Abstract
The chaperonin GroEL is an oligomeric double ring structure that, together with the cochaperonin GroES, assists protein folding. Biochemical analyses indicate that folding occurs in a cis ternary complex in which substrate is sequestered within the GroEL central cavity underneath GroES. Recently, however, studies of GroEL “minichaperones” containing only the apical substrate binding subdomain have questioned the functional importance of substrate encapsulation within GroEL-GroES complexes. Minichaperones were reported to assist folding despite the fact that they are monomeric and therefore cannot form a central cavity. Here we compare directly the folding activity of minichaperones with that of the full GroEL-GroES system. In agreement with earlier studies, minichaperones assist folding of some proteins. However, this effect is observed only under conditions where substantial spontaneous folding is also observed and is indistinguishable from that resulting from addition of the nonchaperone protein α-casein. By contrast, the full GroE system efficiently promotes folding of several substrates under conditions where essentially no spontaneous folding is observed. These data argue that the full GroEL folding activity requires the intact GroEL-GroES complex, and in light of previous studies, underscore the importance of substrate encapsulation for providing a folding environment distinct from the bulk solution.
Chaperonins are large double ring structures that use the energy from ATP hydrolysis to assist protein folding in the cell (1). The best characterized chaperonin is the Escherichia coli protein GroEL, a homo-oligomeric complex composed of 14 58-kDa subunits arranged in two seven-membered rings stacked back to back. Crystallographic studies (2, 3) reveal that the GroEL monomer is composed of three domains: an apical region responsible for binding both substrate and the cochaperonin GroES, an equatorial domain that contains the ATP binding site as well as most of the intra-ring and all of the inter-ring contacts, and an intermediate hinge domain that connects the other domains. GroEL binds a wide variety of nonnative polypeptides in its central cavity. Substrate binding is mediated largely by interactions between exposed hydrophobic sidechains of nonnative polypeptides and hydrophobic residues in the apical domain of GroEL that face the central cavity (4, 5). GroEL alone is sufficient to assist the folding of some substrates. In general, however, GroEL-mediated folding requires the cochaperonin GroES (6). GroES, which is essential for viability, is composed of a single heptameric ring of 10-kDa subunits that binds to the ends of the GroEL cylinder.
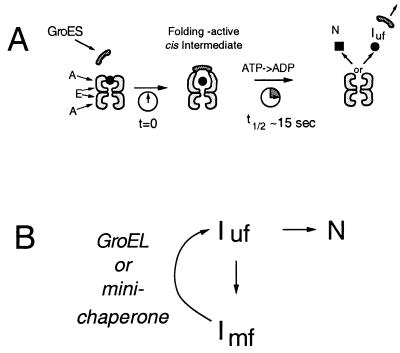
Recently, mechanistic studies have begun to define the GroEL-GroES reaction cycle in detail. For our studies, the critical steps of this folding cycle are as follows (Fig. 1A): (i) Nonnative substrates are bound within the central cavity of GroEL. This binding inhibits both folding and aggregation, and may promote unfolding of misfolded structures (7). (ii) GroES binds to the GroEL-polypeptide complex, forming a cis ternary complex in which the polypeptide is encapsulated within the GroEL-GroES structure (8–10). Binding of GroES obscures the hydrophobic polypeptide recognition regions of the GroEL apical domain thereby releasing the substrate into the central cavity (4, 11). In addition, GroES binding induces a dramatic conformational change in GroEL, doubling the volume of the central cavity (11, 12). As a consequence, the substrate polypeptide in the cis complex is encapsulated within a relatively polar environment that favors formation of the native state. (iii) ATP acts as a timer, giving the substrate 15–30 sec to fold before hydrolysis. Hydrolysis of ATP primes GroEL for GroES release, which allows the exit of substrate to the bulk solution (13). Polypeptides that have not yet folded are rebound by the same or different GroEL complex thereby reinitiating the folding reaction (14, 15). In this “encapsulation” model, the critical role of the GroEL-GroES complex is to provide a protected folding cavity with physical properties that are distinct from that of the bulk solution. The importance of this folding cavity is underscored by the observation that five different substrates [dihydrofolate reductase, green fluorescent protein, rhodanese, RUBISCO, and mitochondrial malate dehydrogenase (mMDH)] have now been shown to fold efficiently and rapidly while sequestered within such cis complexes (9, 10, 13).
Figure 1.
Two models for chaperonin-mediated protein folding. (A) Schematic of the encapsulation model. In the presence of nucleotide, GroES can bind in cis to the polypeptide-GroEL binary complex thereby encapsulating the polypeptide (Left). GroES binding induces a large conformational change in GroEL, doubling the volume of the central cavity and obscuring the hydrophobic polypeptide binding regions (11, 12), thereby initiating a polypeptide folding reaction (9, 10) (Middle). Folding proceeds for ≈15–30 sec after which ATP hydrolysis primes the complex for GroES release (13), giving the polypeptide the opportunity to depart the GroEL complex (Right). The released polypeptide is either committed to fold, or already folded (■) or in an uncommitted state (•) that can rebind the same or a different GroEL complex. This rebinding might also induce the unfolding of misfolded, kinetically trapped species as envisioned in the iterative annealing model (see B). The apical (A) and equatorial (E) domains of GroEL are indicated. (B) Schematic of the iterative annealing model (7, 39). Under nondenaturing conditions, unfolded protein (Iuf) partitions between the native state (N) and kinetically trapped, misfolded intermediates (Imf). Chaperones (either GroEL or minichaperones) bind to such misfolded species and promote their unfolding, thereby giving the polypeptide another opportunity to reach the native state.
