Abstract
The discovery of hyperthermophilic microorganisms and the analysis of hyperthermostable enzymes has established the fact that multisubunit enzymes can survive for prolonged periods at temperatures above 100°C. We have carried out homology-based modeling and direct structure comparison on the hexameric glutamate dehydrogenases from the hyperthermophiles Pyrococcus furiosus and Thermococcus litoralis whose optimal growth temperatures are 100°C and 88°C, respectively, to determine key stabilizing features. These enzymes, which are 87% homologous, differ 16-fold in thermal stability at 104°C. We observed that an intersubunit ion-pair network was substantially reduced in the less stable enzyme from T. litoralis, and two residues were then altered to restore these interactions. The single mutations both had adverse effects on the thermostability of the protein. However, with both mutations in place, we observed a fourfold improvement of stability at 104°C over the wild-type enzyme. The catalytic properties of the enzymes were unaffected by the mutations. These results suggest that extensive ion-pair networks may provide a general strategy for manipulating enzyme thermostability of multisubunit enzymes. However, this study emphasizes the importance of the exact local environment of a residue in determining its effects on stability.
Keywords: hyperthermophile/ion-pair/calorimetry/glutamate dehydrogenase/archaea
The discovery of hyperthermophilic microorganisms that grow optimally at or near 100°C has necessitated a radical revision of ideas concerning protein thermostability. Most of the enzymes from these organisms are stable for many hours at or above 100°C (1), suggesting that these enzymes must embody most of the mechanisms of thermostability that occur in extremely thermostable proteins. Thorough study of these proteins may therefore identify key structural determinants of thermal stability at very high temperatures. Trends commonly associated with elevated thermostability in proteins include relatively small solvent-exposed surface area (2), increased packing density that reduces cavities in the hydrophobic core (3, 4, 5, 6), an increase in core hydrophobicity (7, 8) decreased length of surface loops (6), and hydrogen bonds between polar residues (9). A prominent role for ion pairs in stabilization of proteins at or above 100°C, where the hydrophobic effect is minimal (10), has been inferred from the recently solved structures of several proteins from extreme thermophiles (11–14). This is consistent with earlier suggestions made on the role of ion pairs on stability (15). As yet, there is no general rule that governs amino acid composition of thermostable proteins and methods for predicting and designing stabilizing mutations are not reliable (16, 17, 18). Recent work (19–21) has indicated that significant increments of stability may be achieved in proteins from mesophiles by the inclusion of “rigidifying” mutations; however, we believe that the proteins from the hyperthermophiles may have evolved optimal adaptations of this sort. Our work has focused on homology-based modeling and structure comparison of the hexameric glutamate dehydrogenases (GluDHs) from the hyperthermophiles Pyrococcus furiosus and Thermococcus litoralis, archaea that grow optimally at 100°C (22) and 88°C (23), respectively, and that from a mesophilic organism, Clostridium symbiosum. The two hyperthermophilic enzymes provide a near-ideal comparative experimental system for studying the determinants of exceptional thermostability as they show distinct differences in stability yet are highly homologous. An unusual structural feature of P. furiosus GluDH is a number of extensive networks of buried intersubunit ion pairs (11), which is reduced in extent in homologous, but less thermostable, GluDHs (24, 25). Our strategy has been to identify differences between the ion-pair networks in the P. furiosus and T. litoralis GluDHs and to convert the stability of the T. litoralis GluDH to that of the more thermostable P. furiosus enzyme, to establish a causal relationship between intersubunit ion-pair networks and hyperstability.
MATERIALS AND METHODS
Site-Directed Mutagenesis.
