Summary
Multivesicular bodies (MVBs) are critical intermediates in the trafficking of ubiquitinated receptors and other cargo destined for lysosomes. The formation of MVBs by invagination of the endosomal limiting membrane is catalyzed by the ESCRT complexes, a process that has recently been visualized in three-dimensional detail by electron tomography. Structural and biochemical analysis of the upstream components, Vps27-Hse1, ESCRT-I, and ESCRT-II, shows how these complexes assemble and cluster cargo. Rapid progress has been made in understanding the assembly and disassembly of the ESCRT-III complex and the interactions of its subunits with MIT domain and other proteins. A key role for deubiquitination in the regulation of the system has been demonstrated. One central question remains largely unanswered, which is how the ESCRTs actually promote the invagination of the endosomal membrane.
The lysosome is the eukaryotic cell's main engine for the breakdown of membrane proteins and internalized materials. Cell surface receptors destined for the lysosome arrive through a pathway in which portions of the limiting membrane of endosomes containing these receptors invaginate into the lumen of the endosome [1-3]. At the stage when the endosome becomes filled with intralumenal vesicles (ILVs), it is referred to as a multivesicular body (MVB). Research into MVB biogenesis has exploded following the discovery in yeast of three protein complexes, Endosomal Sorting Complex Required for Transport (ESCRT) I, II, and III [4-6]. The ESCRT pathway is conserved throughout eukaryotes, where it plays essentially the same role in MVB biogenesis as in yeast. In animals, the ESCRT pathway is hijacked by viruses, which use the pathway to bud from the plasma membrane in a reaction that is topologically equivalent to the budding of ILVs [7-9]. Moreover, the ESCRT system is required for a third topologically equivalent process, the membrane scission event during cytokinesis [10,11].
The details of the ESCRT pathway have been described in several excellent reviews [12-17]. Here I summarize the main points (Fig. 1). Cargo is brought into the pathway by the Vps27-Hse1 complex (Table 1). Vps27 is targeted to endosomes via its FYVE domain, which binds to the endosomal lipid phosphatidylinositol 3-phosphate. Vps27 contains P(S/T)XP motifs that recruit the soluble heterotetrameric ESCRT-I complex to the endosomal membrane via the UEV domain of its Vps23 subunit. ESCRTI recruits another soluble heterotetrameric complex, ESCRT-II. All three of the above mentioned complexes contain ubiquitin binding domains that interact with ubiquitinated cargo proteins. The ESCRT-III proteins are soluble monomers until recruited to the endosomal membrane, where they form an insoluble array. Recruitment is thought to be initiated by ESCRT-II binding to Vps20. Cargo is deubiquitinated following assembly of the ESCRT-III lattice. Finally, ILV formation and ESCRT-III disassembly by the ATPase Vps4 take place, either concurrently or sequentially.
Figure 1.
A model for the organization of the ESCRT system. In this scheme, Vps27-Hse1, ESCRT-I, and ESCRT-II are targeted to membranes by lipids (green), where they cluster cargo (red). ESCRT-I and II interact at a 1:1 stoichiometry. The stoichiometry of ESCRT-I binding to Vps27-Hse1 is undefined and for simplicity is shown as 1:1. ESCRT-II recruits ESCRT-III via its Vps20 subunit. Vps20 might nucleate the polymerization of Snf7, which forms filamentous arrays as demonstrated by Hanson, Heuser, and colleagues (personal communication). Other ESCRT-III subunits probably polymerize in a similar manner, and it is not clear whether homo- or heteromeric polymers predominate. Incorporation of Vps2 and Did2 into ESCRT-III initiates the depolymerization of ESCRT-III by Vps4:Vta1. ILVs are thought to be formed at some point during the polymerization/depolymerization cycle. This model is similar to the recently proposed “concentric circle” model [17].
Table 1.
