Abstract
Mitochondria have been proposed to possess base excision repair processes to correct oxidative damage to the mitochondrial genome. As the only DNA polymerase (pol) present in mitochondria, pol γ is necessarily implicated in such processes. Therefore, we tested the ability of the catalytic subunit of human pol γ to participate in uracil-provoked base excision repair reconstituted in vitro with purified components. Subsequent to actions of uracil-DNA glycosylase and apurinic/apyrimidinic endonuclease, human pol γ was able to fill a single nucleotide gap in the presence of a 5′ terminal deoxyribose phosphate (dRP) flap. We report here that the catalytic subunit of human pol γ catalyzes release of the dRP residue from incised apurinic/apyrimidinic sites to produce a substrate for DNA ligase. The heat sensitivity of this activity suggests the dRP lyase function requires a three-dimensional protein structure. The dRP lyase activity does not require divalent metal ions, and the ability to trap covalent enzyme-DNA complexes with NaBH4 strongly implicates a Schiff base intermediate in a β-elimination reaction mechanism.
Human mitochondria possess a circular 16,569-bp genomic element encoding many of the components required for oxidative phosphorylation, and mutation of the mitochondrial genome causes debilitating human diseases stemming from mitochondrial dysfunction (1, 2). The mutation rate of mtDNA is estimated to be ≈10-fold higher than that of nuclear DNA (3), and this difference generally is attributed to increased DNA damage from elevated concentrations of reactive oxygen species produced by oxidative phosphorylation (4, 5). Despite the harsh oxidative environment of the mitochondrial matrix, steady-state levels of oxidative lesions in mtDNA remain low under physiological conditions, implying the existence of mtDNA repair processes (6). Early reports indicated mitochondria cannot perform nucleotide excision repair of UV-induced pyrimidine dimers (7). However, base excision repair (BER) of a variety of lesions has been observed in several mitochondrial systems in vivo (8–11), and repair-associated DNA synthesis in lysates has exhibited the sensitivity to N-ethylmaleimide and resistance to aphidicolin expected for the γ-polymerases (10).
Excision repair of a damaged base requires the concerted activities of a glycosylase to remove the damaged or inappropriate base, a class II apurinic/apyrimidinic (AP) endonuclease to incise the DNA 5′ to the AP site, a lyase activity to remove the 5′ terminal 2-deoxyribose-5-phosphate (dRP) sugar moiety from the downstream DNA, resynthesis, and ligation (12). DNA repair enzymes that have been isolated from mitochondria include uracil-DNA glycosylase (13), AP endonuclease (14), an 8-hydroxydeoxyguanine specific endonuclease (15), and DNA ligase (16). The removal of the dRP moiety can proceed via simple hydrolysis or by enzyme-catalyzed β-elimination (12). Reconstitution experiments with Xenopus laevis mitochondrial proteins localized the dRP lyase function in mitochondrial excision repair to either the mtDNA ligase or DNA polymerase (pol) γ (16). Recently, we cloned, overexpressed, and purified to homogeneity the catalytic subunit of the human pol γ (17, 18). Because pol γ is the only known pol in animal mitochondria (2, 19, 20), we sought to determine the specific roles of pol γ in mitochondrial BER. We used the same approach that successfully identified a Mg2+-independent dRP lyase activity in the conserved helix–hairpin–helix structure within the 8-kDa domain of the nuclear pol β (21–24). In this paper we demonstrate that the catalytic subunit of human pol γ can fill single nucleotide gaps generated as intermediates in the BER pathway, and we report the discovery of 5′ deoxyribose phosphate lyase activity intrinsic to the catalytic subunit.
MATERIALS AND METHODS
Enzymes.
Human AP endonuclease and uracil-DNA glycosylase with an 84-aa truncation at the N terminus were purified as described (25, 26). Human DNA ligase I that had been purified from baculovirus-infected insect cells (27) was a gift from Alan E. Tomkinson (University of Texas Health Science Center, San Antonio). The wild-type, histidine6-tagged wild-type, and 3′→5′ exonuclease-deficient forms of the recombinant catalytic subunit of human pol γ and native HeLa cell pol γ were purified as described (18).
