often to make it impossible to use this measure for indicating optimal locations for topology reporters.
Based on these results, we have extended our previous proof-of-principle study of 12 E. coli inner membrane proteins to an additional 34 proteins. Three technical improvements now make the approach feasible to eventually scale up to the entire E. coli inner membrane proteome: (1) A set of PhoA and GFP fusion vectors with appropriately chosen restriction sites has been constructed; (2) the experimental measurements of PhoA and GFP activities are done in a microtiter plate format; and (3) the latest version of TMHMM (Melén et al. 2003), in which the C terminus can be constrained to an experimentally established location, is used to produce the final topology models, obviating the need for the consensus approach used in our earlier studies (Drew et al. 2002; Kim et al. 2003).
For 31 of the 34 target proteins, the C-terminal location was easily determined because only the GFP or PhoA fusion was active. For the remaining three proteins, both reporters gave a low but detectable level of activity, and the C-terminal locations could thus not be unambiguously determined. These three proteins—roughly 10% of the total—provide an estimate of the limitation of the experimental approach and emphasize the importance of utilizing complementary reporters for a correct interpretation of the activity measurements. If this number scales to the whole inner membrane proteome of E. coli, it indicates that the PhoA/GFP fusion approach should be successful for ~90% of these proteins. Finally, we note that for the two proteins with known 3D structure in our set (BtuC and SdhC), the location of the C terminus was correctly identified by the PhoA/GFP fusions (Table 2).
In summary, we have shown that an approach based on C-terminal reporter fusions to full-length target proteins seems to be the best strategy for obtaining experimental data that can be used to constrain theoretical topology predictions. Using this approach, we have produced 31 additional topology models from an initial set of 34 target E. coli proteins. With the technical improvements reported here, the approach can now in principle be used to produce topology models for the entire E. coli inner membrane proteome.
Materials and methods
Data set of proteins with experimentally determined topologies
An up-to-date, homology-reduced data set of 247 proteins with experimentally determined topologies (L. Käll, A. Krogh, and E.L.L.Sonnhammer, in prep.) was used. Thirteen entries were removed because TMHMM failed to classify them as integral membrane proteins, and one entry was removed because it had no organism name in SwissProt, leaving 233 proteins (129 prokaryotic, 92 eukaryotic, 12 archaeal and viral) for the analysis.
Enzymes and chemicals
Unless otherwise stated, all enzymes and oligonucleotides were from GIBCO BRL. Expand Long Polymerase was from Roche. Alkaline phosphatase antisera was from Abcam, and the GFP-specific antibody was from Santa Cruz Biotechnology. Iodoacetamide (IAA) and the alkaline phosphatase chromogenic substrate PNPP (Sigma 104 phosphatase substrate) were from Sigma-Aldrich.
Strains and plasmids
Cloning was performed in E. coli strain MC1061 (lacX74, araD139, [ara, leu]7697, galU, galK, hsr, hsm, strA) (Dalbey and Wickner 1986). The PhoA assay and expression of the PhoA constructs were performed in CC118 (Δ(ara-leu)7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE(am) recA1) (Lee and Manoil 1994), and the GFP assay and expression in BL21(DE3)pLysS (F− ompT hsdSB(rB−mB−) gal dcm (DE3) pLysS).
PhoA fusion constructs were expressed from a modified version of the pHA-1 plasmid (Sääf et al. 1999), originally derived from the pING1 plasmid (Johnston et al. 1985), by induction with arabinose. Two plasmids, pHA-1 and pHA-4, were used for expression of the PhoA constructs, differing in the combination of restriction enzyme sites in the multiple cloning site (MCS): pHA-1 contains XhoI /KpnI, and pHA-4 contains XhoI/BamHI. In the pHA vectors, the cloned gene is immediately followed by the 17 or 19 amino acid linker sequence (GS)SVPDSYTQVASWTEPFPFC, and then by the phoA gene. The PhoA moiety lacks the 5′ segment coding for the signal sequence and the first five residues of the mature protein.
GFP fusion constructs were expressed from the modified pET28a(+) vector pWaldo (Waldo et al. 1999) by induction with isopropyl-β-D thiogalactoside (IPTG). The pWaldo MCS has two combinations of restriction enzyme sites, namely NdeI/BamHI or NdeI/EcoRI. When the first combination of enzyme sites is used it results in a 12 amino acid-long linker sequence (GSAGSAAG SGEF), while the second combination results in a two amino acid-long linker sequence (EF). The linker is followed by GFP (S65T, F64L + Cycle 3 mutant without the initiator Met). A second GFP vector (pGFPe) was developed from the pWaldo vector and was also used for some of the constructs in this study. pGFPe differs from pWaldo in that it has an extended MCS (5′ XhoI and 3′ EcoRI/KpnI/BamHI), carries a TEV protease site in the linker sequence, and has a His8-tag at the GFP C terminus. The pGFPe vector was only used with the enzyme combination XhoI/KpnI, yielding a 14 amino acid-long linker sequence (SVPGSENLY FQGQF).