We constructed mutants bearing the single Thr 138 replaced with Glu (T138E) and the double T138E/Asp 167 replaced with Thr (D167T) substitutions. Site-directed mutagenesis was performed by using either a modification of the uracil DNA glycosylase method for generating site-directed mutations or a modification of the Eckstein method (QuickChange mutagenesis system, Stratagene). Mutant T138E was constructed by using the uracil DNA glycosylase system and the following primer pairs: p15J (GGA TGA CCA TGG TTG AGC AAG ACC) and pT138-E Rev (U AUC CUC GUA UGG ACU UAU AAC ATC ATA GAT AGC); p16J (AGT GAG GGA TCC TCA CTT CTT GAT CCA TCC) and pT138-E For (A AGU CCA UAC GAG GAU AUU CCA GCT CCA GAC GT). Primers p15J/pT138E Rev and primers p16J/pT138E were used to amplify the 5′ fragment and the 3′ fragment respectively of the gdhA gene. Primers 15J and 16J were designed to anneal to the 5′ and 3′ ends of the gene, respectively, and contain the NcoI and BamHI restriction sites that allow cloning of the gene in the polylinker region of the pET 11-d expression vector (Novagen). The mutagenic primers were designed to be complementary to the mutation site in the gdhA gene, and their 5′ regions contained dU residues in place of dT residues. After amplification (PCR conditions: 95°C, 1′; 55°C, 1′; 72°C, 2′ for 30 cycles), the two gdhA fragments were mixed, uracil DNA glycosylase was added, and the reaction was incubated at 37°C for 12 h. The uracil DNA glycosylase reaction product was then purified (Qiaquick columns, Qiagen), digested with NcoI and BamHI restriction endonucleases, and cloned in pET 11-d plasmid. The ligation product was transformed in DH5a Escherichia coli competent cells, and recombinant clones were identified by direct PCR screening and then sequenced. Mutant D167T was constructed using the QuickChange mutagenesis system using the primer pair: pD167-T For (C TCA AGA AGG AAA ACC CCA TCC TTT GG) and pD167-T Rev (CC AAA GGA TGG GGT TTT CCT TCT TGA G). Following transformation in E. coli XL2 Blue, recombinant clones were selected and the mutated gdhA gene was sequenced. All plasmids containing the mutated genes were transformed in the expression host (E. coli BL21 cells, Novagen) and overexpressed as previously described (26).
Purification of GluDH. E. coli.
BL21(pTGDH) cells were centrifuged and the pellet was washed once in 0.9% NaCl and resuspended in 4 ml (per 0.5 l of original cell culture) of TED buffer (50 mM Tris⋅HCl, pH 7.6/1 mM EDTA/1 mM DTT) (26). A single freezing and thawing step was used to lyse the cells. After the dropwise addition of streptomycin sulfate (to a final concentration of 1%), the lysate was incubated for 1 h at 4°C, and the supernatant was subsequently recovered by centrifugation at 48,000 × g for 20 min at 4°C. The cell extract containing recombinant GluDH was heated for 35 min at 75°C, in 4-ml aliquots. Centrifugation at 48,000 × g for 20 min at 4°C yielded crude GluDH extract. The crude extract was diluted twofold with 50 mM Tris⋅HCl, pH 9.0, and loaded onto an anion exchange column (Q HyperD, 4.6 × 100 mm; Biosepra, Boston, MA), equilibrated with 50 mM Tris⋅HCl, pH 9.0. The column was washed with 50 mM Tris⋅HCl, pH 9.0, followed by 50 mM Tris⋅HCl, 0.2 M NaCl, pH 9.0, and the enzyme was eluted with a 0.2–1.0 M NaCl gradient. GluDH activity started to elute at 0.30 M NaCl. GluDH-containing fractions were pooled, diluted twofold with 10 mM l-glutamate, pH 8.0, and loaded onto an affinity column (Matrix Red A, 1 × 8 cm, Amicon) equilibrated with Buffer A (20 mM Tris⋅HCl, pH 8.0/28 mM NaCl/5 mM l-glutamate). The column was washed with buffer A and followed by buffer B (20 mM Tris⋅HCl, pH 8.0, 28 mM NaCl), and GluDH was eluted by the direct injection of 10 mM NADP. All chromatography steps were carried out at room temperature. Homogeneity of purified GluDH fractions was estimated by using 12% SDS/PAGE. Protein concentrations were determined by the Bio-Rad colorimetric microassay using BSA as a standard (Bio-Rad). GluDH activity was measured by the glutamate-dependent reduction of NADP+ at 80°C in HEPPS [N-hydroxyethyl piperazine-N′-(3-propanesulfonic acid)] buffer, as described (26). In all assays, the reaction was initiated by addition of NADP+. The thermostability of each mutant was determined at 104°C by using an oil bath inside a temperature-controlled oven (Lunaire, Williamsport, PA). For each data set, a protein concentration of 1.0 mg/ml in 700 mM KCl 50 mM diglycine pH 7.0 was used. After incubation at each temperature, residual GluDH activity was measured and the half life measured by using a semi-logarithmic plot of incubation time vs. residual activity.
Calorimetry.