Enzyme Assays.
Reverse-transcriptase activity was measured by the incorporation of [α-32P]dTMP into acid-precipitable poly(A)⋅oligo(dT)12–18 (Pharmacia) as described (18). BER reactions (10 μl) contained 50 mM Hepes-KOH (pH 7.5), 2 mM DTT, 0.2 mM EDTA, 100 μg/ml of BSA, 10% glycerol, 4 mM ATP, 0.3 μM [α-32P]dCTP (3,000 Ci/mmol), and 10 nM of annealed 51-bp synthetic oligodeoxyribonucleotides (Oligos Etc., Wilsonville, OR) with U opposite G at position 22 (28):
 |
Reactions were initiated by incubation with 10 nM uracil-DNA glycosylase for 10 min at 37°C, followed by preincubation with 10 nM AP endonuclease, 15 nM pol γ-p140, and 100 nM DNA ligase I at 4°C for 5 min. Reactions were completed by incubation with 5 mM MgCl2 for 10 min at the indicated temperatures, and products were analyzed by denaturing PAGE and autoradiography as described (28). As indicated, some reactions used the unrelated DNA sequence:
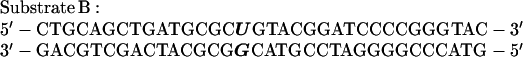 |
The dRP lyase substrate was a 49-bp oligodeoxyribonucleotide containing uracil at position 21 that had been 3′-end labeled with [α-32P]ddATP by terminal deoxynucleotidyl transferase (Promega) and annealed to an unlabeled complementary strand containing a G residue opposite the U (29):
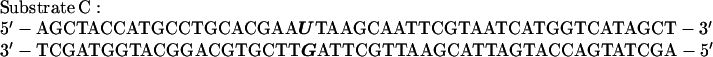 |
Immediately before use, the labeled duplex was treated extensively with uracil-DNA glycosylase and AP endonuclease as described (29) to generate a single nucleotide gap directly upstream from the labeled fragment containing the dRP flap. The dRP lyase activity was measured as described (29), except reactions (10 μl) contained 50 mM Hepes-KOH (pH 7.4), 2 mM DTT, 5 mM MgCl2, 20 nM 3′-end labeled DNA containing a 5′-incised AP site at position 21, 5–100 nM pol γ, and EDTA as indicated in the figure legends. After incubation at 37°C for the indicated times, reaction products were stabilized by incubation with 0.34 M NaBH4 (Sigma) for 30 min at 0°C and analyzed as described above.
Covalent trapping of pol γ-DNA complexes occurred in reactions (10 μl) containing 50 mM Hepes-KOH (pH 7.4), 2 mM DTT, 5 mM EDTA, and 50 nM 3′-end labeled, preincised AP-DNA (substrate C). Reactions were initiated by the simultaneous addition of 20 mM NaBH4/NaCl with 100 nM pol γ or DNA ligase I. After incubation on ice for 10 min, reactions were incubated at 25°C for 20 min and terminated by the addition (10 μl) of SDS/PAGE sample buffer. Samples were boiled for 2 min, and covalent complexes were resolved on a 12% SDS/polyacrylamide gel and visualized by autoradiography.
Sedimentation Analysis.
Protein samples (60 μl) were layered onto linear 10–30% sucrose gradients (5 ml) containing 50 mM Hepes-KOH (pH 7.6), 5% glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.01% Nonidet P-40, and 0.1 M KCl. After centrifugation at 4°C for 16 h at 55,000 rpm in a Beckman SW 55 Ti rotor, fractions (85 μl) were collected from the bottom of each gradient.
RESULTS
Pol γ Participates in BER in Vitro.