A markedly different view of the mechanism of GroEL has emerged from recent studies of a fragment of GroEL that encompasses only the apical domain (16). This “minichaperone” includes the polypeptide binding domain, and consequently recognizes nonnative proteins and can promote unfolding. However, the apical domain does not contain the regions responsible for GroEL oligomerization and is, therefore, unable to form the GroEL central cavity. Surprisingly, monomeric minichaperone has been shown to assist the folding of some polypeptides and its folding activity is not enhanced by GroES. These observations suggest that the critical role of GroEL is not to allow folding within a protected environment, but rather to provide a hydrophobic surface that binds folding intermediates thereby preventing their aggregation and promoting unfolding of misfolded species (Fig. 1B).
It has emerged from a variety of studies that the nature of a GroEL-mediated folding reaction depends critically on the substrate (6, 17, 18). Mechanistic conclusions about the GroE chaperonin system therefore must be made in light of the substrates being studied. In particular, for a number of substrates such as human dihydrofolate reductase and cyclophilin A (CyPA), GroEL alone in the absence of GroES is able to support efficient protein folding (e.g., refs. 6, 17, 19, and 20). Such substrates typically show significant spontaneous folding in vitro and are likely to correspond to the so-called class II proteins (18). For class II substrates only a fraction of the molecules interacts with GroEL in vivo and those that do, interact only transiently (class I proteins completely fail to interact with GroEL in vivo). By contrast, a third set of substrates, such as rhodanese and mMDH, show an absolute requirement for GroEL and GroES under conditions where spontaneous folding does not occur. These substrates are thought to correspond to class III proteins that interact quantitatively with GroEL in vivo.
Because substrate encapsulation requires the concerted action of GroES and GroEL, the fact that class II substrates are folded by GroEL alone indicates that this folding reaction does not depend on encapsulation. These observations suggest a possible resolution of the two models for GroE-mediated folding: the apical domain mimics the GroEL-mediated folding reaction of class II substrates in the absence of GroES, whereas the robust folding of class III substrates depends on encapsulation within the GroEL-GroES complex. To critically test this proposal, we have examined the ability of the apical domain minichaperone to substitute for GroEL and GroES in the folding of both class II and class III substrates.
MATERIALS AND METHODS
General Procedures and Reagents.
All plasmids were verified by DNA sequencing (Applied Biosystems). PCR primers (Operon Technologies, Alameda, CA) were as follows: P1: GGGGGGGGCATATGAAAATCGAAGAAGGTAAACTGGGTATCTGGATTAAC, P2: GGGGGGGGGATCCTTACTTGGTGATACGAGTCTGCGC, P3: CGGATCCGAAGGTATGCAGTTCGACCG, P4: CGAATTCTTAACGGCCCTGGATTGCAGCTTC, P5: TATGCGTGGTTCTCATCATCATCATCATCATGGTCTCGTCCCGCGTG, P6: GATCCACGCGGGACGAGACCATGATGATGATGATGATGAGAACCACGCA.
N-succinyl-Ala-Leu-Pro-Phe-p-nitroanilide was purchased from Bachem, bovine mMDH and α-casein from Sigma.
Plasmid Construction.
The plasmid pSHT345 contains a gene, under control of a T7 promoter, for short histidine tag (sht)-GroEL191–345 that is identical to that described previously (16). pSHT345 was derived from pAED4 (15) by sequential insertion of a fragment encoding the GroEL apical domain and a fragment encoding the amino-terminal sht (16). The fragment encoding GroEL apical domain residues 191–345 was produced by PCR using primers P3 and P4, respectively, and a wild-type GroEL gene as a template. This fragment was then inserted into the BamHI and EcoRI restriction sites of pAED4, and the sht fragment was inserted by ligating the annealed product of P5 and P6 into the NdeI and BamHI restriction sites. The plasmid pMBP5 was constructed by ligating a PCR product encoding the MBP5 gene between the NdeI and BamHI restriction sites of pAED4. pWHC-1 (21), which encodes the MBP Y283D single mutant, was used as the PCR template, and P2 and P1 (which introduced the Val to Gly mutation at residue 8 of MBP) were used as primers.
Protein Expression and Purification.
