Abstract
Elevated levels of the p21WAF1 (p21) cyclin-dependent kinase inhibitor induce growth arrest. We have characterized a panel of monoclonal antibodies against human p21 in an effort to understand the dynamic regulatory interactions between this and other cellular proteins during the cell cycle. The use of these reagents has allowed us to address several important, yet unresolved, issues concerning the biological activity of p21, including the potential kinase activity of complexes that associate with this cyclin-dependent kinase inhibitor. We have found that the kinase activity of cyclin A/Cdk2 associated with p21 is significantly lower than that of cyclin A/Cdk2 free of p21, suggesting that p21 abolishes its activity in vivo, and the use of multiple antibodies has enabled us to begin the study of the molecular architecture of p21 complexes in vivo. In addition, we found that human fibroblasts released from a quiescent state display abundant amounts of p21 devoid of associated proteins (“free” p21), the levels of which decrease as cells approach S phase. Cyclin A levels increase as the amount of monomeric p21 decreases, resulting in an excess of cyclin A/Cdk2 complexes that are not bound to, or inactivated by, p21. Our data strengthen the notion that the G1-to-S phase transition in human fibroblasts occurs when the concentration of cyclin A/Cdk2 surpasses that of p21.
Mammalian cell cycle progression is governed by an interplay between positive and negative regulatory factors. Cyclin-dependent kinases (Cdks), composed of regulatory cyclin and catalytic Cdk subunits, are activated in a periodic manner to promote cell cycle transitions, whereas another family of proteins, the cyclin-dependent kinase inhibitors (CKIs), acts in a negative fashion by extinguishing the activity of Cdks (reviewed in ref. 1). Biochemical studies have shown that one CKI, p21 (also known as WAF1/Cip1/Sdi1/Cap20, henceforth referred to as p21), binds tightly to the G1 and S phase kinases, cyclin E/Cdk2, cyclin D/Cdk4, and cyclin A/Cdk2 and effectively inhibits their activity, whereas p21 is a relatively poor inhibitor of the G2/M phase kinase, cyclin B/Cdc2 (2–4). Additional studies have indicated the presence of proliferating cell nuclear antigen (PCNA), cyclin, and Cdk binding domains in p21 (5–8), and crystallographic studies suggest a mechanism for kinase (and growth) inhibition by the related CKI, p27Kip1 (9). Inhibition of DNA replication through interactions with PCNA is thought to constitute a second mechanism by which p21 can act as a growth suppressor (ref. 1 and references therein). One unexpected result of in vitro studies, however, was the finding that substoichiometric levels of p21 did not abolish kinase activity, but rather facilitated assembly of cyclins with their Cdk partner, whereas higher amounts of p21 were inhibitory (4, 10). Moreover, kinase activity associated with endogenous p21 has also been detected (4, 10, 11).
Nevertheless, a rigorous comparison between the specific activity of endogenous kinase complexes associated with, and devoid of, p21 has been lacking. In an effort to understand how p21 regulates the activity of associated kinases during cell cycle progression, we have begun to characterize a panel of mAbs raised against this protein. These antibodies recognize distinct p21-associated complexes through a diverse array of epitopes on the p21 protein and reveal structural details of such complexes in a more physiological setting than previous analyses. Using these reagents, we have also studied the composition and kinase activity of p21 complexes. These experiments suggest that p21 modulates kinase activity in vivo, and the abundance of this CKI may contribute to the timing of the G1/S transition.
MATERIALS AND METHODS
Antibodies and Epitope Mapping.
Antibodies were produced against recombinant human p21 as described (12). The epitope recognized by each was mapped by using a synthetic peptide array (Pepspots; Jerini BioTools, Berlin, Germany) containing 77 peptides spanning the entire sequence of human p21. All peptides were 13-mers that differed sequentially by two carboxyl-terminal amino acids and were covalently immobilized at their carboxyl terminus to a nitrocellulose membrane as separate spots. Mapping was carried out according to the manufacturer’s instructions. Briefly, the antibody to be mapped was first incubated with the peptide array for 3–5 hr at 4°C. After extensive washes, the bound antibody was transferred to a second piece of nitrocellulose and processed by using standard blotting techniques.