Nomenclature for ESCRT complex subunits
| Complex | Yeast or generic protein | Synonyms (yeast) | Metazoan protein | Synonyms (metazoan) |
|---|---|---|---|---|
| Vps27-Hse1 | Vps27 | VPS27 | HRS | |
| Hse1 | HSE1 | STAM | ||
| ESCRT-I | Vps23 | VPS23 | TSG101 | |
| Vps28 | VPS28 | |||
| Vps37 | VPS37A,B,C,D | |||
| Mvb12 | MVB12A,B | |||
| ESCRT-II | Vps22 | VPS22 | EAP30 | |
| Vps25 | VPS25 | EAP20 | ||
| Vps36 | VPS36 | EAP45 | ||
| ESCRT-III | Vps2 | VPS2A,B | CHMP2A,B | |
| Vps20 | VPS20 | CHMP6 | ||
| Vps24 | VPS24 | CHMP3 | ||
| Snf7 | Vps32 | SNF7A,B,C | CHMP4A, B, C | |
| Vps60 | VPS60 | CHMP5 | ||
| Did2 | DID2 | CHMP1A, B | ||
| Vps4 | V ps4 | VPA4A,B | SKD1 | |
| Vta1 | VTA1 | LIP5 | ||
| Other | Bro1 | Vps31 | BRO1 | ALIX, AIP1 |
The structure of MVBs and the class E compartment
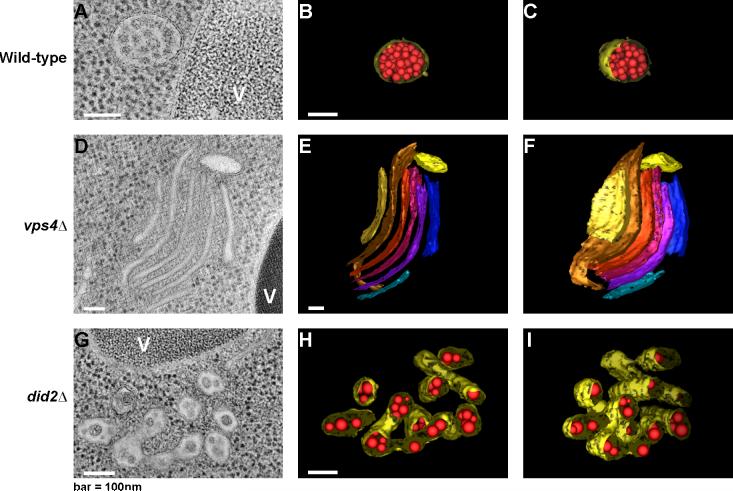
Electron tomography showed that in wild-type yeast, MVBs are roughly spherical, ∼200 nm across, and filled with spherical ∼24 nm ILVs [18] (Fig. 2A-C). In yeast and human [19,20] cells, defects in the ESCRT machinery not only interfere with normal MVB formation, they also manifest a distinctive abnormal subcellular structure, the class E compartment. The class E compartment consists of stacked flat cisternae-like membranes (Fig. 2D-F), which are not connected to each other [18]. It is not known what determines the morphology of the class E compartment. In did2Δ cells, which have an intermediate phenotype, the MVBs are elongated and irregular in shape, and the ILVs are enlarged (Fig. 2G-I) [18].
Figure 2.
Multivesicular bodies and the class E compartment. Genotypes are identified at left. Panels A, D, and G show tomographic sections, and the remaining panels show three-dimensional reconstructions. Panels A, B, C, show normal yeast multivesicular bodies; D, E, F show the class E compartment; and G, H, I show multivesicular bodies under conditions of defective cargo deubiquitination. Reproduced from the Journal of Cell Biology, 2006, 175: 715−720 [18]. Copyright 2006, Rockefeller University Press.
Cargo delivery into the ESCRT pathway
Monoubiquitination [21,22] and Lys-63 linked polyubiquitination [22-24] direct cargo into the endolysosomal pathway. Vps27-Hse1, a key upstream component of the pathway, binds ubiquitin via UIM motifs. While yeast Vps27 contains two UIMs, its human orthologs contains one UIM with a double-sided ubiquitin binding capability [25]. The yeast and human orthologs thus bind cooperatively to multiple ubiquitin moieties by different mechanisms. The Hse1 subunit of the Vps27-Hse1 complex seems to be involved mainly in the recruitment of ubiquitin ligases and deubiquitinating enzymes (DUBs) [30]. The complex heterodimerizes via two intertwined GAT domains [26], linking Vps27 architecturally to the GAT domain-containing GGA and TOM1L trafficking protein families. The GGAs [27] and the TOM1, TOM1L1, and TOM1L2 proteins [28] are like Vps27 in that they are bind to ubiquitin and ESCRT-I. VPS27 appears to concentrate ubiquitinated cargo into microdomains, to which VPS27 recruits clathrin. Clathrin forms a flat coat over these microdomains, rather than the typical curved clathrin coated vesicles [29]. ESCRT-I is thought to either be recruited by VPS27 at the edges of the clathrin coat, or to enter the coat by displacing clathrin in an exchange reaction.