Sht-GroEL191–345 was produced as follows: BL21 cells freshly transformed with pSHT345 were grown in 1 liter 2XYT plus carbenicillin to an OD600 = 0.5–0.8 and expression was induced with the addition of 0.4 mM isopropyl β-d-thiogalactoside. After 3 hr, cells were pelleted and resuspended in 45 ml Tris buffer solution (50 mM Tris, pH 7.4/50 mM NaCl), and disrupted by sonication. Subsequent to centrifugation (20,000 × g, 20 min), the supernatant was applied to a nickel–nitrilotriacetic acid agarose (Qiagen, Chatsworth, CA) column. The protein was eluted by addition of imidazole, and further purified by anion-exchange chromatography (Source15Q, Pharmacia), which was sufficient to remove any detectable PPIase activity.
MBP5 was produced as follows: BL21 cells transformed with pMBP5 were grown and induced as with sht-GroEL191–345. After induction, MBP5 was purified from inclusion bodies by reversed-phase HPLC as described (15). CyPA was produced in BL21 transformed with WISP94–1 (22) and purified as described (23). GroEL and GroES (4) and rhodanese (15) were expressed and purified as described.
Thermal Unfolding.
Temperature denaturation was monitored by circular dichroism spectroscopy at 222 nm. Samples consisted of 8 μM protein in 10 mM Na phosphate (pH 7.0). Measurements were made in a 0.5-cm pathlength cell in an Aviv Model 63D CD Spectrometer.
Refolding Experiments.
CyPA. Denatured CyPA was produced by incubating CyPA (50 μM) in 4.8 M guanidine-HCl, 10 mM DTT, 5 mM Tris, pH 7.8 on ice for an hour. Folding experiments were initiated by diluting denatured CyPA 50-fold (final concentration 1 μM) into buffer A (100 mM potassium phosphate, pH 7.0/10 mM DTT) supplemented with chaperones as indicated in the legend to Fig. 2. Folding was allowed to proceed at 25°C to completion (40 min), and cyclophilin activity was assayed as described (24) by examining the rate of isomerization of the substrate N-succinyl-Ala-Leu-Pro-Phe-p-nitroanilide.
Figure 2.
Characterization of the minichaperone. (A) Thermal denaturation of sht-GroEL191–345 monitored by circular dichroism at 222 nm. The midpoint of thermal denaturation (Tm) for sht-GroEL191–345 is 69°C, as determined by fitting the circular dichroism signal to a two state transition model. Note, throughout the figures sht-GroEL191–345 is indicated by sht345. (B) Effect of various chaperones on cyclophilin (CyPA) folding. CyPA denatured in 4.8 M guanidine-HCl was rapidly diluted (50-fold) into buffer A containing the indicated chaperones. The final concentration of CyPA was 1 μM, and the concentrations of chaperones present were: 0 μM GroEL (Spont.); 7 μM GroEL protomer (EL); 4 μM sht-GroEL191–345 (sht345); 100 μg/ml α-casein (casein). Folding reactions were carried out at 25°C. CyPA activities are reported as the fraction of the input material.
Rhodanese. Denatured rhodanese was produced by solubilizing lyophilized rhodanese in 6 M guanidine-HCl, 10 mM Tris (pH 7.4), 5 mM DTT to 25–40 μM. Folding was initiated by diluting denatured rhodanese to a final concentration of 100 nM in buffer B (25 mM Tris, pH 7.4/20 mM Na2SSO3/12 mM MgCl2/5 mM KCl/5 mM DTT) supplemented with chaperones as indicated in the legend to Fig. 3. The reactions were allowed to proceed to completion (60 min) at the indicated temperature and then quenched by addition of EDTA to 30 mM. Finally, rhodanese enzyme activity was determined as described (25).
Figure 3.
Effect of various chaperones on rhodanese folding. Denatured rhodanese in 6 M guanidine-HCl was rapidly diluted (>200 fold) into buffer B containing the indicated chaperones. For all experiments, the final rhodanese concentration was 100 nM. The final yields of rhodanese activities are reported as the fraction relative to that produced in the presence of GroEL, GroES, and ATP at 25°C, which represents ≈70% of the input material. (A) Rhodanese folding experiments were carried out at 25°C in the presence of the following concentrations of chaperones: 10 μM GroEL and 15 μM GroES protomers (EL/ES/ATP), 10 μM GroEL protomers (EL/ATP); 10 μM sht-GroEL191–345 (sht345); 100 μg/ml α-casein (casein); 100 μg/ml BSA; 0 μM GroEL (Spont.).ATP(5 mM) was present, where indicated. (B) Folding experiments were carried out as in B except that the temperature was 37°C.