Other antibodies used in this study included anti-cyclin A (BF683), anti-cyclin B (GNS1), anti-cyclin E (HE12), and polyclonal anti-cyclin D1, kindly provided by S. Shiff, E. Harlow, and L. Zukerberg (Massachusetts General Hospital, Boston). Additional antibodies against cyclin A (H432), Cdk2 (M2), Cdk4 (C-22), E2F-4, p21 (C-19G), Cdc2 (17), and PCNA (PC10) were purchased from Santa Cruz Biotechnology.
Immunoprecipitations, Kinase Assays, and Western Blotting.
Whole cell extracts were prepared by using ELB buffer containing 50 mM Hepes (pH 7), 250 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.5 mM DTT, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride·HCl, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mM NaF, and 50 mM β-glycerophosphate. Cell extracts (300 μg of total protein) were precleared with protein-A Sepharose and normal rabbit serum and immunoprecipitated by incubation with antibody-conjugated beads. After washes with ELB and kinase buffer, the beads were halved and subjected to Western blot analysis and kinase assays. For immunoprecipitation of 35S-labeled proteins, WI38 cells were incubated for 4 hr in [35S]methionine, lysed in ELB, and precleared before incubation with specific antibody.
Western blots were analyzed by nih image version 1.61. Kinase assays were performed as described (13), and the results were quantitated using a Molecular Dynamics PhosphorImager. We calculated the specific activity of Cdk2 complexes immunoprecipitated by anti-cyclin A and anti-p21 (CP2) as follows: Specific activity of cyclin A/Cdk2 = histone H1 kinase activity (PhosphorImager units)/Cdk2 protein (arbitrary densitometry units). Recombinant human p21 was produced in bacteria and purified as described (14).
Denaturing Versus Native Immunoprecipitation.
To unfold and dissociate all protein complexes, 300 μg of WI38 whole cell extracts were first treated with 8 M urea in ELB buffer for 30 min at room temperature. Samples were then diluted 20-fold with buffer containing indicated antibody-bead conjugates and rocked for 1 hr at 4°C. Control samples either contained a final concentration of 0.4 M urea (to match conditions of refolded samples) or were untreated but normalized for protein concentration. Bound proteins were separated on a 12% SDS/PAGE gel and analyzed by Western blotting using anti-p21 antibody (CP36).
Cell Lines and Cell Cycle Synchronization.
WI38 human fibroblasts and the simian virus 40-transformed derivative WI38-VA13 were obtained from American Type Culture Collection. WI38 cells at a passage number of 15–17, grown to about 70% confluence in DMEM with 10% fetal bovine serum, were serum-starved by incubation in medium containing low serum (0.2%) for 3 days. Under these conditions, more than 85% of the cells were arrested at G0/G1 as determined by fluorescence-activated cell sorter (FACS) analysis (FACScan Analyzer, Becton Dickinson) after propidium iodide staining.
Size Exclusion Chromatography.
A 10 × 300 mm Sephacryl S-300 (Pharmacia) column was used to analyze the size of p21-containing protein complexes. The column was equilibrated in, and developed with, 50 mM Hepes (pH 7.4), containing 250 mM NaCl, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C. The flow rate was 0.5 ml/min, and 0.7 ml fractions were collected. The column was calibrated with the following proteins: cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), BSA (67 kDa), lactate dehydrogenase (140 kDa), catalase (232 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa).
RESULTS AND DISCUSSION
Characterization of Anti-p21 Antibodies.
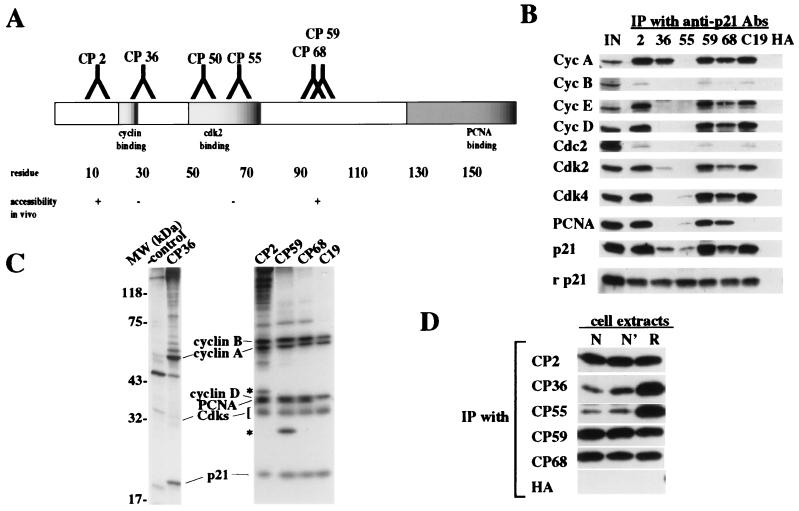
A panel of mAbs raised against native, recombinant human p21 purified from bacteria has been described (12). We mapped precisely the epitopes recognized by each antibody by using a peptide array as described in Materials and Methods (Fig. 1A). Each of these antibodies is specific for p21, although one (CP36) crossreacts weakly with recombinant p27 and p57 on Western blots and immunoprecipitates these recombinant proteins with very low efficiency (12).