How the ESCRT machinery assembles on endosomes
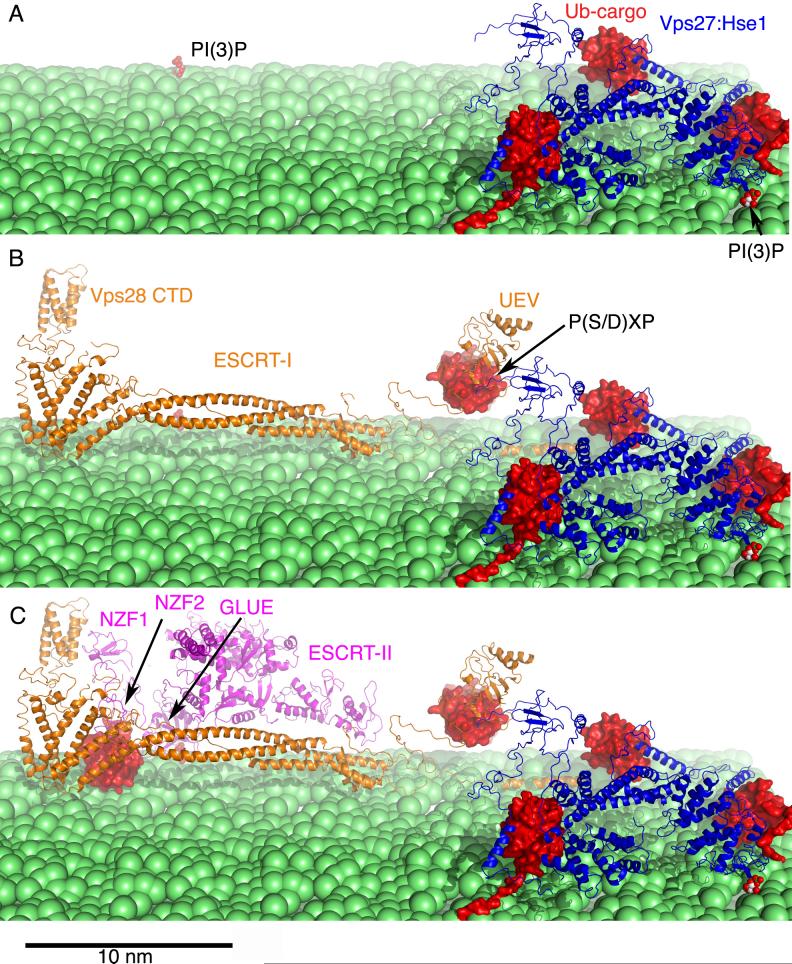
In the conventional model for ESCRT assembly, ESCRT recruitment is initiated by Vps27, and ESCRT-I, II, and III then sequentially recruit one another from the cytosol. A recent ultrastructural study found that ESCRTs are localized to a wide range of endosomal and other membranes within cells, leading to the appearance under optical microscopy that they are cytosolic [31]. ESCRT-I is a heterotetramer of Vps23, Vps28, Vps37, and Mvb12 in both yeast [32-35] and humans [36,37], and the subunits are present in one copy each [35-38]. The binding sites for ubiquitinated cargo and ESCRT-II are at opposite ends of the 25 nm long ESCRT-I complex [35] (Fig. 3), which argues against the concept that ubiquitinated cargo is handed off from one ESCRT complex to another in a “conveyor belt” mechanism. Current thinking favors the idea that ESCRT-I and II co-assemble and cluster multiple ubiquitinated transmembrane proteins for packaging into ILVs [17,35].
Figure 3.
A structural model for the assembly and cargo clustering by A) Vps27-Hse1, B) ESCRT-I, and C) ESCRT-II. The structures shown are described in more detail in references [16,26,35].