Differential scanning microcalorimetry was conducted using a DASM4 instrument (28) operating with a temperature scan rate of 1°C/min from 40°C to 130°C. In control experiments, alteration of the scan rate between 0.5 and 2°C/min had little effect on the position or amplitude of the transitions. The enzyme concentration was 1.0 mg/ml in 700 mM KCl and 50 mM diglycine, pH 7.0. Data reduction was conducted by using microsoft excel.
RESULTS AND DISCUSSION
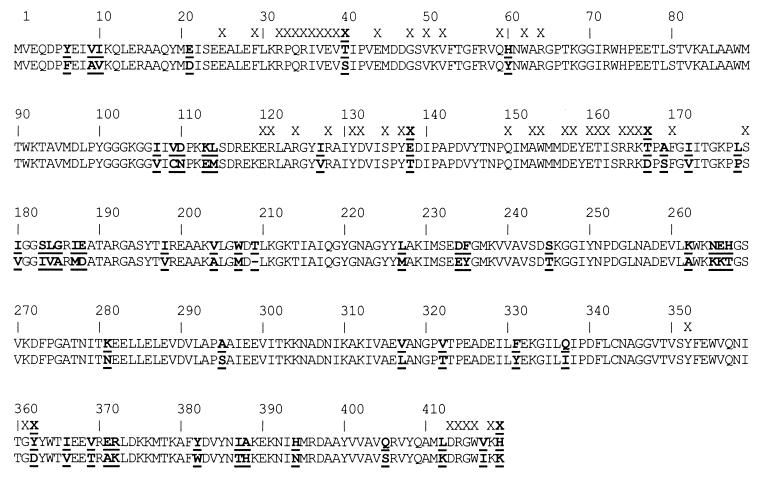
The amino acid sequences of P. furiosus and T. litoralis GluDHs are highly homologous and, following removal of the N-terminal methionine, the mature proteins consist of polypeptides of 419 and 418 amino acids, respectively, which are 87% identical and can be aligned unambiguously (Fig. 1) (unless otherwise stated the following discussion will use the numbering convention of P. furiosus GluDH). The two GluDHs are closely related with respect to reaction kinetics; yet, despite their sequence similarity, the enzyme from P. furiosus is 16-fold more stable at 104°C than the GluDH from T. litoralis (27–29; Table 1). Our initial approach to identifying prospective thermostability determinants before a site-directed mutation study followed a homology-based modeling exercise by using the crystal structure of P. furiosus GluDH (11) to identify changes in the ion-pair networks. This study showed that the most extensive ion-pair networks in the P. furiosus enzyme, which involves 18 residues, appeared to be retained in the T. litoralis GluDH as all the residues involved are conserved in both proteins. The second largest ion-pair network in the P. furiosus enzyme is one of six residues, which includes a string of three basic residues (Arg-Arg-Lys, residues 164–166) and which is centered around a critical glutamic acid residue at position 138, that participates in three ion-pair interactions with Lys and Arg residues in the adjacent subunits (Fig. 2a). In the T. litoralis enzyme, all of the residues in this cluster are conserved with the exception of this critical Glu, which is replaced by Thr. Thus, this single sequence difference necessarily leads to the loss of three ion pairs.
Figure 1.
Sequence alignment of archaeal GluDH from P. furiosus (upper) and T. litoralis (lower) with sequence differences shown in bold and underlined. Intersubunit contacts closer than 3.7 Å within the P. furiosus hexamer are indicated above the sequences (X). Contact residues that are variable (X) include S40T, T138E, D167T, D362Y, and K419H.
Table 1.
Thermostability and Tm of native P. furiosus and recombinant wild-type and mutant T. litoralis GluDHs
| GluDH | t1/2 at 104°C | Tm |
|---|---|---|
| P. furiosus native | 4.6 h | 114.5°C |
| T. litoralis recombinant T138E/D167T | 1.1 h | 111.5°C |
| T. litoralis recombinant wild-type | 0.3 h | 109.0°C |
| T. litoralis recombinant D167T | 0.3 h | 108.5°C |
| T. litoralis recombinant T138E | <0.01 h | 103.5°C |
Half life measurements were conducted at 104°C. The double mutant T138E/D167T of T. litoralis GluDH is ≈4-fold more stable than wild-type.