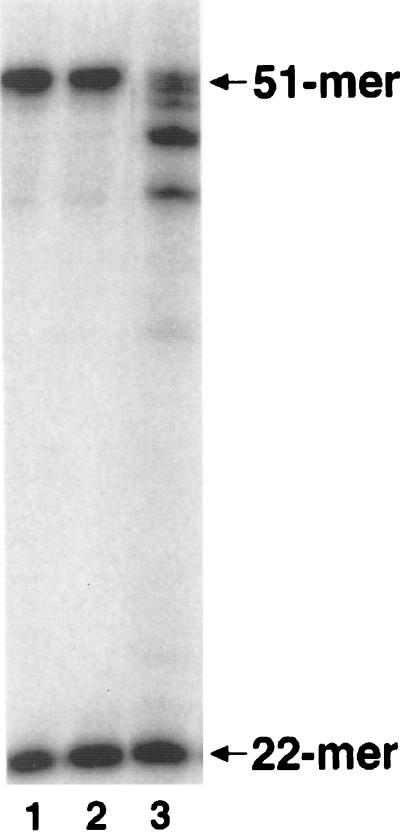
To simulate BER initiated by deamination of cytosine, our reconstitution assay used a synthetic oligonucleotide substrate with a single, centrally located G:U mispair (substrate A). The combined actions of purified uracil-DNA glycosylase and AP-endonuclease on this DNA substrate generated a single nucleotide gap opposite a template G residue immediately upstream from a 5′-deoxyribose phosphate moiety. When presented with this intermediate, the highly purified catalytic subunit of pol γ was able to incorporate a labeled dCMP residue during resynthesis of the single-nucleotide gap, forming the labeled 22-mer (Fig. 1). The subsequent production of labeled 51-mer in reactions with dCTP as the only nucleoside triphosphate indicate the removal of the interfering dRP moiety before ligation. Action of the 3′→5′ exonuclease activity of pol γ on the internally labeled 51-mer was evident at 37°C, but this effect was minimized by performing the reaction at reduced temperatures (Fig. 1). This result strongly suggests that the isolated catalytic subunit of pol γ was able to fill the single nucleotide gap and produced a substrate for DNA ligase I. Identical results were obtained for pol γ from HeLa cells, and the ability of both the native and recombinant forms to use an unrelated DNA substrate (substrate B) indicated that reconstituted BER was independent of DNA sequence context (data not shown).
Figure 1.
Reconstitution of uracil-initiated BER with the purified catalytic subunit of human pol γ. BER reactions (see Materials and Methods) were incubated for 10 min at 15°C, 25°C, or 37°C (lanes 1–3, respectively), and products were analyzed by denaturing PAGE and autoradiography. Incorporation of a single [α-32P]dCMP residue produced a radiolabeled 22-mer intermediate (arrow), and combined dRP lyase and DNA ligase activities generated the 51-mer product (arrow).
Human Pol γ Possesses Intrinsic dRP Lyase Activity.