In yeast, the C-terminal domain (CTD) of the Vps28 subunit of ESCRT-I recruits ESCRT-II [39,40] (Fig. 3). ESCRT-II consists of one molecule each of Vps22 and Vps36, and two molecules of Vps25. The Vps28-CTD domain binds to the NZF1 zinc finger in the N-terminus of the Vps36 subunit of ESCRT-II [38]. Human ESCRT-II lacks the NZF1 domain, leaving the mechanism for the putative human ESCRT-I/II interaction uncertain. The N-termini of Vps36 orthologs contain a pleckstrin homology (PH) domain variant called a “GLUE” domain. The human VPS36 GLUE domain binds to phosphoinositides [41] and ubiquitin [41-43]. The yeast Vps36 GLUE domain binds with moderate affinity and specificity to phosphoinositide-bearing membranes [40], but does not directly bind to ubiquitin. Two zinc fingers, the above-mentioned NZF1, and the ubiquitin-binding NZF2, are inserted within the sequence of the yeast GLUE domain. Thus yeast ESCRT-II binds the same ligands as the human complex through a more complicated set of domains.
Structural and interaction data on Vps27-Hse1, ESCRT-I, and ESCRT-II are sufficient to form a reasonably complete picture of the assembly of the upstream half of the ESCRT machinery (Fig. 3). The individual interactions of the ESCRT complexes with lipids are of moderate to high (∼ 100 nM to low micromolar) affinity, and the interactions with individual ubiquitinated cargo are exceptionally weak (0.1 to 2 mM). However, the assembled network is cross-linked by interactions between the ESCRT complexes, which makes it a multivalent platform that can bind tightly to the membrane and cluster multiple ubiquitinated cargo molecules.
Regulation of ESCRT-III assembly
There are six ESCRT-III or ESCRT-III-like proteins in yeast, Vps2, Vps20, Vps24, Snf7, Did2, and Vps60. Vps20 and Snf7 associate with each other, and probably act an early stage in ESCRT-III assembly. Vps20 binds directly to ESCRT-II, and reportedly to ESCRT-I as well [44]. There is no direct evidence that ESCRT-III comprises the membrane scission “machine” that makes and detaches ILVs. However, the ability of Snf7 and possibly other ESCRT-III subunits to form arrays has made ESCRT-III a prime candidate to fill such a role. The main evidence for array formation comes from SNF7 overexpression studies [45,46]. Direct imaging of the SNF7 array shows that it consists of filaments arranged in a spiral pattern (P. Hanson, pers. comm.). The SNF7 array appears capable of deforming membranes into tubules that project away from the cytoplasm. This observation might be consistent with a direct role for Snf7 in the mechanics of ILV formation. Did2 and Vps2 appear to act at a later stage, where they are responsible for recruiting Vps4 [18,47,48].
ESCRT-III subunits are distinguished by a primarily basic N-terminus and an acidic C-terminus. The crystallization of a VPS24 construct provides most of what we know about ESCRT-III structure [49]. The crystallized VPS24 contains a deletion in its most C-terminal predicted helix, which leads to dimerization both in solution and in the crystal. A flat basic face formed by the two N-terminal helices presents a binding site for acidic phospholipids, while the C-terminal part of the crystallized structure is acidic. The most C-terminal helix, not present in the crystal, has been a topic of investigation in its own right. The C-terminal helix appears to autoinhibit ESCRT-III monomers with respect to array formation [46,50]. Deletion of this helix from VPS24 and SNF7 promotes the formation of insoluble membrane-bound aggregates in vivo that interfere with MVB biogenesis and sorting [46,50]. Furthermore, the C-terminal helices of ESCRT-III proteins play multiple roles in interacting with other proteins. The C-termini of Vps2 and Did2 contain a MIT (microtubule interacting and transport) domain-interacting motif (MIM), defined by six conserved residues that make up a contiguous surface on one face of the helix (Fig. 4) [47,48]. This suggests that binding of other proteins to the C-terminal helices of ESCRT-III monomers could initiate array formation. This might, for example, occur between ESCRT-III proteins Vps20 or Snf7 with ESCRT-II or Bro1, respectively. Reciprocally, ESCRT-III subunits within the array expose their C-terminal helices, making them available to recruit MIT-domain containing proteins such as Vps4.