Figure 1.
Characterization of anti-p21 antibodies. (A) Schematic representation of functional domains of p21 and the epitopes recognized by anti-p21 antibodies. Epitopes were mapped by using a peptide array as described. On the basis of this analysis, we mapped the antibody recognition to the following epitopes: residues 5–13 (CP2), 19–25 (CP36), 35–43 (CP50), 69–78 (CP55), 96–104 (CP59), and 95–102 (CP68). One antibody, CP50, failed to immunoprecipitate endogenous p21 and was not characterized further. (B) Asynchronous WI38 cell extracts (300 μg) were immunoprecipitated with different anti-p21 antibodies as indicated. The first lane (IN) represents 10% of input protein loaded directly. The last lane shows a control using anti-flu hemagglutinin (HA) mAb, 12CA5. For comparison, recombinant p21 (rp21; 3 ng) from bacteria was immunoprecipitated. (C) Immunoprecipitations of extracts of WI38 cells metabolically labeled with [35S]methionine using the indicated antibodies were resolved by SDS/PAGE and visualized by fluorography. The identity of associated proteins was assigned by parallel immunoprecipitations with antibodies against each of the indicated proteins. Asterisks indicate unidentified proteins. (D) The epitopes recognized by CP36 and CP55 are buried in cellular p21-containing complexes. Asynchronous WI38 cell whole extracts (300 μg) were treated without urea (native or N), or with 0.4 M (N′) or 8 M (refolding or R) urea as described. Samples were then diluted 20-fold with buffer (N and R) or 0.4 M urea (N′) and immunoprecipitated and Western blotted. HA is a control using antibody 12CA5.
Initially, we immunoprecipitated whole cell extracts derived from asynchronous W138 human fibroblasts with anti-p21 antibodies and detected p21-associated proteins by Western blotting (Fig. 1B). For the purposes of comparison, we also included in our studies an antibody purchased commercially (C19), which recognizes the overlapping PCNA-binding/carboxyl-terminal cyclin-binding domains of p21 (residues 146–164). We demonstrated that several antibodies (CP2, 59, 68) coprecipitated p21 with the full complement of known cell cycle regulatory proteins, including cyclin A, cyclin B, cyclin D, cyclin E, Cdk2, Cdk4, Cdc2, and PCNA (15). As expected from previous studies, very low levels of cyclin B/Cdc2 associated with p21 in vivo, perhaps as a consequence of the relatively low affinity between this kinase and p21 (2, 4, 16).
Interestingly, several antibodies recognize epitopes that overlap with important functional domains in p21, and they were able to immunoprecipitate subsets of the above complexes (12). For example, CP36 recognizes an epitope in p21 that precisely corresponds to an amino-terminal cyclin-binding domain found in the p21 family of proteins and in the retinoblastoma protein (pRB) family members, p107 and p130 (8, 14, 17). This cyclin-binding domain, referred to variously as the LFG, RXL, or Cy motif (henceforth, Cy domain) is present near the amino- and carboxyl termini of p21. CP36 immunoprecipitates contained cyclin A and Cdk2, but none of the other cyclins, Cdks, or PCNA (Fig. 1B). Significantly, CP36 immunoprecipitates much lower amounts of p21 and Cdk2 than CP2, 59, or 68, although it can retrieve similar amounts of cyclin A, suggesting that the antibody strictly recognizes cyclin A/Cdk2/p21 complexes. Notably, no PCNA is coprecipitated with this complex, suggesting the existence of cyclin A/cdk2/p21 complexes lacking PCNA. We have also been able to distinguish these complexes lacking PCNA from others containing cyclin D/Cdk4, p21, and PCNA by ion exchange chromatography (data not shown). These studies, together with a previous one (8), suggest that cyclins D and E associate with p21 in vivo principally through the amino-terminal cyclin-binding domain recognized by CP36, whereas cyclin A is also able to associate with this inhibitor through the carboxyl-terminal cyclin-binding domain or indirectly by means of a Cdk2 bridge, which can independently associate with p21 (5–8). Because the carboxyl-terminal cyclin-binding (Cy) domain overlaps with the PCNA-binding domain in p21, these results raise the possibility that cyclin A can successfully compete with PCNA for binding to this site in vivo.