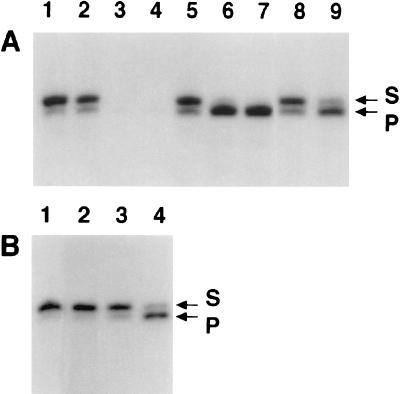
Having indirectly demonstrated removal of the 5′ deoxyribose phosphate moiety to generate a substrate for DNA ligase I, we sought to determine which component of the reconstituted human reaction performed the dRP lyase function. Direct measurement of lyase activity used a 3′-labeled, 5′-preincised AP-DNA substrate (substrate C), and the persistence of the dRP moiety indicated the absence of dRP lyase activity in the uracil-DNA glycosylase and AP endonuclease preparations (Fig. 2 A and B, lane 1). The release of 5′ dRP from the 32P-labeled substrate by the catalytic subunit of human pol γ was determined by the appearance of a radioactive electrophoretic band migrating approximately one-half nucleotide faster than the substrate (Fig. 2). Higher concentrations of pol γ resulted in loss of label, presumably because of 3′→5′ exonucleolytic removal of the 3′-dideoxynucleotide label (Fig. 2A, lanes 3 and 4). Because lyase activity does not require divalent metal ions (30), we inhibited the nuclease activity with EDTA and reassessed lyase activity (Fig. 2A, lanes 5–7). Under these conditions lyase activity was roughly proportional to enzyme concentration, confirming a metal-independent reaction mechanism. We previously constructed an exonuclease-deficient form of pol γ by site-directed mutagenesis of two invariant acidic residues in the Exo I region of the pol γ gene (18). As expected, pol γ rendered exonuclease deficient also exhibited lyase activity in the absence of added EDTA (Fig. 2A, lanes 8 and 9). In addition, pol γ that had been heat-treated could not perform the lyase function (Fig. 2B, compare lanes 3 and 4), excluding the formal possibility that simple chemistry incidental to buffer components or the primary sequence of the enzyme was mimicking lyase activity as detected, for example, with polypeptides containing Lys-Trp-Lys sequences (31). Human DNA ligase I used in the base excision assay in Fig. 1 did not have detectable dRP lyase activity (Fig. 2B, lane 2).
Figure 2.
dRP lyase activity of human pol γ was determined as described in Materials and Methods, and the relative mobilities of DNA substrate (S) and product (P) bands are indicated. (A) Lane 1, no enzyme; lanes 2 and 5, 10 nM His-p140; lanes 3 and 6, 50 nM His-p140; lanes 4 and 7, 100 nM His-p140; lane 8, 5 nM Exo−p140; lane 9, 20 nM Exo−p140. Reactions 5–7 also contained 5 mM EDTA. Incubation times were 30 min for reactions 1–7 and 60 min for reactions 8 and 9. (B) Lane 1, no enzyme; lane 2, 100 nM DNA ligase I; lane 3, 50 nM His-p140 heated at 100°C for 5 min before the reaction; lane 4, 50 nM untreated His-p140. Reactions were incubated for 15 min.
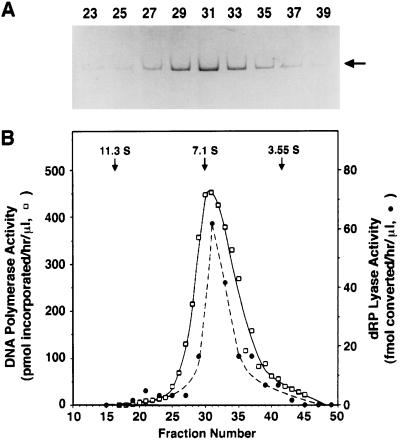
To ensure that the dRP lyase activity was intrinsic to the catalytic subunit and not from a contaminant we analyzed our preparation of recombinant human pol γ by band sedimentation on sucrose gradients. This analysis demonstrated physical association of dRP lyase and pol activities with the monomeric pol γ polypeptide (Fig. 3), proving the dRP lyase activity was intrinsic to the catalytic subunit of human pol γ.
Figure 3.
Pol and dRP lyase activities cosediment. Homogenous, recombinant, His-tagged pol γ catalytic subunit (29 μg) was subjected to band sedimentation through 10–30% sucrose gradients as described in Materials and Methods. (A) The indicated gradient fractions (16 μl) were resolved by electrophoresis through 4–20% SDS/polyacrylamide gels and stained with Coomassie brilliant blue. The arrow indicates the position of the pol γ catalytic subunit. (B) Pol activity (□) and dRP lyase activity (•) were determined as described in Materials and Methods on 2-μl samples of the indicated gradient fractions. The peak positions of standard proteins (catalase, S20,w = 11.3 S; γ-globulin, S20,w = 7.1 S; ovalbumin, S20,w = 3.55 S) sedimented in a parallel gradient are shown.