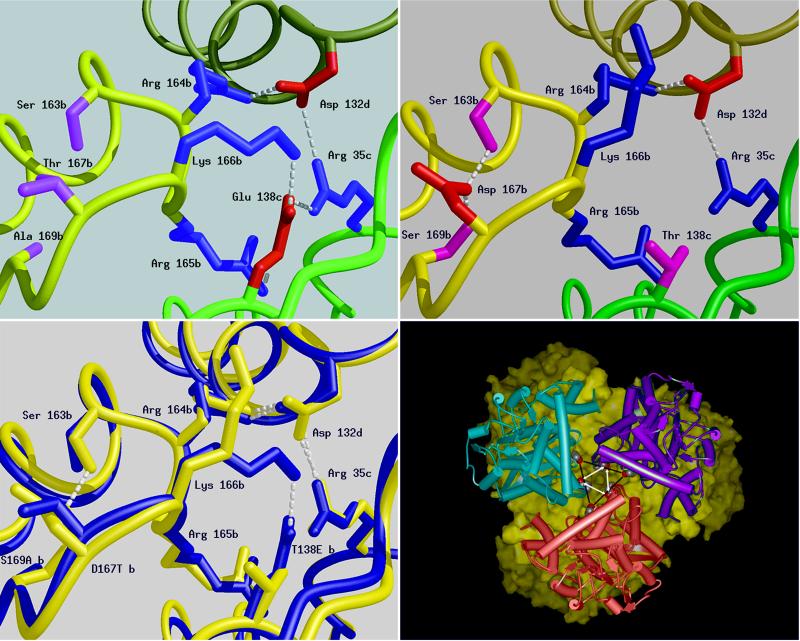
Figure 2.
Schematic diagrams of a six residue-charged cluster in the structure of the GluDHs formed between residues from three adjacent subunits produced by the program midas (31). The main chain of each subunit is shown as a smooth ribbon with b, c, and d subunits colored yellow, green, and khaki, respectively. The positively charged side chains are shown in blue, the negatively charged ones in red, and hydrogen bonds shown as dashed lines. (Upper Left) P. furiosus GluDH: Glu-138, subunit c can be seen to form triple ion-pair interactions to Arg-35, subunit c; Arg-165, subunit b; and Lys-166, subunit b. (Upper Right) The same cluster in T. litoralis GluDH, highlighting the difference caused by the substitution in this enzyme of Glu-138 by Thr (shown in pink). The main consequence of this cluster is the reduction in the size of the charged network from six residues to three. The transposition of residues 138 and 167 leads to very little change in the position of neighboring residues with the notable exception of K166, which in T. litoralis is found extended toward the inter-trimer interface rather than between the subunits of the trimer. D167 (T. litoralis) is unshielded and its carboxyl groups are buried away from the lumen on the threefold axis. (Lower Left) A superimposition of the structures of the GluDHs from T. litoralis (blue) and P. furiosus (red). This diagram highlights the difference between the two structures in the region of the sequence change at position 138. Subtle displacement of the main chain occurs with the two backbone loops of the P. furiosus GluDH further apart at position 167. (Lower Right) View along the threefold axis of T. litoralis GluDH hexamer was generated by using msi WebLab. One trimer is depicted as a solid surface (yellow) and the other as three monomers (pink, blue, and purple). The mutation sites side chains are indicated by the vertices of the central triangles (T138 black bars, D167 white arrows). The corresponding sites on the surface of the other trimer are juxtaposed.
We postulated that the lower thermostability and lower melting temperature of T. litoralis GluDH compared with the P. furiosus GluDH results from the breakdown of this ion pair network, and these observations suggested that these ionic interactions could be installed by a single replacement of Glu at position 138 in the structure. Therefore, we replaced the Thr residue at position 138 in T. litoralis GluDH with Glu, and purified the recombinant enzyme, which we expected would be stabilized as a result of the mutation.
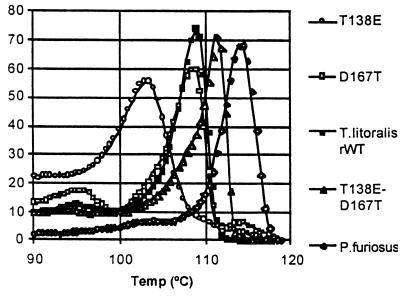
The results of differential scanning microcalorimetry and thermostability measurements of the wild-type and mutant GluDHs are summarized in Table 1 and Fig. 3. Both the P. furiosus and T. litoralis wild-type GluDHs show single, highly cooperative melting transitions with Tm = 114.5° and Tm = 109°C, respectively (Table 1; Fig. 3). The T138E mutant enzyme had a specific activity at 80°C of 150 units/mg equivalent to that of the recombinant wild-type T. litoralis GluDH. However, contrary to our initial expectations, the T. litoralis T138E mutant enzyme was significantly destabilized, with T1/2 too short to determine at 104°C (Table 1) and a Tm at 103.5°C with a gradual rise in the heat capacity compared with either of the wild-type enzymes (Table 1, Fig. 3). We consider that the loss of cooperativity is highly significant because it has not been observed in previous studies on the melting transitions of hyperstable GluDHs.