Another antibody, CP55, recognizes an epitope in p21 (residues 69–78) corresponding to a region in p27KIP1 known to form a 310 helix that is buried deeply in the catalytic cleft of Cdk2 in the cyclin A/Cdk2/p27 ternary complex, the structure of which was solved crystallographically (9). Intriguingly, immunoprecipitates of CP55 were devoid of all other cyclin and Cdk proteins as detected by Western blotting (Fig. 1B). Occasionally, small amounts of PCNA and Cdk4 were coprecipitated with p21, but in no cases were any cyclins detected. These and other experiments (described below) suggest that CP55 is able to recognize a form of p21 we have termed free p21.
To visualize the purity of complexes that copurify with p21, we immunoprecipitated p21 from exponentially growing WI38 cells that were metabolically labeled with [35S]methionine. In these experiments, immunoprecipitates were highly enriched for the same p21-associated proteins revealed by Western blotting (Fig. 1C). Few contaminating proteins were detected in most immunoprecipitates, although certain ones contained unidentified proteins that await further characterization. Importantly, these experiments confirmed the specificity of our anti-p21 antibodies for this CKI family member, since we did not observe the coprecipitation of either p27 or p57 from WI38 cell lysates (data not shown). The purity of these complexes and the absence of the two p21-related proteins was an important consideration for subsequent functional studies concerning the kinase activity of p21 complexes (see below), and they suggest the usefulness of these antibodies in purifying distinct complexes to homogeneity for future activity studies.
The inability of CP36 and CP55 to recognize a subset of p21-associated proteins was not due to reduced affinities between these antibodies and p21, because each of the five mAbs immunoprecipitated equal amounts of purified, recombinant p21 produced in bacteria (Fig. 1B). Furthermore, when we examined the ability of each of our antibodies to recognize native versus denatured p21-containing complexes, we noticed distinct differences between the two groups of antibodies (Fig. 1D). Whereas CP2, CP59, and CP68 recognized equivalent amounts of p21 before and after denaturation of cell lysates with 8 M urea, both CP36 and CP55 immunoprecipitated significantly more p21 after denaturation and renaturation in the presence of antibody (compare lanes N and R).
Taken together, these data suggest that regions recognized by CP36 (Cy motif) and CP55 (residues that potentially form a Cdk inhibitory helix) are not exposed in vivo. These experiments for the first time (to the best of our knowledge) begin to elucidate the molecular architecture of p21 complexes in vivo and are in agreement with previous in vitro and structural studies.
Cyclin/Cdk Complexes Associated with p21 Are Inactive.
One important unresolved issue regarding p21 concerns the apparent ability of this cyclin-dependent kinase inhibitor to associate with active cyclin/Cdk complexes in vitro and in vivo (4, 10, 11). Counterbalancing these studies are others suggesting that depletion of p21-containing complexes does not decrease the overall histone H1 kinase activity associated with cyclin A (16). It has been postulated that the presence or absence of p21-associated kinase activity depends on the stoichiometry of p21 molecules bound to the kinase; low levels of p21 actually stimulate assembly of associated kinases, whereas incorporation of a second molecule inhibits activity (4, 10, 11).