Catalytic Efficiency and Reaction Mechanism.
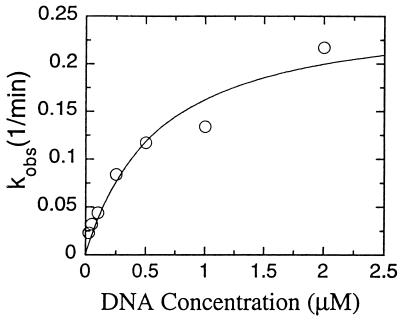
Many basic proteins such as histones and polyamines are able to perform dRP lyase activity with very low efficiency (32). The dRP lyase activity of human pol γ was assayed under steady-state conditions in the presence of EDTA on preincised AP-DNA to determine the catalytic efficiency of the reaction (Fig. 4). The apparent KM for this DNA and the kcat were 0.6 μM and 0.26 min−1, respectively, demonstrating the reaction was indeed catalytic. Compared with values for pol β on preincised AP-DNA (KM 0.5 μM and kcat 4.5 min−1) (29), values for pol γ show a similar KM but a 17-fold lower kcat. Considering this rate of dRP removal and the kinetics of nucleotide insertion on poly(A)⋅oligo(dT)12–18 by recombinant pol γ (KM = 7 μM, kcat = 4.5 sec−1, data not shown), we propose that single nucleotide gap filling precedes dRP removal during mitochondrial BER in vitro, as has been shown for pol β in nuclear BER reactions (28, 29). The observed accumulation of 22-mer intermediate under conditions of excess DNA ligase I (Fig. 1) support this proposed reaction sequence for pol γ.
Figure 4.
Steady-state kinetic analysis of the dRP lyase activity of pol γ. The release of dRP from preincised AP-DNA was measured at the indicated substrate concentrations as described in Materials and Methods. Reactions contained 50 nM pol γ and were incubated at 37°C for 15 min. Apparent activity was fitted to the Michaelis equation by a nonlinear least-squares algorithm.
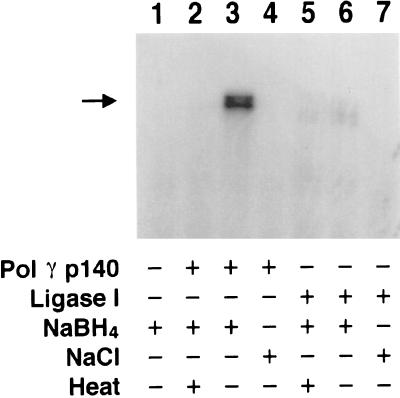
Collectively, the observations presented above suggested a β-elimination reaction mechanism potentially using a Schiff base nucleophilic intermediate (22), like that deduced for pol β (21, 29). To test this idea, dRP lyase reactions were initiated and interrupted by the addition of NaBH4 to trap reaction intermediates. Analysis by SDS/PAGE and autoradiography revealed a doublet of radiolabeled oligonucleotide covalently crosslinked to the catalytic subunit of the human pol γ (Fig. 5, lane 3). Similar results were obtained with substrate B (data not shown). The inability of the heat-denatured enzyme to crosslink demonstrated that the three-dimensional structure of the human pol γ and not the simple primary sequence was required for Schiff base formation (Fig. 5, compare lanes 2 and 3). Additionally, substitution of NaCl for NaBH4 did not result in crosslinking, supporting the notion that the covalent intermediate is a Schiff base and subject to borohydride reduction (Fig. 5, lane 4). Human DNA ligase I used throughout this study showed negligible trapping by NaBH4 (Fig. 5, lanes 5 and 6). Thus, our results demonstrate that the highly purified catalytic subunit of human pol γ can catalyze the removal of deoxyribose phosphate via a Schiff base intermediate.