Figure 4.
MIT domains and MIM motifs coordinate the assembly and disassembly of ESCRT-III. The MIM of DID2B (red) bound to the MIT domain of VPS4A (orange) [48] illustrates the recognition of the C-terminal MIM regions of ESCRT-III subunits by Vps4. The structure of the C-terminally truncated dimer of VPS24 [49] is shown to illustrate the putative membrane binding “tiles” of the ESCRT-III array. The structure is colored according to electrostatic potential, with blue electropositive, and red electronegative.
Deubiquitination: the final signal for cargo entry into ILVs?
In yeast, the key DUB associated with the ESCRT pathway is Doa4. Doa4 is targeted to endosomes via an N-terminal predicted helical region [51]. Doa4 localization depends on the assembly of ESCRT-III, while its enzymatic activation is promoted by Bro1 [52]. Doa4 is not critical for MVB biogenesis, but in its absence, ILVs are fewer and smaller [52]. This is consistent with a lighter cargo load as ubiquitinated cargoes such as Cps1 and Gap1 are retained at the limiting membrane [51-53].
The major human DUBs implicated in the MVB pathway are AMSH and UBPY. Knockdown or catalytic inactivation of these enzymes markedly slows the degradation of EGF receptor [54-58]. AMSH and UBPY are recruited to endosomes by interactions between their N-terminal MIT domains and late-acting ESCRT-III subunits [54-57,59]. Remarkably, UBPY is catalytically activated by the HSE1 subunit of the early-acting VPS27-HSE1 complex [57]. UBPY thus appears to function at both early and late stages of MVB biogenesis. The findings in human cells are in good agreement with the yeast data, and support the concept that cargo deubiquitination is a critical late-stage signal for cargo entry into ILVs. This concept represents a significant change in thinking: the complete ubiquitination/deubiquitination cycle is considered essential, not just the initial ubiquitination of the cargo.
Regulation of ESCRT-III disassembly
The main thermodynamic driving force for MVB biogenesis is thought to be the consumption of ATP by Vps4. A complex of Vps4 with another protein, Vta1, [60-63] appears to disassemble the ESCRT-III lattice. In isolation, Vta1 is a rod-like homodimer, and it binds to the C-terminal β domain of Vps4 through a short conserved VSL region at its C-terminus [63]. Vta1 accelerates the ATPase activity of Vps4, and promotes the assembly of Vps4 into its functional from, a double hexameric ring. Vta1 directly binds to the ESCRT-III subunit Did2 [62]. Since Vps4 also directly binds to a subset of ESCRT-III subunits that includes Did2 [47,48], the multiplicity of binding sites in the Vps4:Vta1 complex suggests a mechanism for high cooperativity in binding to the ESCRT-III lattice. Current evidence favors a model in which Vps4 is an ATP-powered engine that pumps ESCRT-III monomers through the central pore of the double ring and into the cytosol [64]. A key question is whether Vps4 is involved directly in the mechanics of ILV scission, or indirectly by cleaning up the ESCRT-III lattice afterwards to enable additional rounds of scission.
Concluding remarks
The past two years have seen an explosion of structural and mechanistic insights into the targeting, assembly, and disassembly of the ESCRT machinery. The role of ubiquitination/deubiquitination circuits as regulatory elements is emerging in considerable detail, and there are hints that other modifications, such as phosphorylation, will also be important [37]. Advances in molecular structural analysis by x-ray crystallography and NMR have helped propel this field for the past few years, and EM tomography is now beginning to make equally important contributions at the level of subcellular structure. In vitro reconstitution of the ESCRT reaction should in principle be possible, given the identification and purification of many, if not all, of the factors involved. The field now appears ready to tackle the single most pressing issue head on, the mechanism of ILV formation and scission.