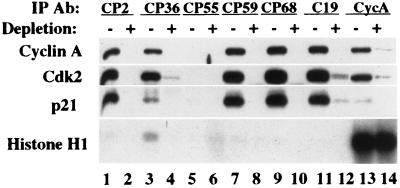
In an effort to resolve these contradictory observations, we have measured the histone H1 kinase activity associated with p21 in vivo by using our panel of antibodies. We reasoned that this approach had the following advantages over previous analyses. First, we employed at least six different mAb-recognizing epitopes spanning the p21 protein. This reduces the likelihood that kinase activity was altered by antibody binding to the complex. Second, the use of antibodies that immunoprecipitate all known associated cyclin/Cdk complexes as well as subsets of complexes (for example, CP36), suggests that we can study both the total Cdk2 kinase activity of all associated complexes as well as that of the cyclin A/Cdk2 complex in isolation. Third, the use of high-affinity antibodies results in immunoprecipitates that have been greatly enriched for p21-containing complexes (Fig. 1C) and are thus less likely to contain contaminating kinase activities. Finally, a meaningful estimate of p21-associated kinase activity requires not only a measurement of absolute kinase activity but also the specific activity of cyclin A/Cdk2 complexes in a free state and when bound to p21.
Fig. 2 shows that anti-p21 immunoprecipitates displayed very low levels of histone H1 kinase activity detectable only upon prolonged autoradiographic exposure. CP36 immune complexes had the highest amounts of associated kinase activity (12), but even in this case, the specific activity of the ternary complex (see Materials and Methods) was significantly reduced relative to cyclin A/Cdk2 that was immunoprecipitated directly from extracts or from p21-depleted extracts (lanes 13 and 14; for quantitation, see Fig. 3F). Moreover, cyclin A immunoprecipitates had similar amounts of histone H1 kinase activity whether or not p21 had been previously depleted, in agreement with a recent report (16). Similar results were obtained when extracts were immunoprecipitated with anti-Cdk2 antibodies or an unrelated anti-cyclin A mAb, rendering unlikely the possibility that anti-cyclin A antibodies artificially caused the activation of kinase complexes (data not shown). We have also observed similar differences in the specific activities of binary and ternary complexes in the human breast cancer cell line MCF-7 (data not shown).
Figure 2.
The p21/cyclin A/Cdk2 ternary complex is significantly less active than the cyclin A/Cdk2 complex in vivo. Whole cell extracts from exponentially growing WI38 cells were split into two equivalent samples. One half was left untreated, whereas the other was treated with three rounds of CP2-conjugated beads to deplete p21-containing complexes. Immunoprecipitations were performed on both depleted (+) and nondepleted (−) samples using different antibodies as indicated. The immunoprecipitated samples were split again and tested by Western blotting using anti-p21, anti-cyclin A, and anti-Cdk2 antibodies and in histone H1 kinase assays.
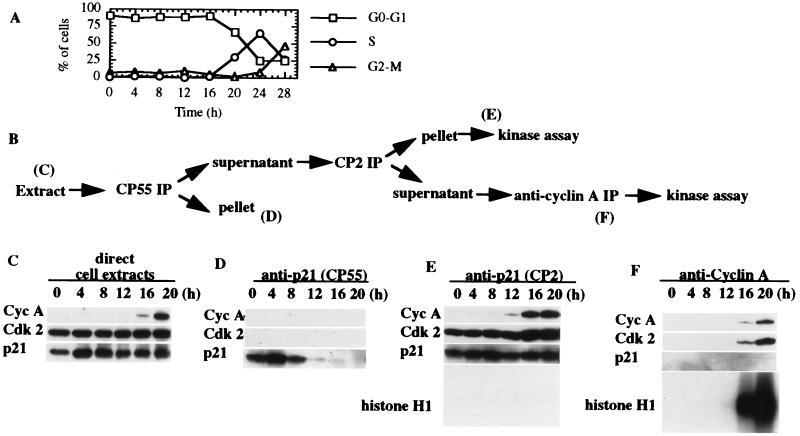
Figure 3.
Excess free p21 exists exclusively during early G1 phase. (A) WI38 cells were synchronized by serum deprivation, and the cell cycle profile determined by FACS analysis is shown. (B) Whole cell extracts derived from WI38 cells were analyzed directly or by sequential immunoprecipitation according to the scheme shown. (C) Extract (20 μg) from each stage was loaded directly and detected by Western blotting to reveal the total amounts of cyclin A, Cdk2, and p21. (D) Protein (300 μg) was immunoprecipitated by using CP55 to detect free p21 at each cell cycle time point (the first round of three consecutive depletions is shown). (E) Supernatants after three rounds of depletion of free p21 by CP55 were subjected to CP2 immunoprecipitation to detect cyclin A/Cdk2/p21 ternary complexes during the cell cycle. Histone H1 kinase activity was measured in parallel. (F) Supernatants after the immunoprecipitation in E were subjected to immunoprecipitation using anti-cyclin A antibody-conjugated beads to detect the remaining cyclin A/Cdk2 complex. Relative Western blot exposure times are the same for each part of the figure. Histone H1 kinase activity was measured in parallel.