Figure 5.
Covalent crosslinking of pol γ-DNA complexes by sodium borohydride. The dRP lyase reactions were performed as described in Materials and Methods, and products were resolved on 12% SDS/polyacrylamide gel and visualized by autoradiography. NaBH4 (20 mM) was added to the reactions containing 100 nM His-p140 (lanes 2 and 3) or 100 nM DNA ligase I (lanes 5 and 6). His-p140 pol γ and DNA ligase I also were heat-denatured by boiling for 5 min before assembly of some reactions (lanes 2 and 5). NaCl (20 mM) was added in place of NaBH4 to the reactions in lanes 4 and 7. The relative mobility of pol γ His-p140 crosslinked to 3′-labeled DNA is indicated by the arrow.
DISCUSSION
As the sole pol in animal mitochondria, the role of pol γ in replication of the mitochondrial genome is implicit. Although immunocytochemical detection of 5-bromo-2-deoxyuridine incorporation indicated preferential synthesis of mtDNA in the perinuclear region of tissue culture cells (33), our immunofluorescence studies established the presence of pol γ in all mitochondria of the cell (34). Such maintenance of pol γ activity in nonreplicating, peripheral mitochondria led us to investigate the proposed additional role of pol γ in mitochondrial BER, and we chose uracil-initiated BER as our model system. We demonstrate here that the native and recombinant catalytic subunit of pol γ can participate in BER in vitro. Pol γ was able to fill a single nucleotide gap on preincised AP-DNA and produced a ligatable substrate that required the removal of the 5′-deoxyribose phosphate. Because no other enzyme in our system contained an activity to remove the dRP moiety, we tested the capacity for pol γ catalytic subunit to carry out this function. As demonstrated by direct dRP lyase assay, the human pol γ readily removed the dRP moiety with reasonable catalytic efficiency, and this activity was intrinsic to the catalytic subunit.
The dRP lyase activity of pol γ did not require divalent metal ions and could be interrupted by NaBH4 crosslinking, suggesting a β-elimination reaction and the formation of a Schiff base intermediate. The active site for this reaction mechanism in human pol β has been localized to a helix–hairpin–helix structural motif (21, 23, 24, 29, 35, 36). Interestingly, a comparison between the human pol γ and human pol β using the GCG bestfit program identified a 40-aa region of pol γ with significant similarity to the relevant pol β helix–hairpin–helix structure. Corresponding to amino acids 353–391, this region of human pol γ lies between exonuclease motifs 2 and 3 and is predicted to be a helix–hairpin–helix structure (17). Lys-371 of this region is invariant within the gamma family of pols, and it may be functionally equivalent to the Lys-72 catalytic residue of pol β that acts as the predominant Schiff base nucleophile in the dRP lyase β-elimination mechanism (23, 24).
Presenting evidence that a replicative eukaryotic pol possesses dRP lyase activity, this report assigns two primary roles to pol γ: replication of the mitochondrial genome and DNA synthesis during BER. Therefore, pol γ is vital to maintaining the integrity of both replicating and nonreplicating mitochondrial genomes. In addition to the nuclear “short patch” or simple BER pathway in which pol β replaces a single nucleotide, a second pathway for BER has been reported recently in which nuclear enzymes synthesize 2–6 nucleotides in “long patch” BER tracts (37–39). The long-patch pathway required proliferating cell nuclear antigen and relied on the activity of DNaseIV (FEN1) to produce a ligatable species (37). Our reconstitution experiments were performed without the addition of accessory factors, indicating the dRP lyase and pol functions of pol γ are both necessary and sufficient for “short patch” or simple BER in mitochondria. It is unclear what effects accessory proteins or other protein factors might have on the repair capabilities of pol γ, and the existence of alternate pathways for repair of abasic sites in mitochondria is uncertain. Further studies with human mitochondrial proteins could help to identify other human factors involved in recognition and repair of altered bases.