Figure 3.
Relative heat capacity of GluDHs between 90°C and 130°C. The thermal stability of the double mutant T138E/D167T is elevated relative to T. litoralis wild-type and approaches that of the native P. furiosus GluDH. The single mutants T138E and D167T are relatively less stable, acutely so in the case of T138E, which has an extremely gradual transition.
To investigate the possible causes of this destabilization we crystallized and determined the three dimensional structure of the T. litoralis GluDH to 2.5 Å resolution (ref. 30 and T.J.S., K.L.B., D.W.R., F.T.R., M. W. Adams, and K. Ma, unpublished results). Comparison of this structure with that of the P. furiosus enzyme revealed that the overall structures are very closely related [rmsd 0.38 Å and 0.55 Å for 208 and 209 overlap α-carbon atoms of residues in domains 1 (1–182, 394–419 in P. furiosus GluDH) and 2 (183–207, 210–393 in P. furiosus GDH), respectively]. As expected, this analysis confirmed the breakdown of the six residue ion-pair cluster centered on residue 138 in T. litoralis GluDH following the Glu to Thr substitution between the two enzymes (Fig. 2b). However, close inspection of the structures revealed that in a number of sites the substitutions between the two enzymes gave rise to subtle shifts in the relative positions in the both side chain and main chain elements of the structure, which appear to move to maximize packing following the sequence changes (T.J.S., K.L.B., D.W.R., F.T.R., M. W. Adams, and K. Ma, unpublished results). In particular, our attention was drawn to one of these substitutions in the vicinity of Thr-138, namely Asp-167, which is substituted by Thr-167 in the P. furiosus enzyme. The latter residue is situated on the loop connecting helix α5 to βF and lies at the contact surface between subunits, in close proximity to the six-residue ion-pair cluster (T.J.S., K.L.B., D.W.R., F.T.R., M. W. Adams, and K. Ma, unpublished results). Detailed comparison of the two hyperthermophilic GluDH structures revealed small movements in position, which affected both residue 138 and the basic string of residues (164–166), which partake in the six-residue ion-pair network (Fig. 2c) and the shift and partial disordering of Lys-166. Although these shifts were often small (typically <0.5 Å movements), we wondered what influence such second site substitutions might have on the primary mutation at position 138. Indeed, one of the features of the ion-pair networks in the P. furiosus enzyme is an almost optimal hydrogen-bonding arrangement between the ion-pair partners, and such interactions might well demand critical positioning of the residues in order for the interactions to maintain an optimum stabilizing effect.
The T. litoralis GluDH was therefore altered to contain D167T, both singly and as a double mutant in combination with T138E. Both the mutant and wild-type T. litoralis GluDHs had a specific activity at 80°C of 150 units/mg, as previously reported. The introduction of the D167T mutation alone results in a small reduction in stability of the mutant with respect to the wild-type T. litoralis enzyme (Table 1; Fig. 3). The double substitution T138E/D167T was considered to be a minimal change that effectively transposes Thr from position 138 to position 167. Surprisingly, this variant has properties unlike the single mutants or the wild-type enzyme, displaying substantially increased thermostability at 104°C and a Tm of 111.5°C, intermediate between T. litoralis and P. furiosus GluDHs (Table 1). The second-site change of D167T is therefore a single alteration that raises the Tm of the T138E mutant enzyme by 8°C and restores the highly cooperative melting kinetics that are characteristic of wild-type, hyperstable enzymes (28, 32, and T.J.S., K.L.B., D.W.R., F.T.R., M. W. Adams, and K. Ma, unpublished results).