We conclude that in an asynchronous population of human fibroblasts, cyclin A/Cdk2 complexes associated with p21 are inactive. Here, we have, to the best of our knowledge, for the first time compared the specific activities (in contrast to absolute kinase activity alone) of native cyclin A/Cdk2 complexes by immunoprecipitation with multiple antibodies. Although these experiments do not allow us to discriminate between complexes that may contain one or more molecules of p21–the latter being the proposed inhibitory form of p21 (10)–they nevertheless indicate the complete loss of activity of Cdks upon association with p21 in vivo. Recent experiments with p21−/− mouse embryo fibroblasts, in which Cdk2 (but not Cdk4) activity is enhanced relative to the wild-type control, support such an in vivo inhibitory activity for p21 (18).
Dynamics of p21 Interactions with Cyclin/Cdk2 During the Cell Cycle.
In light of the above findings with asynchronous cells suggesting the near-complete inhibition of Cdk2 associated with p21, we considered the possibility that p21 complexes with cyclin-dependent kinases might display activity at specific stages of the cell cycle and that a survey of an asynchronous population would preclude a measurement of such activity. To this end, WI38 cells were made quiescent by serum starvation and restimulated to enter the cell cycle by the addition of 10% serum. Serum deprivation resulted in the arrest of a majority of cells (>85%) as determined by FACS analysis (Fig. 3A). The cells reentered the cell cycle upon serum addition, entering S phase between 16 and 20 hr after restimulation. The largest fraction of cells were in S phase 24 hr after restimulation. Extracts of samples collected at each time point during serum deprivation and restimulation were subjected to direct Western blotting to determine the total amounts of cyclin A, Cdk2, and p21 present during cell cycle reentry (Fig. 3C). As expected from previous studies (19–21), the abundance of p21 protein does not fluctuate dramatically during the time course, although p21 levels did increase by approximately 2-fold upon serum stimulation (compare 0 and 4 hr time points). In contrast, cyclin A levels rose sharply as cells entered S phase, peaking between 20–24 hr poststimulation (data not shown).
To estimate the level of cyclin A/Cdk2 kinase activity associated with p21 as cells exited G0 and progressed through G1 and S phases, we performed the sequential immunoprecipitations outlined in Fig. 3B. Western blotting was performed at each stage to estimate the levels of each protein of interest. Because the ultimate goal of these experiments was to determine the kinase activity of cyclin A/Cdk2/p21 complexes and cyclin A/Cdk2 that was completely devoid of p21, we initially performed immunoprecipitations on extracts from each time point with CP55 to inspect free p21 levels and to remove this factor from further activity considerations. As expected from our initial studies, CP55 immunoprecipitated p21 to the exclusion of both cyclin A and Cdk2 (Fig. 3D). Remarkably, we observed a vast excess of free p21 in quiescent and early G1 phase cells. Between 8 and 12 hr poststimulation, however, free p21 levels dropped precipitously, although the total amount of p21 during this time remained largely unchanged (compare 4–12 hr time points in C and D). Furthermore, this cell cycle-dependent immunoprecipitation of p21 by CP55 indicates that this antibody is not simply disrupting p21 complexes and thereby artificially creating free p21.
After depletion of free p21 from extracts at each time point with CP55, we subsequently immunoprecipitated the same extracts with CP2, which recognizes all known p21-associated cyclin/Cdk complexes (see Fig. 1). Although robust amounts of p21 and Cdk2 were precipitated throughout the time course, cyclin A was not detected until approximately 12 hr after serum addition, as expected from Western blots of total cell lysate (Fig. 3E). However, association between this newly synthesized cyclin A/cdk2 and p21 resulted in negligible levels of histone H1 kinase activity, again suggesting that Cdk2 complexes are inhibited by association with p21 in vivo, irrespective of cell cycle position.