Acknowledgements
I thank B. Wendland, E. Conibear, Y. Ye, and J. Bonifacino for comments on the manuscript, P. Hanson for sharing unpublished data, G. Odorizzi for providing the images used in Fig.2, W. Sundquist and J. Skalicky for sharing structural coordinates prior to release, and D. Yang and Y.-G. Kim for assistance with figures. Work in my laboratory is supported by the NIH NIDDK intramural program and the NIH IATAP program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 2.Piper RC, Luzio JP. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 2007;19:459–465. doi: 10.1016/j.ceb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell MRG, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 5.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 6.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 7.Morita E, Sundquist WI. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 8.Demirov DG, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 10.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [**This paper, along with [11], reveals a third role for the ESCRTs in membrane scission, in addition to their known roles in MVB formation and viral budding. This adds to the body of evidence that the ESCRTs act directly in membrane scission.] [DOI] [PubMed] [Google Scholar]
- 11.Morita E, Sandrin V, Chung HY, Morham SG, Gygi S, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 13.Babst M. A Protein's Final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 14.Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends In Cell Biology. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomolec. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 17.Nickerson DP, Russell DW, Odorizzi G. A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 2007;8:644–650. doi: 10.1038/sj.embor.7401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [**With [62], the poorly understood ESCRT-III-like protein Did2 is shown to play a key role in recruiting Vps4 and thereby initiating ESCRT-III disassembly. What distinguishes this paper, along with [52], is the use of electron tomography to visualize the effects of defects in the ESCRT pathway on MVB morphology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyotte A, Russell MRG, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian ‘Class E’ compartment: a multicisternal early endosome with multiple sorting defects. J. Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- 20.Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 22.Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang FT, Kirkpatrick D, Jiang XJ, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, Luzio JP, Lehner PJ. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. Embo J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nature Structural & Molecular Biology. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 26.Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, Bonifacino JS, Hurley JH. The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell. 2007;12:973–986. doi: 10.1016/j.devcel.2007.04.013. [*The structure and 1:1 stoichiometry of the Vps27-Hse1 core complex provide a key missing link in understanding the assembly of the upstream components of the ESCRT machinery.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattera R, Puertollano R, Smith WJ, Bonifacino JS. The trihelical bundle subdomain of the GGA proteins interacts with multiple partners through overlapping but distinct sites. J. Biol. Chem. 2004;279:31409–31418. doi: 10.1074/jbc.M402183200. [DOI] [PubMed] [Google Scholar]
- 28.Puertollano R. Interactions of Tom1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- 29.Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J. Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [*This paper provides insight into the role of clathrin, and suggests parallels between the GGA adaptors and VPS27 (Hrs) as ubiquitin-binding clathrin adaptors functioning upstream of the ESCRTs.] [DOI] [PubMed] [Google Scholar]
- 30.Ren J, Kee Y, Huibergtse JM, Piper RC. Hse1, a Component of the Yeast Hrs-STAM Ubiquitin Sorting Complex, Associates with Ubiquitin Peptidases and a Ligase to Control Sorting Efficiency into Multivesicular Bodies. Mol Biol Cell. 2007;18:324–335. doi: 10.1091/mbc.E06-06-0557. [*Reveals that the less-studied half of the Vps27-Hse1 complex has multiple roles in recruiting deubiquitinating enzymes and a ubiquitin ligase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welsch S, Habermann A, Jager S, Muller B, Krijnse-Locker J, Krausslich HG. Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic. 2006;7:1551–1566. doi: 10.1111/j.1600-0854.2006.00489.x. [*This study suggests the concept that the major pool of “inactive” ESCRTs is cytosolic needs to be reconsidered.] [DOI] [PubMed] [Google Scholar]
- 32.Oestreich AJ, Davies BA, Payne JA, Katzmann DJ. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol Biol Cell. 2006;18:646–657. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol Biol Cell. 2006;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu T, Sun J, Saksena S, Emr SD. New component of ESCRT-I regulates endosomal sorting complex assembly. J. Cell Biol. 2006;175:815–823. doi: 10.1083/jcb.200608053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostelansky MS, Schluter C, Tam YYC, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [**An essentially complete model of the 1:1:1:1 ESCRT-I heterotetramer was obtained, thanks to the identification of Mvb12 as the fourth subunit of the complex, as independently described in [32-34]. The 25 nm long rod-like structure is roughly on the same size scale as a yeast ILV. The ubiquitin and ESCRT-II binding sites are 25 nm apart, arguing against a direct hand-off mechanism, and favoring a cargo clustering mechanism.