Our study indicates that the manipulation of intersubunit ion-pair interactions can contribute significantly to thermostability at temperatures above 100°C and would appear to suggest that the area close to residue 138 is an important localized hotspot for subunit dissociation at very high temperatures. Interestingly, of the 53 differences in the amino acid sequences of the two hyperthermophilic enzymes, the two mutation sites identified in this study are both involved in intra-trimer intersubunit contacts (Fig. 1). This observation suggests that the critical event preceding irreversible denaturation is a highly cooperative dissociation of the hexamer to monomers. However, the study also clearly shows that the manipulation of these ion-pair networks to gain stability is far from straight forward as both single mutants were destabilizing. The free energy of stabilization of cellular proteins is relatively small and of the order of 50 kJ/mol. Furthermore, the additional increase in stability necessary to stabilize a hyperthermophilic protein operating at temperatures of 100°C is of the same order of magnitude (32 and T.J.S., K.L.B., D.W.R., F.T.R., M. W. Adams, and K. Ma, unpublished results). Because a single salt bridge can contribute 13–22 kJ/mol to the free energy of folding (32) and, unlike hydrophobic interactions (10), they are relatively unaffected at extremely high temperatures, our results suggest that they can play an important role in maintaining enzyme stability at extreme temperatures. However, because the overall stabilization energies are equivalent only to a small number of weak interactions, it is easy to see how subtle structural changes can have a profound effect on the properties of the proteins. In our experiments, the successful enhancement of thermostability was achieved only by maintaining a local net charge identical to P. furiosus GluDH. Consequently, the region in the structure of T. litoralis GluDH had to be replaced en bloc with the equivalent residues from P. furiosus GluDH. This strongly suggests that the exact context in which a residue occurs within a structure is of crucial importance. At this stage, it is hard to assess the subtle influence of small movements in the structure, induced by second site substitutions as seen around residue 138 (Fig. 2 Lower Left), on the strength of the ion pairs. Furthermore, we cannot discount the possibility that the mechanism by which the Asp residue at position 167 in the single mutant (T138E) destabilizes the enzyme is via an unfavorable electrostatic interaction even though the carboxyl group of D167 lies some 10 Å from the center of the ion-pair network. It is possible that the dynamic properties of the structure during flexing movements are extremely important in determining when subunit dissociation and rapid denaturation commence. Unfortunately, in common with previous studies on the structures of hyperthermostable proteins, our structural data are restricted to a static view of the molecule confined in a crystal lattice and at a temperature far lower than the normal operating temperature of the enzyme. Indeed, we know that there must be some conformational differences between the “cold” and “hot” structures of these enzymes because of the reversible changes in heat capacity that accompany temperature scanning during differential scanning microcalorimetry experiments (34). The nature of these is not currently understood, but the enzyme is known to undergo significant conformational changes between an open and closed state during catalysis (35), and thus the changes seen by differential scanning microcalorimetry may relate to a modification of the domain orientation as a function of temperature.
The useful enhancements to stability achieved through the manipulation of ion-pair networks in GluDH are consistent with the prediction that such interactions may play a major role in enzyme stabilization at high temperatures (11, 15); although, as noted in a recent review (36), several adaptive mechanisms may contribute to stability. The studies of Bogin et al. (19) and Kawamura et al. (37, 38) show that the structures of mesophilic proteins can be stabilized, using sequence alterations predicted by reference to homologous, thermostable proteins. Van den Burg et al. (19) introduced a relatively large number of rigidifying mutations (such as Gly → Ala and Ala → Pro and the installation of a disulfide bridge) designed to reduce the entropy of the unfolded state to achieve substantial thermostability. These studies on a monomeric thermolysin-like protease resulted in eightfold enhanced thermostability by a combination of eight mutations that affected local unfolding hotspots (19), indicating that the stabilization of enzymes can be mediated by different molecular mechanisms. The major difference between these studies and our work is the deliberate placement of a buried ion-pair network in a position which stabilizes cooperative intersubunit interactions in a large, hexameric enzyme. In this respect, it would appear that subunit interactions may be critical for overall stability. Interestingly, the modifications on the thermolysin-like protease result in retention of full activity at low temperatures (19). Similarly, the ion-pair mutations in T. litoralis GluDH, which improve stability at high temperatures, do not result in any reduction of low temperature activity. However, we note that the mutant and wild-type T. litoralis GluDHs all show similar levels of activity at 40°C, which is significantly below that observed at the optimum temperature for activity (26, 27).