Successive rounds of immunoprecipitation with CP2 led to depletion of p21 from extracts, as determined by Western blotting (Figs. 2 and Fig. 3F). Next, the use of these depleted extracts allowed us to estimate the abundance and kinase activity of cyclin A/Cdk2 complexes lacking p21 as cells traversed the G1 phase. Immunoprecipitation of such extracts with anti-cyclin A antibodies revealed the presence of cyclin A/Cdk2 16 and 20 hr after serum stimulation, and as expected this kinase was not associated with p21 (Fig. 3F). Thus, most, but not all, of the cyclin A/Cdk2 is associated with p21 in these cells. When we compared the specific kinase activity of residual cyclin A/Cdk2 not associated with p21 (Fig. 3F, 16 and 20 hr time points) with that of the cyclin A/Cdk2/p21 ternary complex (Fig. 3E, 16 and 20 hr time points), we noticed a striking difference. Although there was considerably less cyclin A and Cdk2 in binary complexes as compared with the amount present in the ternary complex obtained by immunoprecipitation with CP2, the amount of kinase activity in the former complex was much greater. Indeed, when we calculated the specific Cdk2 activity of both complexes by performing PhosphorImager analysis and densitometric scanning of corresponding autoradiograms and Western blots (see Materials and Methods), we found that the activity of the cyclin A/Cdk2 complex was 166- and 158-fold higher than that of the ternary complex at 16 and 20 hr poststimulation, respectively.
We were concerned about the possibility that the above findings may have resulted from artificially elevated levels of p21 induced by serum stimulation of quiescent cells (19, 20, 22). To alleviate these concerns, we induced quiescence in WI38 cells by a second method, namely, contact inhibition. This treatment resulted in growth arrest of a substantial fraction of cells (>80%), and high levels of free p21 similar to those described above were observed exclusively at early time points after release from contact inhibition (data not shown). This suggests that the existence of free p21 may be an intrinsic feature of early G1 cells, although confirmation will require the testing of additional cell lines using similar and unrelated synchronization techniques.
From these experiments we can draw several conclusions. First, although total p21 levels do not change dramatically during the cell cycle, the amount of free p21 (i.e., p21 that is not associated with cyclin/Cdk complexes) is high in early G1, but drops sharply as cells progress toward S phase. From these data, we surmise that excess p21 ensures that cyclin/Cdk complexes are inactive in early G1. As cells progress through G1, cyclin A levels rise, and the levels of cyclin A/Cdk2 surpass those of p21, providing the impetus for the onset of S phase. Thus, our data support a threshold hypothesis for p21, which had been proposed based on in vitro experiments, in which the ratio of Cdk to p21 determines enzymatic activity (4). However, in contrast with previous findings, we do not detect significant histone H1 kinase activity associated with p21 during cell cycle progression from G0 to S phase, when cyclin A/Cdk2 and cyclin A/Cdk2/p21 complexes are compared.
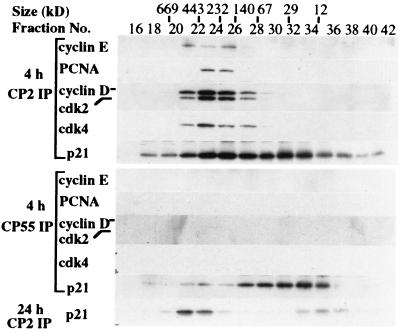
Estimating the Size of Free p21.
The preceding experiments indicated that one antibody, CP55, was uniquely able to recognize p21 to the exclusion of all other cell cycle regulators. To rule out the possibility that CP55 immunoprecipitated a complex of other proteins not detected in our Western blotting analysis, we estimated the size of p21 complexes in early G1 (4 hr after serum restimulation) and S phases (24 hr after serum stimulation) by subjecting extracts of WI38 cells to S-300 gel filtration chromatography (Fig. 4). Extracts from early G1 cells contained both free p21, which migrated at a position (between the 12 and 29 kDa markers) predicted for a monomer, and larger complexes (between the 67 and 443 kDa markers) that contained p21 in association with cyclin D/Cdk4, cyclin E/Cdk2, and PCNA. Of these two pools, CP55 was solely capable of immunoprecipitating the low molecular fraction between 12 and 29 kDa in size. The p21 fraction immunoprecipitated by CP55 comigrates exactly with recombinant p21 purified from bacteria (data not shown), suggesting that a form of p21 free of interacting proteins was recognized by this antibody. In striking contrast, p21 is largely confined to high molecular weight complexes in extracts from S phase cells.