DNA techniques
The genes encoding the 34 E. coli proteins were amplified from E. coli strains MG1655 or MC1061 using Expand Long Polymerase, and were cloned using primer-introduced sites 5′ XhoI /3′ KpnI, 5′ XhoI /3′ BamHI, 5′ NdeI /3′ BamHI, or 5′ NdeI /3′ EcoRI into the previously constructed pHA-1, pHA-4 (Whitley et al. 1994), pWaldo, and pGFPe plasmids. All plasmid constructs were confirmed by DNA sequencing from the 5′ and 3′ ends using Big Dye (Applied Biosystems).
Activity of GFP fusion proteins
GFP emission was measured in the E. coli strain BL21(DE3)pLysS transformed with a C-terminal GFP fusion vector (Waldo et al. 1999) carrying the appropriate GFP-fusion constructs under control of the T7 promoter. One-milliliter cultures were grown overnight at 37°C in Luria Broth (LB) medium containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol. The overnight cultures were diluted 1:50 in 15 mL of fresh medium with antibiotics and grown at 37°C, 250 rpm (we have later confirmed that 5 mL cultures can be successfully grown in 24-well plates for the GFP assay). When the OD600 reached 0.4–0.6, cells were grown for another 4 h in the presence of 0.4 mM IPTG. Before harvesting, 500 μL of cells were taken to measure the final optical density of the cultures at OD600 (ranging from 0.6 to 1.4) and another 4 mL of culture was removed for Western blotting (see below). The remaining 10.5 mL of cells were used for the GFP assay. These cells were harvested and resuspended in 280 μL of buffer containing 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 15 mM EDTA, and were incubated for 30 min at room temperature. Two hundred microliters were transferred into a black 96-well Nunc plate (Nunclone) and analyzed for GFP fluorescence emission using a FLOUstar microtiter plate reader, excitation filter 490 nm, and emission filter 520 nm (BMG LabTechnologies). GFP emission from each sample was normalized against its OD600 and background cell fluorescence was subtracted. Mean activity values for each sample was obtained from at least two (but in general five) independent measurements.
Activity of PhoA fusion proteins
The alkaline phosphatase activity assay was performed in the CC118 strain transformed with the pHA-1 or pHA-4 plasmid carrying the appropriate PhoA-fusion constructs. Cells were inoculated in a 2.2-mL 96-well growth plate (ABgene) containing 1 mL Luria Broth (LB) medium supplemented with 100 μg/mL ampicillin and grown overnight at 37°C. Overnight cultures were diluted 1:100 into two separate 2.2-mL 96-well growth plates containing 1 mL of fresh medium with antibiotics and grown at 37°C, 250 rpm. When the OD600 reached 0.13–0.18 cells were induced with arabinose (to a final concentration of 0.2%) and grown until the bacteria had reached a final density of 0.3–0.6. To prevent the spontaneous activation of PhoA that is localized to the cytoplasm, 1 mM of iodoacetamide was added to the cultures 10 min prior to harvesting and to all buffers used subsequently (Derman and Beckwith 1995). One of the 96-well growth plates was used for the PhoA assay and the other plate was used for Western blotting (see below). The activity assay was carried out as described (Manoil 1991). The time of the assay was set to 90 min for all samples to facilitate the assay for a 96-well layout. Mean activity values were obtained from at least two (but in general five) independent measurements.
Expression of PhoA and GFP fusion proteins
Western blotting was carried out to detect the proteins fused to the PhoA/GFP moiety. Whole-cell Western blot samples were centrifuged and resuspended in a volume of SDS-PAGE loading buffer corresponding to 0.1 OD units/5 μL loading buffer and boiled at 95°C before being loaded onto a gel. 0.1 OD units of whole cells samples were loaded onto a standard 8% SDS-polyacrylamide gel (PhoA samples) or a 10% SDS-polyacrylamide gel (GFP samples). Proteins were transferred from the gel to a PVDF membrane using a semidry Trans-Blot apparatus. To detect PhoA- or GFP-containing proteins, blots were decorated with PhoA-specific antibody and GFP-specific antibody, respectively. Blots were developed using the ECL detection system (according to the instructions of the manufacturer, Pharmacia).
Sequence data
Gene sequences were downloaded from the E. coli genome databank (Blattner et al. 1997) at http://bmb.med.miami.edu/EcoGene/EcoWeb/.
Electronic supplemental material
The detailed topology predictions produced by TMHMM with the location of the C terminus fixed to the experimentally determined location for each of the 31 proteins analyzed here are available as Supplemental Material.