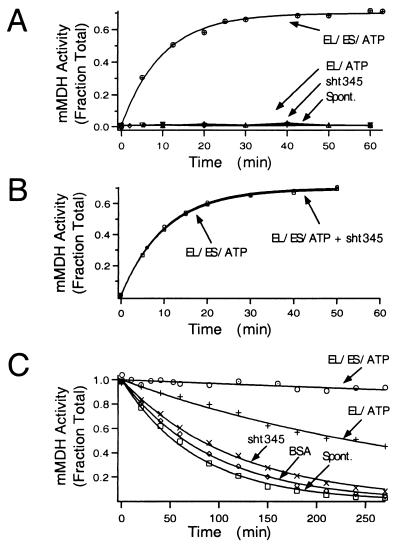
mMDH. mMDH was denatured by incubating 50 μM mMDH in 3 M guanidine-HCl and buffer C (25 mM Tris, pH7.4/5 mM KCl/10 mM MgCl2/5 mM DTT) for 1 hr on ice. mMDH folding was initiated by rapidly diluting denatured mMDH 100-fold into buffer C containing chaperones as indicated in legend to Fig. 4. Thermal denaturation was initiated by diluting native mMDH into buffer C containing indicated chaperones. At the indicated times, mMDH activities were measured as described (26). The concentration of mMDH was 0.5 μM and the temperature was 37°C.
Figure 4.
Minichaperone does not assist the folding of mMDH. (A) Effect of minichaperone on folding of mMDH. mMDH denatured in 3 M guanidine-HCl was rapidly diluted (100-fold) into buffer C containing the indicated chaperones. The concentration of mMDH in the folding reactions was 0.5 μM. At the indicated times, an aliquot was removed and the yield of mMDH activity measured. mMDH activities are reported as the fraction of the input material. The following concentrations of chaperones were present in the reactions: 28 μM GroEL and 42 μM GroES protomer (EL/ES/ATP); 28 μM GroEL protomer (EL/ATP); 28 μM sht-GroEL191–345 (sht345); 0 μM GroEL (Spont.). Where indicated, 10 mM ATP was present. All mMDH experiments were carried out at 37°C. (B) Effect of sht-GroEL191–345 minichaperone on GroEL-GroES-mediated folding of mMDH. GroEL-GroES folding reactions were carried out as in A in the presence or absence of 28 μM minichaperone as indicated. (C) The effect of chaperones on thermostability of native mMDH. Native mMDH was diluted into buffer C containing the indicated chaperones. The final concentration of mMDH is 0.5 μM. The concentration of BSA was 100 μg/ml, and the concentrations of the other chaperones and ATP were as in A.
MBP5. Denatured MBP5 was produced by dissolving lyophilized MBP5 in 6 M guanidine-HCl to a concentration of 25 μM. Folding reactions were initiated by dilution (>50 fold) of denatured MBP5 into buffer D (20 mM Tris, pH 7.4/0.2 M KCl/5 mM MgCl2) containing chaperones at 37°C. Unfolding reactions were initiated by diluting purified native MBP5 into buffer D and 4 M urea in the presence or absence of 10 mM maltose at 37°C. Pure native MBP5 was produced by first folding denatured MBP5 in the presence of GroEL, GroES, and ATP for 60 min. Native MBP5 was then purified from GroEL and GroES by gel-filtration chromatography (Superdex75, Pharmacia) in buffer D.
RESULTS
The Minichaperone Assists Folding of the Class II Substrate CyPA.
For the present studies, the chaperone activities of a previously described GroEL fragment, referred to as sht-GroEL191–345, were examined (16). Sht-GroEL191–345 consists of a sht followed by GroEL apical domain residues 191–345. Several observations argued that our GroEL apical domain fragment had a native-like structure with physical properties closely resembling those of apical fragments used previously. (i) Circular dichroism spectroscopy demonstrated that the isolated apical domain folds into an ordered structure. This structure was resistant to thermal denaturation, showing a cooperative folding transition with a midpoint of denaturation (Tm) of 69°C (Fig. 2A).(ii) As in previous studies (5, 16), our preparations of apical domain readily formed diffraction quality protein crystals (data not shown). (iii) Far Western blot analysis indicated that, as noted (16), the apical domain recognized denatured substrates (data not shown).
We next examined the chaperonin-mediated folding of the substrate protein CyPA. CyPA is a monomeric 18-kDa protein that catalyzes peptidyl-prolyl cis-trans isomerization. Upon dilution of denatured CyPA into folding buffer, ≈30%of the isomerase activity was regained in the absence of chaperone (Fig. 2B). By contrast, in the presence of GroEL essentially 100% of the activity was recovered. This dramatic enhancement in folding yield required neither GroES nor ATP. Similarly, in excellent agreement with earlier studies (16), the presence of minichaperone also supported quantitative recovery of CyPA activity. To test the specificity of this chaperone activity, we examined whether a nonchaperone protein α-casein could also assist the folding of CyPA. α-Casein has no sequence or structural similarities to the GroEL chaperonin but it does expose hydrophobic surfaces to the solvent, allowing it to interact with some unfolded proteins. Significantly, addition of α-casein also supported quantitative refolding of CyPA (Fig. 2B).
The Apical Domain Shows Little or No Chaperone Activity for Three Different Class III Substrates.