ABBREVIATIONS
- BER
base excision repair
- dRP
2-deoxyribose-5-phosphate
- AP
apurinic/apyrimidinic
- pol
DNA polymerase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Wallace D C. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 2.Shadel G S, Clayton D A. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 3.Brown W M, George M J, Wilson A C. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter C, Park J W, Ames B N. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shigenaga M K, Hagen T M, Ames B N. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegler J, Bittner D, Boiteux S, Epe B. Carcinogenesis. 1993;14:2309–2312. doi: 10.1093/carcin/14.11.2309. [DOI] [PubMed] [Google Scholar]
- 7.Clayton D A, Doda J N, Friedberg E C. Proc Natl Acad Sci USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeDoux S P, Wilson G L, Beecham E J, Stevnsner T, Wassermann K, Bohr V A. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 9.Anson R M, Croteau D L, Stierum R H, Filburn C, Parsell R, Bohr V A. Nucleic Acids Res. 1998;26:662–668. doi: 10.1093/nar/26.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryoji M, Katayama H, Fusamae H, Matsuda A, Sakai F, Utano H. Nucleic Acids Res. 1996;24:4057–4062. doi: 10.1093/nar/24.20.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croteau D L, Bohr V A. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 13.Domena J D, Mosbaugh D W. Biochemistry. 1985;24:7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- 14.Tomkinson A E, Bonk R T, Linn S. J Biol Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 15.Croteau D L, ap Rhys C M, Hudson E K, Dianov G L, Hansford R G, Bohr V A. J Biol Chem. 1997;272:27338–27344. doi: 10.1074/jbc.272.43.27338. [DOI] [PubMed] [Google Scholar]
- 16.Pinz K G, Bogenhagen D F. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ropp P A, Copeland W C. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 18.Longley M J, Ropp P A, Lim S E, Copeland W C. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M E, Clayton D A. Curr Opin Genet Dev. 1993;3:769–774. doi: 10.1016/s0959-437x(05)80097-8. [DOI] [PubMed] [Google Scholar]
- 20.Fry M, Loeb L A. Animal Cell DNA Polymerases. Boca Raton, FL: CRC; 1986. [Google Scholar]
- 21.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 22.Piersen C E, Prasad R, Wilson S H, Lloyd R S. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 23.Prasad R, Beard W A, Chyan J Y, Maciejewski M W, Mullen G P, Wilson S H. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Kim K, Katz D S, Feng J. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- 25.Strauss P R, Beard W A, Patterson T A, Wilson S H. J Biol Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 26.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle N M, Haug T, Levine D W, Krokan H E. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y C, Burkhart W A, Mackey Z B, Moyer M B, Ramos W, Husain I, Chen J, Besterman J M, Tomkinson A E. J Biol Chem. 1994;269:31923–31928. [PubMed] [Google Scholar]
- 28.Srivastava D K, Vande Berg B J, Prasad R, Molina J T, Beard W A, Tomkinson A E, Wilson S H. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 29.Prasad R, Beard W A, Strauss P R, Wilson S H. J Biol Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 30.Graves R J, Felzenszwalb I, Laval J, O’Connor T R. J Biol Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 31.Behmoaras T, Toulme J J, Helene C. Nature (London) 1981;292:858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- 32.Bailly V, Verly W G. Biochem J. 1988;253:553–559. doi: 10.1042/bj2530553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis A F, Clayton D A. J Cell Biol. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis A F, Ropp P A, Clayton D A, Copeland W C. Nucleic Acids Res. 1996;24:2753–2759. doi: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelletier H, Sawaya M R, Wolfle W, Wilson S H, Kraut J. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 36.Doherty A J, Serpell L C, Ponting C P. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klungland A, Lindahl T. EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortini P, Pascucci B, Parlanti E, Sobol R W, Wilson S H, Dogliotti E. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 39.Biade S, Sobol R W, Wilson S H, Matsumoto Y. J Biol Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]