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audhya A, McLeod IX, Yates JR, Oegama K. MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLOS ONE. 2007;2:e956. doi: 10.1371/journal.pone.0000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host and Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, Williams RL. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. Embo J. 2007;26:600–612. doi: 10.1038/sj.emboj.7601501. [*This structure provides essential insights into how yeast ESCRT-I and II assemble together, although it seems clear that different principles will apply to the metazoan complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, Emr SD, Hurley JH. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–126. doi: 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teo HL, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 41.Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiano D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [*The discovery of the GLUE domain in mammalian ESCRT-II explains how this complex is recruited to membranes by phosphoinositides and how it binds to ubiquitinated cargo. This findings were expanded to yeast ESCRT-II and elucidated structurally in [40].] [DOI] [PubMed] [Google Scholar]
- 42.Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nature Structural & Molecular Biology. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- 43.Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nature Structural & Molecular Biology. 2006;13:1031–1032. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- 44.Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7−1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [*The homo-oligomeric SNF7 array described here underpins current thinking about how ESCRT-III might mediate membrane scission.] [DOI] [PubMed] [Google Scholar]
- 46.Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-terminal Domain. Traffic. 2007 doi: 10.1111/j.1600-0854.2007.00584.x. 10.1111/j.1600−0854.2007.00584.x. [* see [50].] [DOI] [PubMed]
- 47.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [** See [48].] [DOI] [PubMed] [Google Scholar]
- 48.Stuchell-Brereton M, Skalicky J, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [**The structural characterization of MIT/ESCRT-III recognition are very timely, given an explosion of information on MIT domain proteins. These studies are consistent with the identification of Did2 as a Vps4 coupling factor, and show that Vps2 also has this function.] [DOI] [PubMed] [Google Scholar]
- 49.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [**The first and so far only structure of an ESCRT-III protein reveals a flattened tile-like dimer with a basic membrane-binding face, suggesting a mechanism for array assembly.] [DOI] [PubMed] [Google Scholar]
- 50.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [With [46], suggests a model for allosteric regulation of ESCRT-III.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amerik A, Sindhi N, Hochstrasser M. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J. Cell Biol. 2006;175:825–835. doi: 10.1083/jcb.200605134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. Embo J. 2007;26:2454–2464. doi: 10.1038/sj.emboj.7601692. [*Together with [51,53], shows deubiquitination is important for the sorting of ubiquitinated cargoes into ILVs in yeast, and goes on to characterize the effects of reducing the cargo load on ILV density and MVB size.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikko E, Andre B. Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic. 2007;8:566–581. doi: 10.1111/j.1600-0854.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- 54.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 55.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 2006;281:23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 56.Ma YM, Boucrot E, Villen J, Affar el B, Gygi SP, Gottlinger H, Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor degradation. J. Biol. Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 57.Row PE, Lui H, Hayes S, Welchman R, Charalabous P, Hofmann K, Claugue MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient EGF receptor degradation. J. Biol. Chem. 2007 doi: 10.1074/jbc.M704009200. in press. [*With [54-56,58]highlights the role of deubiquitination in the sorting of ubiquitinated cargo into MVBs in mammalian cells, and goes on to add a detailed characterization of the allosteric regulation of, and the identificatio of a MIT domain, in UBPY.] [DOI] [PubMed] [Google Scholar]
- 58.Kyuuma M, Kikuchi K, Kojima K, Sugawara Y, Sato M, Mano N, Goto J, Takeshita T, Yamamoto A, Sugamura K, et al. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Structure And Function. 2006;31:159–172. doi: 10.1247/csf.06023. [DOI] [PubMed] [Google Scholar]
- 59.Tsang HTH, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Haas TJ, Sliwinski MK, Martinez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell. 2007;19:1295–1312. doi: 10.1105/tpc.106.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vajjhala PR, Catchpoole E, Nguyen CH, Kistler C, Munn AL. Vps4 regulates a subset of protein interactions at the multivesicular endosome. Febs Journal. 2007;274:1894–1907. doi: 10.1111/j.1742-4658.2007.05736.x. [DOI] [PubMed] [Google Scholar]
- 62.Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta 1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [*With [60-62], sheds light on the role of Vta1 as a partner with Vps4 in ESCRT-III disassembly, and adds new information on the role of Vta1 in allosterically activating Vps4.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, et al. Structural and mechanistic studies of VPS4 proteins. Embo J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]