Rhodanese is a monomeric 33-kDa protein that is one of the best characterized GroEL substrates (e.g., refs. 9, 17, 18, and 27). In vitro rhodanese has been shown to fold within the GroEL-GroES cavity in a reaction that requires GroES. Similarly, folding of rhodanese in vivo is strongly dependent on GroEL and GroES. Previous work demonstrating that sht-GroEL191–345 assists rhodanese folding, therefore, provided a particularly strong argument against the functional role of encapsulation within GroEL-GroES complexes. Those studies, however, were conducted under conditions (25°C, 100nM substrate) where significant spontaneous folding of rhodanese also occurs. Under such conditions, the major reason for the failure to fold appears to be the rapid rate of off-pathway aggregation events (28). Consequently, the yield of folded protein is dependent on the precise details of the folding reaction [e.g., the presence of the nonspecific protein serum albumin or detergents (29, 30)].
Here we characterized the ability of minichaperone to assist rhodanese folding under a broader range of folding conditions. In initial experiments, we observed that addition of minichaperone increased the rhodanese yield for a folding reaction carried out at 25°C and 100nM substrate. To maximize this activity, we examined the effect of minichaperone concentration on the yield of native rhodanese. Optimal folding was observed between 5–20 μM apical domain, with the rhodanese yield being insensitive to minichaperone concentration in this range (data not shown). For all subsequent experiments, 10 μM minichaperone was used. In agreement with Zahn et al. (16), sht-GroEL191–345 resulted in a modest increase in rhodanese folding, 20 ± 2% (Fig. 3A). This folding yield is somewhat lower than that reported earlier (≈40%) but nonetheless represents a significant increase over that observed in a spontaneous folding reaction or in the presence of the nonspecific protein BSA (≈10%). This quantitative difference in folding yield might result from differences in substrates preparation because the folding yield is highly dependent on rhodanese concentration under these conditions. Intriguingly, as with CyPA, we found that addition of the nonchaperone protein α-casein resulted in yield (22 ± 2%) that was comparable to that resulting from the optimal concentration of apical domain. We next examined folding at the physiological temperature, 37°C. Under these conditions no significant folding was observed unless GroEL, GroES, and ATP were present (Fig. 3B). Thus, under stringent folding conditions neither the apical domain nor α-casein have significant chaperone activity.
To determine if the requirement for intact GroEL and GroES is a general feature of chaperonin-mediated folding reactions under stringent folding conditions, we examined the folding of two other GroEL-GroES dependent substrates. The first, mMDH, is a 35-kDa dimeric protein that is a physiological chaperonin substrate. At 37°C mMDH folding is completely dependent on GroEL, GroES, and ATP (6, 26, 31). As noted previously, we observed that GroEL and GroES efficiently supported the refolding of chemically denatured mMDH. By contrast, no detectable mMDH activity was regained in the presence of GroEL alone or in the presence of minichaperone (Fig. 4A). Normally during GroE-mediated folding, dimerization competent mMDH monomers are released from GroEL and undergo spontaneous dimerization (32). A failure to regain mMDH activity could result if the minichaperone inhibits oligomerization even if it were able to assist folding of the mMDH monomer. If this were the case, then addition of minichaperone would inhibit the GroEL-GroES-mediated folding of mMDH. We observed no such inhibition (Fig. 4B). Thus, the failure to produce native mMDH in the presence of minichaperone alone is due specifically to an inability of minichaperone to assist the folding of denatured mMDH to a dimerization competent, native-like monomer.
In addition to assisting the folding of newly synthesized proteins, GroEL protects native proteins against thermal denaturation. At 37°C, mMDH undergoes spontaneous unfolding with a half time of 55 min. The presence of GroEL dramatically slowed the rate of inactivation of mMDH. By contrast, the presence of sht-GroEL191–345 minichaperone did not provide significant protection from thermal denaturation (Fig. 4C). We conclude that for mMDH, both folding and protection from thermal unfolding are strictly dependent on the intact GroEL oligomer.
The final GroEL substrate examined was a folding-impaired derivative of maltose binding protein (MBP). MBP is a 41-kDa protein that binds maltose tightly in its native state. MBP has several properties that make it desirable as a folding substrate: (i) It is monomeric. (ii) The in vitro folding reaction of MBP has been extensively characterized (e.g., 21, 33). Finally, formation of the native state results in a dramatic enhancement in intrinsic tryptophan fluorescence, making it possible to continuously monitor the formation of native protein, as both GroEL and GroES are devoid of tryptophans. Wild-type MBP, however, is not a suitable GroEL model-substrate as it folds efficiently in the absence of chaperones. Previously, Buchner and coworkers (34) showed that the rate of folding of a point mutant of MBP, which renatures slowly in the absence of GroEL, is significantly enhanced by the addition of GroEL, GroES, and ATP. We reasoned that further destabilization of the native MBP conformation might yield a substrate whose folding is strictly dependent on GroEL and GroES.