The fact that both single mutations in our study are directly detrimental to stability would suggest that manipulating electrostatic interactions is fraught with difficulties. Indeed it is clear that, without reference to the sequence of both proteins and the structure of the more stable P. furiosis GluDH as a blue print, any attempt at rational design to introduce ion-pair interactions into the T. litoralis structure would have been very difficult. Our results also have significant implications for strategies of directed evolution, in which individual mutations are required to provide increments of improvement to a phenotype or at least not to be severely detrimental. Our analysis would imply that the screening of combinations of mutants could be necessary to identify improved phenotypes. The burgeoning database of microbial genomes will provide many opportunities for the application of rational protein design by using comparative sequences and structural analysis. This study provides a model for future alterations of stability of multisubunit enzymes by using strategies that do not affect the catalytic properties of the enzymes and indicates that the manipulation of ion pairs provides an important mechanism to adapt hyperstable enzymes for use as industrial biocatalysts.
Acknowledgments
We are grateful to Sipho Hlati and Madhu Chauhan for technical assistance and to John Tainer, Roberto Poljak, and Dale Oxender for helpful discussions. We acknowledge support from the European Union, Biotechnology and Biological Sciences Research Council, and the New Energy and Industrial Technology Development Organization (to D.W.R.), Foundation for Research Development (RSA) (to D.L.M. and H.H.K.) and the National Science Foundation, U.S. Department of Energy, and the U.S. Department of Commerce (Advanced Technology Program) (to F.T.R.). The Krebs Institute is a designated BBSRC Biomolecular Science Centre and a member of the North of England Structural Biology Centre. This paper is contribution number 475 from the Center of Marine Biotechnology.
ABBREVIATIONS
- GluDH
glutamate dehydrogenase EC 1.4.1.3
- T138E
Thr 138 replaced with Glu
- D167T
Asp 167 replaced with Thr
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates for Thermococcus litoralis GluDH have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1BVU).
References
- 1. Adams M W. Annu Rev Microbiol. 1993;47:627–658. doi: 10.1146/annurev.mi.47.100193.003211. [DOI] [PubMed] [Google Scholar]
- 2.Chan M K, Mukund S, Kletzin A, Adams M W, Rees D C. Science. 1995;267:1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D E, Hurley J H, Nicholson H, Baase W A, Matthews B W. Protein Sci. 1993;2:1285–1290. doi: 10.1002/pro.5560020811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley J H, Baase W A, Matthews B W. J Mol Biol. 1992;224:1143–1159. doi: 10.1016/0022-2836(92)90475-y. [DOI] [PubMed] [Google Scholar]
- 5.Britton K L, Baker P J, Borges K M, Engel P C, Pasquo A, Rice D W, Robb F T, Scandurra R, Stillman T J, Yip K S P. Eur J Biochem. 1995;229:688–695. doi: 10.1111/j.1432-1033.1995.tb20515.x. [DOI] [PubMed] [Google Scholar]
- 6.Russell R J M, Hough D W, Danson M J, Taylor G L. Structure. 1994;2:1157–1167. doi: 10.1016/s0969-2126(94)00118-9. [DOI] [PubMed] [Google Scholar]
- 7.Spassov V Z, Karshikoff A D, Ladenstein R. Protein Sci. 1995;4:1516–1527. doi: 10.1002/pro.5560040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumann J, Bohm G, Schumacher G, Rudolph R, Jaenicke R. Protein Sci. 1993;10:1612–1620. doi: 10.1002/pro.5560021007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner J J, Hecht R M, Krause K L. Biochemistry. 1996;35:2597–2609. doi: 10.1021/bi951988q. [DOI] [PubMed] [Google Scholar]