Figure 4.
Free p21 in WI38 cells 4 hr after serum restimulation revealed by sizing column chromatography. An S-300 column was used to separate p21 and its complexes as described. The elution of calibration standards is indicated at the top. Protein (500 μg) derived from extracts of cells 4 or 24 hr after serum stimulation were loaded onto the column. Fractions were collected, halved, and immunoprecipitated with either CP2 or CP55 as labeled. The samples were analyzed by Western blotting to reveal p21 and its complexes.
We tested whether mixing extracts from cells stimulated for 4 hr would inhibit cyclin A/Cdk2 kinase activity found in 24 hr cell extracts, and this was indeed the case (data not shown), suggesting that this CKI was at least partly responsible for the inhibition of kinase activity in early G1 cells.
In summary, we have described a set of antibodies raised against p21 that have allowed us to document the low specific kinase activity associated with this CKI in vivo. These findings contrast with previous ones, some of which relied on reconstitution of ternary complexes with recombinant proteins, to show that kinase complexes with p21 retained activity (4, 10). Our experiments differ in several important ways. First, we have used multiple mAbs to study endogenous p21-associated histone H1 kinase activity, whereas previous studies relied on polyclonal sera. Second, we have examined the specific activity of p21-associated kinase complexes, rather than absolute kinase activity, and compared it with cyclin A/Cdk2 complexes lacking p21. It is important to point out that we have not addressed the specific kinase activity of p21 complexes with cyclin D and cyclin E. Such experiments may be informative in light of recent data with a p27-inducible cell line in which it was shown that Cdk2, but not Cdk4, was inhibited by elevated p27 (23). Nevertheless, our findings regarding free p21 in early G1 phase lend physiological support to a threshold hypothesis for cell cycle regulation by the p21 protein.
Acknowledgments
B.D.D. is indebted to E. Harlow, in whose laboratory the anti-p21 antibodies were produced and for the gift of various additional antibodies. We are grateful to Y. Kleyner for excellent technical help and to K. Engel, J. Ross, and I. Sánchez for comments on the manuscript. We thank S. Shiff for anti-cyclin B antibodies. This work was supported by Department of the Army Breast Cancer Research Grant DAMD17-96-1-6092. B.D.D. was also supported by a Damon Runyon Scholar Award (DRS-01), generously donated by E. and K. Langone, during the initial phase of this work.
ABBREVIATIONS
- Cdk
cyclin-dependent kinase
- CKI
cyclin-dependent kinase inhibitor
- PCNA
proliferating cell nuclear antigen
- HA
hemagglutinin
References
- 1. Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 2.Gu Y, Turck C W, Morgan D O. Nature (London) 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 3.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 4.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L-H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goubin F, Ducommun B. Oncogene. 1995;10:2281–2287. [PubMed] [Google Scholar]
- 6.Nakanishi M, Robetorye R S, Adami G R, Pereira-Smith O M, Smith J R. EMBO J. 1995;14:555–563. doi: 10.1002/j.1460-2075.1995.tb07031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fotedar R, Fitzgerald P, Rouselle T, Cannella D, Doree M, Messier H, Fotedar A. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 8.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo A R, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Nature (London) 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Hannon G J, Beach D. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 11.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 12.Dynlacht B D, Ngwu C, Winston J, Swindell E C, Elledge S J, Harlow E, Harper J W. Methods Enzymol. 1997;283:230–244. doi: 10.1016/s0076-6879(97)83019-4. [DOI] [PubMed] [Google Scholar]
- 13.Dynlacht B D, Flores O, Lees J A, Harlow E. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Harlow E, Dynlacht B D. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Xiong Y, Beach D. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulic V, Stein G H, Far D F, Reed S I. Mol Cell Biol. 1998;18:546–557. doi: 10.1128/mcb.18.1.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brugarolas J, Bronson R T, Jacks T. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Jenkins C W, Nichols M A, Xiong Y. Oncogene. 1994;9:2261–2268. [PubMed] [Google Scholar]
- 20.Macleod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Hannon G J, Beach D, Stillman B. Curr Biol. 1996;6:189–199. doi: 10.1016/s0960-9822(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Y X, el-Deiry W S. Oncogene. 1996;12:1557–1564. [PubMed] [Google Scholar]
- 23.Blain S W, Montalvo E, Massague J. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]