For the present work, we have produced a mutant form of MBP (termed MBP5) in which two previously described destabilizing mutations in MBP (V8G and Y283D) (21) are combined. As monitored by fluorescence, at 37°C MBP5 rapidly refolds (T1/2 = ≈5 min) in the presence of GroEL, GroES, and ATP (Fig. 5B). The increase in fluorescence is likely to represent formation of native structure as the rate of denaturation of refolded MBP5 is dramatically slowed by the presence of the ligand maltose (Fig. 5A). The maltose binding site within MBP is located at the interface of two subdomains, thus the ability of the renatured MBP5 to bind maltose provides strong evidence that the protein had regained its complete native structure. As with mMDH and rhodanese, renaturation of MBP5 was strictly dependent on the presence of both GroES and GroEL at 37°C (Fig. 5B). Moreover, as with the other chaperonin dependent substrates, the shtGroEL191–345 minichaperone had no observable effect on MBP5 folding. Importantly, the addition of GroEL and GroES promoted folding even for MBP5 that had been preincubated with the minichaperone for an extended period (1,200 sec, Fig. 5C). Thus the failure of minichaperone to assist folding was not due to the formation of irreversible MBP aggregates.
Figure 5.
Minichaperone does not assist the folding of MBP5. Denatured MBP5 in 6 M guanidine-HCl was rapidly diluted (>50-fold) into buffer D containing the indicated chaperones. All reactions were carried out at 37°C and folding was monitored by changes in intrinsic tryptophan fluorescence intensity (excitation: 295 nm, emission: 344 nm). The fluorescence intensity is reported as the fraction relative to native MBP5. (A) The effect of maltose on stability of MBP5. Native MBP5 was produced by folding in the presence of GroEL, GroES, and ATP followed by purification on a gel filtration column. Purified MBP5 was then diluted into buffer D plus 4 M urea. Shown is the time course of denaturation, as monitored by UV fluorescence, in the presence or absence of 10 mM maltose. (B) Folding of MBP5 in the presence of various chaperones. The concentration of MBP5 in folding reactions was 60 nM. The following concentrations of chaperones were present in the reactions: 1.75 μM GroEL and 1.75 μM GroES protomer (EL/ES/ATP); 1.75 μM GroEL protomer (EL/ATP); 1.75 μM sht-GroEL191–345 (sht345); 0 μM GroEL (Spont.). We also failed to detect any significant recovery of native MBP5 in the presence of a 5-fold higher or 5-fold lower concentrations of minichaperone (data not shown). 2.5 mM ATP was present, where indicated. (C) Rescue of an MBP5-sht345 folding reaction by GroEL-GroES. The course of the reaction is indicated schematically by the time line: At time zero, denatured MBP5 was diluted into buffer D containing 1.75 μM sht-GroEL191–345. After 1,200 sec, 3.5 μM GroEL, 3.5 μM GroES and 2.5 mM ATP were added.
DISCUSSION
GroEL-Mediated Protein Folding Requires Formation of an Intact Central Cavity.
The chaperonin GroEL, together with the cochaperonin GroES, constitute a highly conserved and essential protein folding machine. Structural and biochemical studies have now begun to define the mechanism underlying this remarkable activity (1). The critical and perhaps most surprising feature of this reaction cycle is that a polypeptide folds while encapsulated within a GroEL-GroES complex (9, 10). This encapsulation creates a protected environment in which protein folding can proceed without the possibility of aggregation. The mechanism of the distantly related group II chaperonins found in archaea and eukaryotes, which do not have cochaperonins, is less well understood. Recent crystallographic studies, however, reveal that group II chaperonins contain a helical protrusion that extends over the central cavity, forming a dome-like structure with a striking similarity to GroES (35, 36). Thus despite the absence of cochaperonins for group II chaperonins, it is possible that the basic mechanism of encapsulation and folding within a polar cavity is conserved.
The present studies provide strong evidence that minichaperone molecules composed solely of the apical domain of GroEL are unable to functionally mimic the full GroEL-GroES folding machine. (i) Although the folding of the class II substrate CyPA is assisted by the minichaperone, this effect is not different from that resulting from the addition of a nonspecific protein α-casein. It remains to be seen whether minichaperone and α-casein assist folding by similar mechanisms. (ii) Even under conditions where there is significant spontaneous folding of the monomeric protein rhodanese, addition of minichaperone results in only a modest enhancement of folding, again at a level no greater than that produced by α-casein. Moreover, under conditions where essentially no spontaneous folding occurs, only the full GroEL-GroES system is able to assist folding of rhodanese. (iii) Minichaperone fails to fold the chaperonin substrate mMDH. This failure results directly from an inability to convert denatured mMDH to a folded monomer, as folded monomers efficiently oligomerize in the presence of minichaperone. Minichaperone is also unable to protect mMDH from thermal denaturation. (iv) Minichaperone fails to support folding of MBP5, a derivative of the monomeric protein MBP, whose folding is dependent on GroEL and GroES. Addition of GroEL and GroES leads to the rapid refolding of MBP5 that had been preincubated with minichaperone for an extended period of time. Thus, as with mMDH, the failure of minichaperone to support folding of MBP5 results from an inability to promote the conversion of soluble unfolded protein to the native state.