Acknowledgments
This work was supported by grants from the Swedish Research Council and the Marianne and Marcus Wallenberg Foundation to G.v.H.; by an EMBO Long-Term Fellowship to D.O.D.; by a grant from the Research School of Medical Bioinformatics funded by Swedish Knowledge Foundation to K.M.; and by a center grant from the Foundation for Strategic Research to Stockholm Bioinformatics Center. We are indebted to Dr. Lukas Käll, Center for Genomics and Bioinformatics, Karolinska Institutet, Stockholm, for help with the data set of proteins with experimentally determined topologies.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material: see www.proteinscience.org
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03553804.
References
- Blattner, F.R., Plunkett, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277 1453–1462. [DOI] [PubMed] [Google Scholar]
- Canfield, V.A. and Levenson, R. 1993. Transmembrane organization of the Na,K-ATPase determined by epitope addition. Biochemistry 32 13782–13786. [DOI] [PubMed] [Google Scholar]
- Chang, X.B., Hou, Y.X., Jensen, T.J., and Riordan, J.R. 1994. Mapping of cystic fibrosis transmembrane conductance regulator membrane topology by glycosylation site insertion. J. Biol. Chem. 269 18572–18575. [PubMed] [Google Scholar]
- Chen, C.P., Kernytsky, A., and Rost, B. 2002. Transmembrane helix predictions revisited. Protein Sci. 11 2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey, R.E. and Wickner, W. 1986. The role of the polar, carboxyl-terminal domain of Escherichia coli leader peptidase in its translocation across the plasma membrane. J. Biol. Chem. 261 13844–13849. [PubMed] [Google Scholar]
- Derman, A.I. and Beckwith, J. 1995. Escherichia coli alkaline phosphatase localized to the cytoplasm slowly acquires enzymatic activity in cells whose growth has been suspended: A caution for gene fusion studies. J. Bacteriol. 177 3764–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, D., Sjöstrand, D., Nilsson, J., Urbig, T., Chin, C.N., de Gier, J.W., and von Heijne, G. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. 99 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilmeier, B.J., Iseminger, G., Schroeder, D., Webber, H., and Phillips, G.J. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182 4068–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S., Lee, J.H., and Ray, D.S. 1985. High-level expression of M13 gene II protein from an inducible polycistronic messenger RNA. Gene 34 137–145. [DOI] [PubMed] [Google Scholar]
- Kerppola, R.E. and Ames, G.F. 1992. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease—Comparison with other members of the family. J. Biol. Chem. 267 2329–2336. [PubMed] [Google Scholar]
- Kim, H., Melén, K., and von Heijne, G. 2003. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and prediction. J. Biol. Chem. 278 10208–10213. [DOI] [PubMed] [Google Scholar]
- Kimura, T., Ohnuma, M., Sawai, T., and Yamaguchi, A. 1997. Membrane topology of the transposon 10-encoded metal- tetracycline/H+ antiporter as studied by site-directed chemical labeling. J. Biol. Chem. 272 580–585. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. 2001. Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Lee, E. and Manoil, C. 1994. Mutations eliminating the protein export function of a membrane-spanning sequence. J. Biol. Chem. 269 28822–28828. [PubMed] [Google Scholar]
- Locher, K.P., Lee, A.T., and Rees, D.C. 2002. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science 296 1091–1098. [DOI] [PubMed] [Google Scholar]
- Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell Biol. 34 61–75. [DOI] [PubMed] [Google Scholar]
- Manoil, C. and Beckwith, J. 1986. A genetic approach to analyzing membrane protein topology. Science 233 1403–1408. [DOI] [PubMed] [Google Scholar]
- Melén, K., Krogh, A., and von Heijne, G. 2003. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327 735–744. [DOI] [PubMed] [Google Scholar]
- Nilsson, J., Persson, B., and von Heijne, G. 2000. Consensus predictions of membrane protein topology. FEBS Lett. 486 267–269. [DOI] [PubMed] [Google Scholar]
- Rudd, K. 2000. Ecogene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sääf, A., Johansson, M., Wallin, E., and von Heijne, G. 1999. Divergent evolution of membrane protein topology: The Escherichia coli RnfA and RnfE homologues. Proc. Natl. Acad. Sci. 96 8540–8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady, G.E. and Simon, I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850. [DOI] [PubMed] [Google Scholar]
- Waldo, G.S., Standish, B.M., Berendzen, J., and Terwilliger, T.C. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17 691–695. [DOI] [PubMed] [Google Scholar]
- Whitley, P., Nilsson, I., and von Heijne, G. 1994. De novo design of integral membrane proteins. Nat. Struct. Biol. 1 858–862. [DOI] [PubMed] [Google Scholar]
- Wilkinson, B.M., Critchley, A.J., and Stirling, C.J. 1996. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J. Biol. Chem. 271 25590–25597. [DOI] [PubMed] [Google Scholar]
- Yankovskaya, V., Horsefield, R., Törnroth, S., Luna-Chavez, C., Miyoshi, H., Léger, C., Byrne, B., Cecchini, G., and Iwata, S. 2003. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299 700–704. [DOI] [PubMed] [Google Scholar]