- 10.Privalov P L, Gill S J. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 11.Yip K S P, Stillman T J, Britton K L, Artymiuk P J, Baker P J, Sedelnikova S E, Engel P C, Pasquo A, Chiaraluce R, Consalvi V, et al. Structure. 1995;3:1147–1158. doi: 10.1016/s0969-2126(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 12.Korndörfer I, Steipe B, Huber R, Tomschy A, Jaenicke R. J Mol Biol. 1995;246:511–521. doi: 10.1006/jmbi.1994.0103. [DOI] [PubMed] [Google Scholar]
- 13.Russell R J M, Ferguson J M C, Hough D W, Danson M J, Taylor G L. Biochemistry. 1997;36:9983–9994. doi: 10.1021/bi9705321. [DOI] [PubMed] [Google Scholar]
- 14.Korolev S, Nayal M, Barnes W M, Di Cera E, Waksman G. Proc Natl Acad Sci USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perutz M F, Raidt H. Nature (London) 1975;255:256–259. doi: 10.1038/255256a0. [DOI] [PubMed] [Google Scholar]
- 16.Creighton T E. J Phys Chem. 1985;89:2452–2559. [Google Scholar]
- 17.Bohm G, Jaenicke R. Int J Pept Protein Res. 1994;43:97–106. doi: 10.1111/j.1399-3011.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 18.Moult J. Curr Opin Biotechnol. 1996;7:422–427. doi: 10.1016/s0958-1669(96)80118-2. [DOI] [PubMed] [Google Scholar]
- 19.Van den Burg B, Vriend G, Veltman O R, Venema G, Eijsink V G H. Proc Natl Acad Sci USA. 1998;95:2056–2060. doi: 10.1073/pnas.95.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogin O, Peretz M, Hacham Y, Korkhin Y, Frolow F, Kalb A J, Burstein Y. Protein Sci. 1998;7:1156–1163. doi: 10.1002/pro.5560070509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X J, Baase W A, Shoichet B K, Wilson K P, Matthews B W. Protein Eng. 1995;8:1017–1022. doi: 10.1093/protein/8.10.1017. [DOI] [PubMed] [Google Scholar]
- 22.Fiala G, Stetter K O. Arch Microbiol. 1985;145:56–61. [Google Scholar]
- 23.Neuner A, Jannasch H W, Belkin S, Stetter K O. Arch Microbiol. 1990;153:205–207. [Google Scholar]
- 24.Knapp S, de Vos W M, Rice D W, Ladenstein R. J Mol Biol. 1997;11:916–932. doi: 10.1006/jmbi.1996.0900. [DOI] [PubMed] [Google Scholar]
- 25.Yip K S P, Britton K L, Stillman T J, Lebbink J, de Vos W M, Robb F T, Vetriani C, Maeder D, Rice D W. Eur J Biochem. 1998;255:336–346. doi: 10.1046/j.1432-1327.1998.2550336.x. [DOI] [PubMed] [Google Scholar]
- 26.Di Ruggiero J, Robb F T. Appl Environ Microbiol. 1995;61:159–164. doi: 10.1128/aem.61.1.159-164.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma K, Robb F T, Adams M W. Appl Environ Microbiol. 1994;60:562–568. doi: 10.1128/aem.60.2.562-568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiRuggiero J, Robb F T, Jagus R, Klump H H, Borges K M, Kessel M, Adams M W. J Biol Chem. 1993;268:17767–17744. [PubMed] [Google Scholar]
- 29.Robb F T, Park J-B, Adams M W. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 30.Sedelnikova S E, Yip K S P, Stillman T J, Kesen M A, Adams M W W, Robb F T, Rice D W. Acta Crystallogr D. 1996;53:122–124. doi: 10.1107/S0907444996007421. [DOI] [PubMed] [Google Scholar]
- 31.Ferrin T E, Huang C C, Jarvis L E, Langridge R. J Mol Graphics. 1988;6:13–27. .Yip, K. S. P., et al. manuscript in preparation. [Google Scholar]
- 32.Jaenicke R. FEMS Microbiol Rev. 1996;18:215–224. doi: 10.1111/j.1574-6976.1996.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 33.Anderson D E, Becktel W J, Dahlquist F W. Biochemistry. 1990;29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 34.Klump H, DiRuggiero J, Kessel M, Adams M W W, Robb F T. J Biol Chem. 1992;267:22681–22685. [PubMed] [Google Scholar]
- 35.Stillman T J, Baker P J, Britton K L, Rice D W. J Mol Biol. 1993;234:1131–1139. doi: 10.1006/jmbi.1993.1665. [DOI] [PubMed] [Google Scholar]
- 36.Robb F T, Maeder D L. Curr Opin Biotechnol. 1998;9:288–291. doi: 10.1016/s0958-1669(98)80061-x. [DOI] [PubMed] [Google Scholar]
- 37.Kawamura S, Tanaka I, Yamasaki N, Kimura M. J Biochem (Tokyo) 1997;121:448–455. doi: 10.1093/oxfordjournals.jbchem.a021609. [DOI] [PubMed] [Google Scholar]
- 38.Kawamura S, Kakuta Y, Tanaka I, Hikichi K, Kuhara S, Yamasaki N, Kimura M. Biochemistry. 1996;35:1195–1200. doi: 10.1021/bi951581l. [DOI] [PubMed] [Google Scholar]