Our results are in excellent agreement with the recent work of Hartl, Georgopoulos, and coworkers (F. Weber, F. Keppel, C. Georgopoulos, M. K. Hayer-Hartl, and F. U. Hartl, personal communication), who find that in vitro minichaperones fail to assist the folding of rhodanese and mMDH as well as another GroE substrate, citrate synthase (F. U. Hartl, personal communication). Importantly, these workers also show that expression of minichaperones will not complement the loss of GroEL protein in vivo that is consistent with the view that the essential role of GroEL in vivo is to assist folding of Class III substrates. Previously it had been shown that formation of a cis ternary GroEL-GroES-polypeptide complex is both necessary and sufficient for the folding of GroES dependent substrates (8–10). Taken together with the present findings demonstrating that minichaperone does not functionally substitute for the full GroEL-GroES system, these observations underscore the importance of protein folding in the central cavity of GroEL-GroES complexes.
Although our studies demonstrate that apical domains alone cannot fully substitute for the intact GroEL-GroES chaperonin system, minichaperones are nonetheless a valuable tool for both mechanistic studies and potential practical applications. In particular, minichaperones could provide a model system for studying the folding of substrates that do not require encapsulation within GroEL-GroES complexes. It will be interesting to determine whether for such substrates GroEL and/or minichaperones are functioning solely by inhibiting aggregation or if they also promote unfolding of misfolded species (7, 16). In addition, the isolated apical domains should facilitate efforts to characterize substrate recognition (5). Finally, minichaperones attached to agarose beads have been shown to enhance renaturation of a number of substrates (37).
Is There a Role for Catalyzed Unfolding in a GroEL-Mediated Folding Reaction?
A major challenge facing a folding protein is that the folding intermediates are highly aggregation prone. Folding within the GroE cavity provides a general mechanism for enhancing protein folding yields by providing a protected environment in which folding can occur without the possibility of forming inappropriate aggregates (38). In addition to preventing aggregation, however, GroEL appears to alter folding reactions more actively. For example, GroEL and GroES have been shown to rescue kinetically trapped folding intermediates of both RUBISCO and mMDH that only slowly form irreversible aggregates but nonetheless do not reach the native state in the absence of chaperonin (14, 31).
As well as assisting protein folding, GroEL is also able to promote the unfolding of some substrates. These and other considerations have suggested an “iterative annealing” model in which GroEL actively promotes folding by unfolding kinetically trapped protein folding intermediates (7, 39). Consistent with this model, the GroEL-mediated folding of class III substrates such as mMDH and RUBISCO typically proceeds through multiple rounds of unfolding and refolding of substrate. However, Rye et al. (13)recently demonstrated that folding of mMDH and RUBISCO proceeds efficiently even in the absence of rounds of substrate binding and release. These observations suggest that the environment of the central cavity rather than cycles of binding and release may be responsible for the enhancement in folding rates. However, it also remains possible that the enhanced folding rates result from disruption of low order aggregates and inhibition of their further formation (32). In summary, although there is strong evidence that GroEL can promote protein unfolding, it remains to be established rigorously whether for any GroEL substrate such catalyzed unfolding actually enhances the rate or yield of formation of the native state.
Acknowledgments
We gratefully acknowledge L. Randall for plasmid pWHC-1, and C. Hill for plasmid WISP94–1. We thank members of the Weissman laboratory, C. Gross, and K. Yamamoto for helpful discussions. We thank F. U. Hartl for communicating results before publication. This work was supported by a Howard Hughes Medical Institute Predoctoral Fellowship (J.D.W.), a National Science Foundation predoctoral fellowship (M.D.M.), and grants from Howard Hughes Medical Institute, National Institutes of Health, the David and Lucile Packard Foundation, and the Searle Scholars Program.
ABBREVIATIONS
- CyPA
cyclophilin A
- MBP
maltose binding protein
- mMDH
mitochondrial malate dehydrogenase
Note Added in Proof
Chatellier et al. (40) recently reported that a slightly shorter form of minichaperone, consisting of residues 193–335, enhanced colony formation in cells containing limiting amounts of GroEL, although none of the minichaperones could replace GroEL in cells in which the chromosomal groEL gene has been deleted. We did not observe any differences in the folding activities in vitro between the various forms of minichaperones.
References
- 1. Bukau B, Horwich A L. Cell. 1998;92:351–66. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 2.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 3.Boisvert D C, Wang J, Otwinowski Z, Horwich A L, Sigler P B. Nat Struct Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 4.Fenton W A, Kashi Y, Furtak K, Horwich A L. Nature (London) 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 5.Buckle A M, Zahn R, Fersht A R. Proc Natl Acad Sci USA. 1997;94:3571–3575. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt M, Buchner J, Todd M J, Lorimer G H, Viitanen P V. J Biol Chem. 1994;14:10304–10311. [PubMed] [Google Scholar]
- 7.Zahn R, Perrett S, Stenberg G, Fersht A R. Science. 1996;271:642–645. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
- 8.Weissman J S, Hohl C M, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil H R, Fenton F A, Horwich A L. Cell. 1995;83:577–588. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 9.Weissman J S, Rye H S, Fenton W A, Beechem J M, Horwich A L. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 10.Mayhew M, da Silva A C, Martin J, Erdjument-Bromage H, Tempst P, Hartl F U. Nature (London) 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z, Horwich A L, Sigler P B. Nature (London) 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Nature (London) 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 13.Rye H S, Burston S G, Fenton W A, Beechem J M, Xu Z, Sigler P B, Horwich A L. Nature (London) 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 14.Todd M J, Viitanen P V, Lorimer G H. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 15.Weissman J S, Kashi Y, Fenton W A, Horwich A L. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 16.Zahn R, Buckle A M, Perrett S, Johnson C M, Corrales F J, Golbik R, Fersht A R. Proc Natl Acad Sci USA. 1996;93:15024–15029. doi: 10.1073/pnas.93.26.15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin J, Langer T, Boteva R, Schramel A, Horwich A L, Hartl F U. Nature (London) 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 18.Ewalt K L, Hendrick J P, Houry W A, Hartl F U. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 19.Viitanen P V, Donaldson G K, Lorimer G H, Lubben T H, Gatenby A A. Biochemistry. 1991;30:9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- 20.Corrales F J, Fersht A R. Proc Natl Acad Sci USA. 1996;93:4509–4512. doi: 10.1073/pnas.93.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun S Y, Strobel S, Bassford P, Jr, Randall L L. J Biol Chem. 1993;268:20855–20862. [PubMed] [Google Scholar]
- 22.Yoo S, Myszka D G, Yeh C, McMurray M, Hill C P, Sundquist W I. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Albers M W, Chen C M, Schreiber S L, Walsh C T. Proc Natl Acad Sci USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer G, Bang H, Berger E, Schellenberger A. Biochim Biophys Acta. 1984;791:87–97. doi: 10.1016/0167-4838(84)90285-1. [DOI] [PubMed] [Google Scholar]
- 25.Tandon S, Horowitz P M. J Biol Chem. 1989;264:9859–9866. [PubMed] [Google Scholar]
- 26.Staniforth R A, Cortes A, Burston S G, Atkinson T, Holbrook J J, Clarke A R. FEBS Lett. 1994;344:129–135. doi: 10.1016/0014-5793(94)00348-3. [DOI] [PubMed] [Google Scholar]
- 27.Mendoza J A, Lorimer G H, Horowitz P M. J Biol Chem. 1991;266:16973–16976. [PubMed] [Google Scholar]
- 28.Horowitz P, Bowman S. J Biol Chem. 1987;262:5587–5591. [PubMed] [Google Scholar]
- 29.Jarabak R, Westley J, Dungan J M, Horowitz P. J Biochem Toxicol. 1993;8:41–48. doi: 10.1002/jbt.2570080107. [DOI] [PubMed] [Google Scholar]
- 30.Zardeneta G, Horowitz P M. J Biol Chem. 1992;267:5811–6. [PubMed] [Google Scholar]
- 31.Peralta D, Hartman D J, Hoogenraad N J, Hoj P B. FEBS Lett. 1994;339:45–49. doi: 10.1016/0014-5793(94)80381-1. [DOI] [PubMed] [Google Scholar]
- 32.Ranson N A, Dunster N J, Burston S G, Clarke A R. J Mol Biol. 1995;250:581–586. doi: 10.1006/jmbi.1995.0399. [DOI] [PubMed] [Google Scholar]
- 33.Ganesh C, Shah A N, Swaminathan C P, Surolia A, Varadarajan R. Biochemistry. 1997;36:5020–5028. doi: 10.1021/bi961967b. [DOI] [PubMed] [Google Scholar]
- 34.Sparrer H, Rutkat K, Buchner J. Proc Natl Acad Sci USA. 1997;94:1096–1100. doi: 10.1073/pnas.94.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klumpp M, Baumeister W, Essen L O. Cell. 1997;91:263–70. doi: 10.1016/s0092-8674(00)80408-0. [DOI] [PubMed] [Google Scholar]
- 36.Ditzel L, Lowe J, Stock D, Stetter K O, Huber H, Huber R, Steinbacher S. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 37.Altamirano M M, Golbik R, Zahn R, Buckle A M, Fersht A R. Proc Natl Acad Sci USA. 1997;94:3576–3578. doi: 10.1073/pnas.94.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saibil H R, Zheng D, Roseman A M, Hunter A S, Watson G M F, Chen S, auf der Mauer A, O’hara B P, Wood S P, Mann N H, et al. Curr Biol. 1993;3:265–273. doi: 10.1016/0960-9822(93)90176-o. [DOI] [PubMed] [Google Scholar]
- 39.Todd M J, Lorimer G H, Thirumalai D. Proc Natl Acad Sci USA. 1996;93:4030–4035. doi: 10.1073/pnas.93.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatellier J, Hill F, Lund P A, Fersht A R. Proc Natl Acad Sci USA. 1998;95:9861–9866. doi: 10.1073/pnas.95.17.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]